Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle
Abstract
:1. Introduction
2. Results
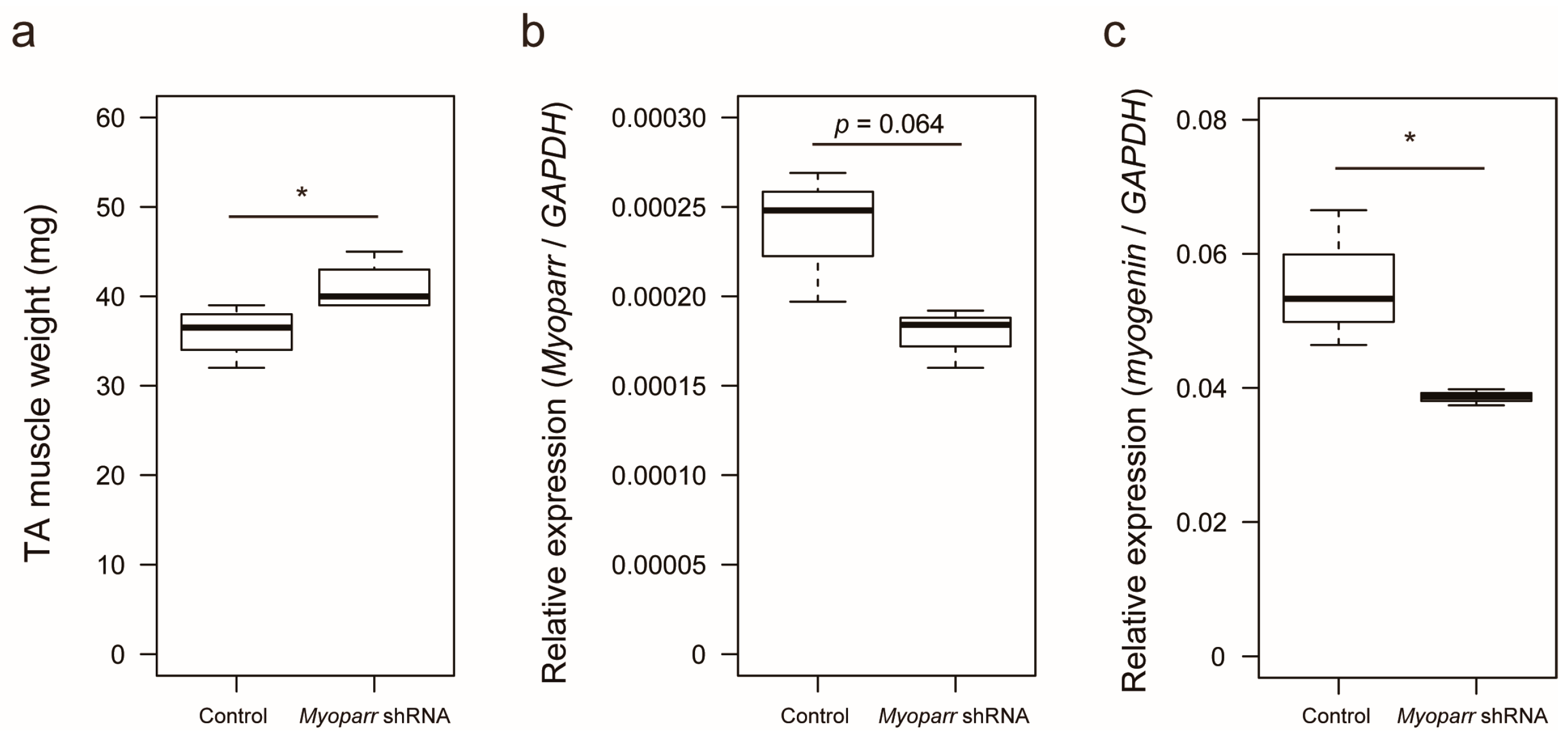
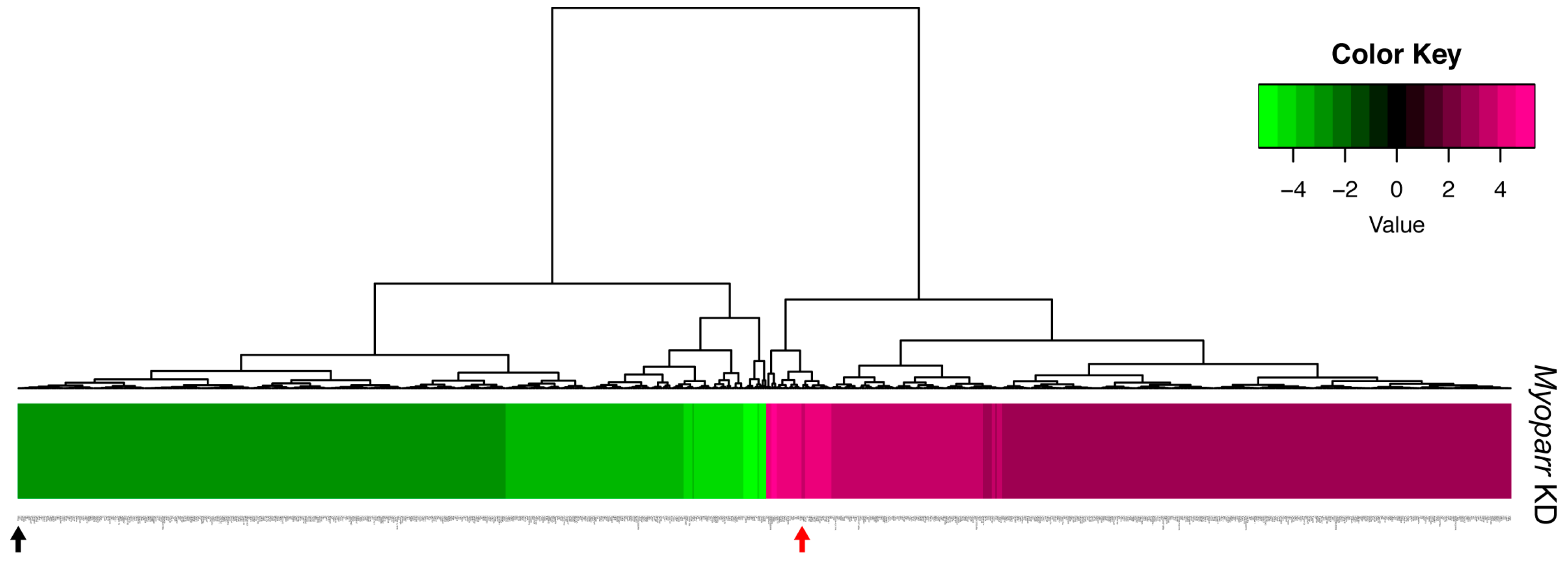
2.1. Knockdown of Myoparr Affected Global Gene Expression in Denervated Mouse Skeletal Muscles
2.2. Knockdown of Myoparr Increased the Expression Level of Growth/Differentiation Factor 5
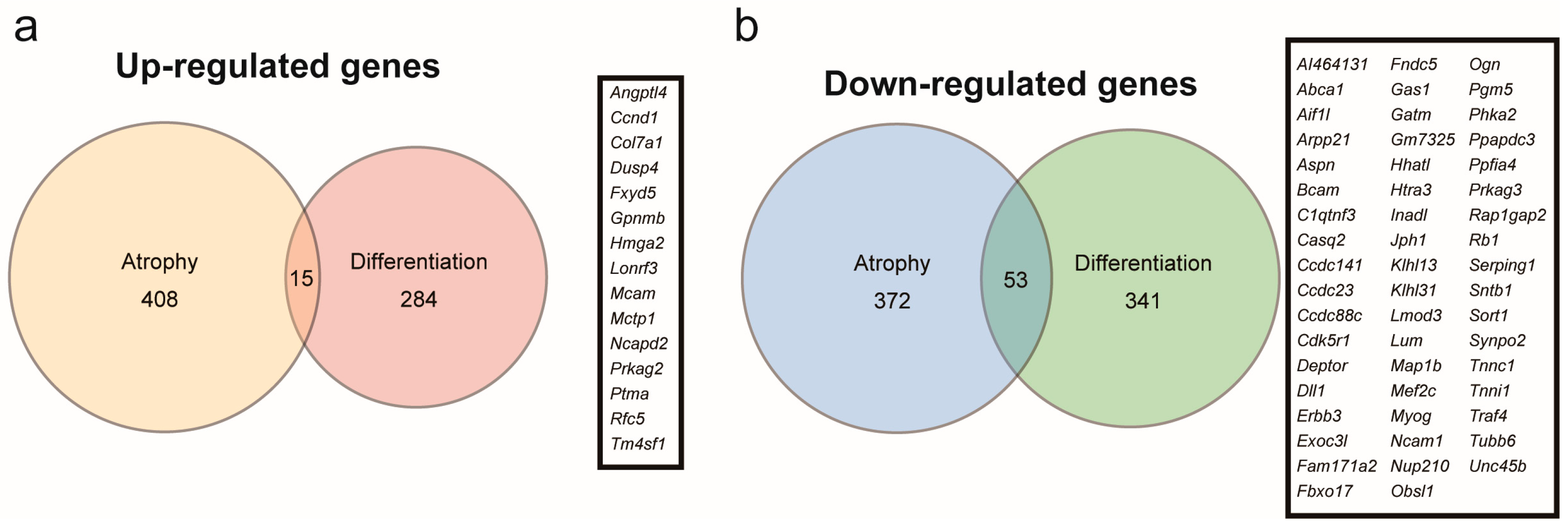
2.3. Genes Regulated by Myoparr Knockdown Differed Between Myogenic Differentiation and Muscle Atrophy
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. RNA Isolation, Quantitative Reverse Transcription Polymerase Chain Reaction, Library Construction, Sequencing, and Data Analysis
4.3. Protein Extraction and Western Blot Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell. Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Hirose, T.; Mishima, Y.; Tomari, Y. Elements and machinery of non-coding RNAs: toward their taxonomy. EMBO Rep. 2014, 15, 489–507. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2018, 7, 877. [Google Scholar] [CrossRef]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinformatics 2017, 18, 558–576. [Google Scholar] [CrossRef]
- Marahrens, Y.; Panning, B.; Dausman, J.; Strauss, W.; Jaenisch, R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997, 11, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Davidow, L.S.; Warshawsky, D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999, 21, 400–404. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef]
- Feil, R.; Walter, J.; Allen, N.D.; Reik, W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1994, 120, 2933–2943. [Google Scholar]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Amândio, A.R.; Necsulea, A.; Joye, E.; Mascrez, B.; Duboule, D. Hotair is dispensible for mouse development. PLoS Genet. 2016, 12, e1006232. [Google Scholar] [CrossRef]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2013, e01749. [Google Scholar] [CrossRef]
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Considerations when investigating lncRNA function in vivo. Elife 2014, 3, e03058. [Google Scholar] [CrossRef]
- Han, X.; Luo, S.; Peng, G.; Lu, J.Y.; Cui, G.; Liu, L.; Yan, P.; Yin, Y.; Liu, W.; Wang, R.; et al. Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J. Mol. Cell Biol. 2018, 10, 175–178. [Google Scholar] [CrossRef]
- Faralli, H.; Dilworth, F.J. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp. Funct. Genomics 2012, 2012, 836374. [Google Scholar] [CrossRef]
- Hitachi, K.; Nakatani, M.; Takasaki, A.; Ouchi, Y.; Uezumi, A.; Ageta, H.; Inagaki, H.; Kurahashi, H.; Tsuchida, K. Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 2019, 20, e47468. [Google Scholar] [CrossRef]
- Sartori, R.; Schirwis, E.; Blaauw, B.; Bortolanza, S.; Zhao, J.; Enzo, E.; Stantzou, A.; Mouisel, E.; Toniolo, L.; Ferry, A.; et al. BMP signaling controls muscle mass. Nat. Genet. 2013, 45, 1309–1318. [Google Scholar] [CrossRef]
- Schertzer, J.D.; Plant, D.R.; Lynch, G.S. Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 2006, 13, 795–803. [Google Scholar] [CrossRef]
- Kaspric, N.; Picard, B.; Reichstadt, M.; Tournayre, J.; Bonnet, M. ProteINSIDE to easily investigate proteomics data from ruminants: Application to mine proteome of adipose and muscle tissues in bovine foetuses. PLoS ONE 2015, 10, e0128086. [Google Scholar] [CrossRef]
- Tono-Oka, S.; Tanase, S.; Miike, T.; Tanaka, H. Transient expression of collagen type XIV during muscle development and its reappearance after denervation and degeneration. J. Histochem. Cytochem. 1996, 44, 907–918. [Google Scholar] [CrossRef]
- Brandan, E.; Fuentes, M.E.; Andrade, W. Decorin, a chondroitin/dermatan sulfate proteoglycan is under neural control in rat skeletal muscle. J. Neurosci. Res. 1992, 32, 51–59. [Google Scholar] [CrossRef]
- Svensson, A.; Norrby, M.; Libelius, R.; Tågerud, S. Secreted frizzled related protein 1 (Sfrp1) and Wnt signaling in innervated and denervated skeletal muscle. J. Mol. Histol. 2008, 39, 329–337. [Google Scholar] [CrossRef]
- Moresi, V.; Williams, A.H.; Meadows, E.; Flynn, J.M.; Potthoff, M.J.; McAnally, J.; Shelton, J.M.; Backs, J.; Klein, W.H.; Richardson, J.A.; et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 2010, 143, 35–45. [Google Scholar] [CrossRef]
- Lee, S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef]
- Winbanks, C.E.; Chen, J.L.; Qian, H.; Liu, Y.; Bernardo, B.C.; Beyer, C.; Watt, K.I.; Thomson, R.E.; Connor, T.; Turner, B.J.; et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013, 203, 345–357. [Google Scholar] [CrossRef]
- Macpherson, P.C.D.; Farshi, P.; Goldman, D. Dach2-Hdac9 signaling regulates reinnervation of muscle endplates. Development 2015, 142, 4038–4048. [Google Scholar] [CrossRef]
- Tang, H.; Goldman, D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA 2006, 103, 16977–16982. [Google Scholar] [CrossRef]
- Méjat, A.; Ramond, F.; Bassel-Duby, R.; Khochbin, S.; Olson, E.N.; Schaeffer, L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 2005, 8, 313–321. [Google Scholar] [CrossRef]
- Cao, Y.; Kumar, R.M.; Penn, B.H.; Berkes, C.A.; Kooperberg, C.; Boyer, L.A.; Young, R.A.; Tapscott, S.J. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006, 25, 502–511. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Martinet, C.; Monnier, P.; Louault, Y.; Benard, M.; Gabory, A.; Dandolo, L. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 2016, 143, 962–971. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef]
- Sun, L.; Si, M.; Liu, X.; Choi, J.M.; Wang, Y.; Thomas, S.S.; Peng, H.; Hu, Z. Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle 2018, 9, 962–974. [Google Scholar] [CrossRef]
- Jin, J.J.; Lv, W.; Xia, P.; Xu, Z.Y.; Zheng, A.D.; Wang, X.J.; Wang, S.S.; Zeng, R.; Luo, H.M.; Li, G.L.; et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA 2018, 115, E9802–E9811. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 2018, 26, 2669–2680. [Google Scholar] [CrossRef]
- Liang, T.; Zhou, B.; Shi, L.; Wang, H.; Chu, Q.; Xu, F.; Li, Y.; Chen, R.; Shen, C.; Schinckel, A.P. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2018, 32, 377–389. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef]
- Chen, X.; He, L.; Zhao, Y.; Li, Y.; Zhang, S.; Sun, K.; So, K.; Chen, F.; Zhou, L.; Lu, L.; et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017, 3, 17002. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, K.; Zhao, Y.; Zhang, S.; Wang, X.; Li, Y.; Lu, L.; Chen, X.; Chen, F.; Bao, X.; et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015, 6, 10026. [Google Scholar] [CrossRef]
- Militello, G.; Hosen, M.R.; Ponomareva, Y.; Gellert, P.; Weirick, T.; John, D.; Hindi, S.M.; Mamchaoui, K.; Mouly, V.; Döring, C.; et al. A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 2018, 10, 102–117. [Google Scholar] [CrossRef]
- Neppl, R.L.; Wu, C.L.; Walsh, K. lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 2017, 216, 3497–3507. [Google Scholar] [CrossRef]
- Moreno-Miralles, I.; Schisler, J.C.; Patterson, C. New insights into bone morphogenetic protein signaling: Focus on angiogenesis. Curr. Opin. Hematol. 2009, 16, 195–201. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Nakatani, M.; Tsuchida, K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int. J. Biochem. Cell Biol. 2014, 47, 93–103. [Google Scholar] [CrossRef]


| Gene Symbol | Protein ID | Entrez ID | log2FoldChange | z-score |
|---|---|---|---|---|
| Fabp5 | Q05816 | 16592 | 0.374131436 | 4.002252172 |
| Lgals3 | P16110 | 16854 | 0.373718827 | 3.997828189 |
| Gdf5 | P43027 | 14563 | 0.361753963 | 3.869541224 |
| Pf4 | Q9Z126 | 56744 | 0.352996303 | 3.775641818 |
| Timp1 | P12032 | 21857 | 0.34793676 | 3.721393528 |
| Csf1 | P07141 | 12977 | 0.3124471 | 3.340874304 |
| Cfp | P11680 | 18636 | 0.293360391 | 3.136227099 |
| Pxdn | Q3UQ28 | 69675 | 0.273907602 | 2.927654795 |
| Nrcam | Q810U4 | 319504 | 0.273420533 | 2.922432453 |
| Thbs4 | Q9Z1T2 | 21828 | 0.273376805 | 2.921963603 |
| Gars | Q9CZD3 | 353172 | 0.272354163 | 2.910998862 |
| Egfl7 | Q9QXT5 | 353156 | 0.269565054 | 2.881094106 |
| F7 | P70375 | 14068 | 0.258828912 | 2.765981467 |
| Angptl4 | Q9Z1P8 | 57875 | 0.25297163 | 2.70317984 |
| Emilin2 | Q8K482 | 246707 | 0.247003963 | 2.639194667 |
| Col7a1 | Q63870 | 12836 | 0.245381068 | 2.621794029 |
| Sdc4 | O35988 | 20971 | 0.244800608 | 2.615570352 |
| Cd9 | P40240 | 12527 | 0.244138059 | 2.608466518 |
| Gene Symbol | Protein ID | Entrez ID | log2FoldChange | z-score |
|---|---|---|---|---|
| Olfml2b | Q3V1G4 | 320078 | −0.481025783 | −5.166721595 |
| Nid1 | P10493 | 18073 | −0.449335456 | −4.826938722 |
| Gpx3 | P46412 | 14778 | −0.448922444 | −4.822510417 |
| C3 | P01027 | 12266 | −0.445172583 | −4.782304504 |
| C1qtnf3 | Q9ES30 | 81799 | −0.445104367 | −4.781573094 |
| Srpx2 | Q8R054 | 68792 | −0.436334685 | −4.687544788 |
| Lifr | P42703 | 16880 | −0.429117336 | −4.610160558 |
| Sorl1 | O88307 | 20660 | −0.414098638 | −4.449130463 |
| Col14a1 | Q80X19 | 12818 | −0.386271994 | −4.1507739 |
| Postn | Q62009 | 50706 | −0.370968623 | −3.986691548 |
| Svep1 | A2AVA0 | 64817 | −0.370316314 | −3.979697508 |
| Pi16 | Q9ET66 | 74116 | −0.367955107 | −3.954380707 |
| Pltp | P55065 | 18830 | −0.364986609 | −3.922552548 |
| Fndc5 | Q8K4Z2 | 384061 | −0.362929143 | −3.90049245 |
| F13a1 | Q8BH61 | 74145 | −0.344575054 | −3.703700378 |
| Dcn | P28654 | 13179 | −0.333557655 | −3.585572109 |
| Hspg2 | Q05793 | 15530 | −0.324036042 | −3.483481619 |
| Sfrp1 | Q8C4U3 | 20377 | −0.306352312 | −3.293877119 |
| Cilp | Q66K08 | 214425 | −0.301045081 | −3.236973125 |
| Serping1 | P97290 | 12258 | −0.295270029 | −3.175053165 |
| Lum | P51885 | 17022 | −0.293171855 | −3.152556597 |
| Col19a1 | Q0VF58 | 12823 | −0.28991971 | −3.117687182 |
| Htra3 | Q9D236 | 78558 | −0.288268162 | −3.099979327 |
| Mfap4 | Q9D1H9 | 76293 | −0.282983368 | −3.043315901 |
| Aspn | Q99MQ4 | 66695 | −0.279272444 | −3.003527469 |
| Ogn | Q62000 | 18295 | −0.277371455 | −2.983145114 |
| Apoe | P08226 | 11816 | −0.276102919 | −2.969543903 |
| Adamts2 | Q8C9W3 | 216725 | −0.274377529 | −2.951044316 |
| Islr | Q6GU68 | 26968 | −0.273173486 | −2.938134598 |
| Nucb2 | P81117 | 53322 | −0.272713326 | −2.933200774 |
| Lama2 | Q60675 | 16773 | −0.271718749 | −2.922536945 |
| St3gal1 | P54751 | 20442 | −0.262186218 | −2.820329392 |
| Col1a1 | P11087 | 12842 | −0.262170298 | −2.820158698 |
| Hsd17b11 | Q9EQ06 | 114664 | −0.261915887 | −2.81743091 |
| Igsf10 | Q3V1M1 | 242050 | −0.260696015 | −2.804351474 |
| Cp | Q61147 | 12870 | −0.259206576 | −2.788381747 |
| Gas6 | Q61592 | 14456 | −0.256647441 | −2.760942767 |
| Serpine1 | P22777 | 18787 | −0.252661608 | −2.718206768 |
| Serpine2 | Q07235 | 20720 | −0.250764111 | −2.697861854 |
| Comp | Q9R0G6 | 12845 | −0.246462973 | −2.651745163 |
| Fgl2 | P12804 | 14190 | −0.241340354 | −2.596820573 |
| Lox | P28301 | 16948 | −0.240518253 | −2.588006027 |
| Timp3 | P39876 | 21859 | −0.240498779 | −2.587797228 |
| Qsox1 | Q8BND5 | 104009 | −0.239576044 | −2.577903687 |
| Galnt1 | O08912 | 14423 | −0.239367349 | −2.575666064 |
| Cxcl14 | Q9WUQ5 | 57266 | −0.238871098 | −2.570345274 |
| Timp2 | P25785 | 21858 | −0.234496758 | −2.523443712 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hitachi, K.; Nakatani, M.; Tsuchida, K. Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle. Non-Coding RNA 2019, 5, 33. https://doi.org/10.3390/ncrna5020033
Hitachi K, Nakatani M, Tsuchida K. Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle. Non-Coding RNA. 2019; 5(2):33. https://doi.org/10.3390/ncrna5020033
Chicago/Turabian StyleHitachi, Keisuke, Masashi Nakatani, and Kunihiro Tsuchida. 2019. "Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle" Non-Coding RNA 5, no. 2: 33. https://doi.org/10.3390/ncrna5020033
APA StyleHitachi, K., Nakatani, M., & Tsuchida, K. (2019). Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle. Non-Coding RNA, 5(2), 33. https://doi.org/10.3390/ncrna5020033






