Abstract
Skeletal muscle is a highly plastic tissue and decreased skeletal muscle mass (muscle atrophy) results in deteriorated motor function and perturbed body homeostasis. Myogenin promoter-associated long non-coding RNA (lncRNA) Myoparr promotes skeletal muscle atrophy caused by surgical denervation; however, the precise molecular mechanism remains unclear. Here, we examined the downstream genes of Myoparr during muscle atrophy following denervation of tibialis anterior (TA) muscles in C57BL/6J mice. Myoparr knockdown affected the expression of 848 genes. Sixty-five of the genes differentially regulated by Myoparr knockdown coded secretory proteins. Among these 65 genes identified in Myoparr-depleted skeletal muscles after denervation, we focused on the increased expression of growth/differentiation factor 5 (GDF5), an inhibitor of muscle atrophy. Myoparr knockdown led to activated bone morphogenetic protein (BMP) signaling in denervated muscles, as indicated by the increased levels of phosphorylated Smad1/5/8. Our detailed evaluation of downstream genes of Myoparr also revealed that Myoparr regulated differential gene expression between myogenic differentiation and muscle atrophy. This is the first report demonstrating the in vivo role of Myoparr in regulating BMP signaling in denervated muscles. Therefore, lncRNAs that have inhibitory activity on BMP signaling may be putative therapeutic targets for skeletal muscle atrophy.
1. Introduction
Long non-coding RNAs (lncRNAs), which exceed 200 nucleotides in length, are derived from intergenic regions, intronic regions, and cis-regulatory regions (enhancers and promoters) [1], and show tissue-specific expression patterns compared to protein-coding RNAs [2,3]. Divergent species of lncRNAs have been identified from mammalian genomes, including the human genome. Although the nucleic acid sequences of lncRNAs among different species are poorly conserved, their biological function tends to be conserved between species, possibly because of their similar secondary structure [4].
Gain-of-function and loss-of-function of lncRNAs in cell culture systems have revealed that lncRNAs exhibit multiple biological roles, such as epigenetic regulation, transcriptional regulation, translational regulation, and functioning as structural cores, among others [5,6]. Correlations between mutations and dysregulation of lncRNAs and human diseases have been identified [7,8,9]. In addition, in vivo functions of lncRNAs have been established in dosage compensation and genome imprinting processes [10,11,12,13]. Other in vivo functions of several lncRNAs have been experimentally demonstrated using knockout mice. For instance, Fendrr regulates heart and body wall development [14] and Neat1 regulates corpus luteum formation and pregnancy [15]. Furthermore, Hotair is essential for repression of Hoxd expression in cultured cells [16], however, the in vivo role of Hotair remains controversial [17]. Several genetic studies have also raised questions about the roles of multiple lncRNAs in the development of mice [18,19,20]. Thus, in vivo analysis of the molecular function of lncRNAs is essential in order to reveal the physiological roles of lncRNAs in differentiation, development, and various diseases.
Skeletal muscle is a highly plastic tissue that is required not only for motion but also for maintaining body homeostasis. During embryonic development, the number of proliferating mononuclear myoblasts determines the size of future skeletal muscle tissue. As development progresses, myoblasts cease growing and undergo differentiation to form multinuclear myotubes. This process, which is called myogenesis, has been studied as a model system of cell differentiation. The myogenin gene product is one of the major transcriptional regulators in myogenesis [21]. We previously identified Myoparr, a lncRNA expressed in the opposite direction from the promoter region of myogenin [22]. Myoparr promotes myogenic differentiation through the activation of neighboring myogenin expression. Intriguingly, Myoparr expression also increases during skeletal muscle atrophy caused by denervation and promotes muscle atrophy in mice. While Myoparr activates the expression of myogenin in vivo, the exact molecular mechanism by which Myoparr promotes skeletal muscle atrophy caused by denervation remains to be elucidated.
In the current study, we aimed to examine changes in the expression profiles of genes in denervated tibialis anterior (TA) muscles of mice following Myoparr knockdown. Myoparr knockdown caused the upregulation or downregulation of hundreds of genes at an earlier time-point after denervation. We focused on 65 genes that coded for potential secretory proteins in which expression was altered by Myoparr knockdown, and examined the involvement of these genes in skeletal muscle atrophy. We showed the importance of elevated growth/differentiation factor 5 (GDF5) expression—which is also known as bone morphogenetic protein (BMP) 14 and an inhibitor of muscle atrophy in mice [23]—for preventing muscle atrophy in Myoparr-depleted muscles under denervation. Therefore, our findings reveal that Myoparr promotes skeletal muscle atrophy by repressing GDF5/BMP signaling in denervated skeletal muscles and may help identify putative therapeutic targets for preventing and treating skeletal muscle atrophy.
2. Results
2.1. Knockdown of Myoparr Affected Global Gene Expression in Denervated Mouse Skeletal Muscles
We previously showed that small hairpin RNA (shRNA)-mediated knockdown of Myoparr in skeletal muscle decreases myogenin expression and attenuates skeletal muscle atrophy 7 days post sciatic nerve transection [22]. To identify the early response genes by Myoparr knockdown other than myogenin, we examined skeletal muscle mass at 3 days post denervation. Increased skeletal muscle mass was observed in response to Myoparr knockdown after denervation compared with that of the control side of the mice (Figure 1a). Thus, Myoparr knockdown attenuated muscle atrophy at an earlier time point than previously reported.
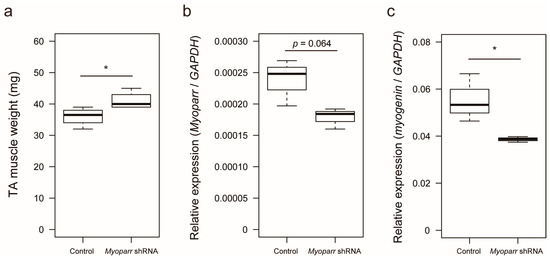
Figure 1.
Knockdown of Myoparr attenuated skeletal muscle atrophy caused by denervation. (a) Box-and-whisker plots showing the weights of denervated tibialis anterior (TA) muscles of C57BL/6J mice electroporated with either control short hairpin RNA (shRNA) against LacZ or Myoparr-specific shRNA. TA muscle weights were measured 3 days post denervation (n = 4 per group). * p < 0.05, unpaired two-tailed Student’s t-test. Central black bar indicates median; lower and upper box limits are 25th and 75th percentiles, respectively; whiskers show maximum and minimum values. (b) The level of Myoparr expression in denervated TA muscles electroporated either with control or Myoparr shRNA (n = 3 per group) was measured by quantitative reverse transcription polymerase chain reaction (qRT–PCR). p = 0.064, unpaired two-tailed Student’s t-test. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. (c) Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) showing decreased myogenin expression by Myoparr knockdown 3 d post denervation (n = 3 per group). * p < 0.05, unpaired two-tailed Student’s t-test. Data were normalized to GAPDH expression.
To better characterize the in vivo role of Myoparr, we examined global gene expression in Myoparr-depleted TA muscles using RNA-sequencing (RNA-seq) analysis. Tibialis anterior (TA) muscles were transfected with either Myoparr-specific or control shRNA and the sciatic nerve was then transected. Three days after denervation, the total RNA was extracted from the muscles and used for RNA-seq library construction. Before RNA-seq analysis, Myoparr expression was quantified by quantitative reverse transcription polymerase chain reaction (qRT–PCR). Although statistical significance (p < 0.05) was not observed, the level of Myoparr expression tended to decrease by Myoparr knockdown (Figure 1b). Instead, we observed that Myoparr knockdown significantly decreased myogenin expression, which we previously identified as a Myoparr target gene [22] (Figure 1c), indicating that electroporation-mediated Myoparr knockdown in TA muscles altered the expression of the downstream gene of Myoparr. RNA-seq analysis showed that knockdown of Myoparr affected global gene expression, although the rates of changes were relatively mild, possibly as a result of unaltered gene expression in the remaining untransfected myofibers. Therefore, we converted the log2 values of the RNA-seq results to z-score values. In this study, the z-score threshold was set to >2.5 or <−2.5. The gene expression dynamics in Myoparr-depleted muscles were shown by cluster analysis based on their z-score values. As illustrated by the heatmap in Figure 2, Myoparr knockdown altered the expression level of 848 genes in denervated TA muscles with 423 genes being upregulated and 425 genes being downregulated (Table S1). It is of note that the expression of the myogenin gene was also decreased as indicated by the black arrow in Figure 2. These results indicated that Myoparr knockdown globally affected gene expression in denervated muscles in mice.
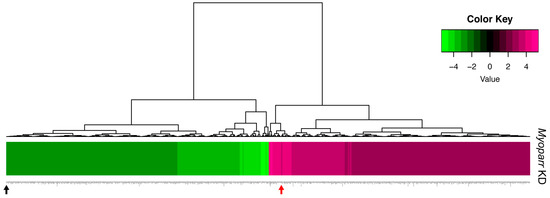
Figure 2.
Knockdown of Myoparr altered global gene expressions in denervated skeletal muscle. The heatmap displayed the changes in expression for 848 genes altered by Myoparr knockdown in denervated TA muscle of C57BL/6J mice (values are z-scores). The black and red arrow indicate the decreased expression of myogenin and increased expression of Gdf5, respectively.
2.2. Knockdown of Myoparr Increased the Expression Level of Growth/Differentiation Factor 5
The efficiency of transgene expression is approximately 30% of total myofibers at 3 days following electroporation [24], indicating that shRNA inhibits Myoparr expression in 30% of denervated TA muscle myofibers. However, we observed significant increases in weight of TA muscles following Myoparr knockdown. Based on these observations, we hypothesized that Myoparr knockdown altered the expression of secretory proteins and also affected the size of untransfected myofibers. To investigate this possibility, we screened the genes coding for secretory proteins among the 848 genes differentially regulated by Myoparr knockdown using ProteINSIDE analysis [25]. We identified genes coding for secretory proteins by screening for proteins that had been annotated as secretory proteins. The ProteINSIDE analysis extracted 65 genes with secreted annotation from the 848 genes regulated by Myoparr knockdown. Of these 65 genes, 18 were upregulated (Table 1) and 47 were downregulated (Table 2). Notably, RNA-seq data showed that Myoparr knockdown decreased the expression levels of Col14a1, Dcn, and Sfrp1, which are essential for muscle homeostasis and reported to increase following denervation [26,27,28]. This supported the notion that Myoparr knockdown attenuated muscle atrophy caused by denervation.

Table 1.
Upregulated genes encoding secretory proteins following Myoparr knockdown in denervated tibialis anterior (TA) muscles of C57BL/6J mice.

Table 2.
Downregulated genes encoding secretory proteins following Myoparr knockdown in denervated TA muscle of C57BL/6J mice.
Among the various genes coding for secretory proteins, our attention was drawn to the Gdf5 gene that has previously been shown to prevent muscle atrophy caused by denervation [23]. Thus, we examined the expression level of GDF5 in Myoparr-depleted TA muscles by western blot. At 3 days post denervation, Myoparr knockdown largely increased the expression level of GDF5 compared with that in the muscles transfected with control shRNA (Figure 3a,b). In agreement with the previous report that Gdf5 is required for the activation of BMP signaling in denervated muscles [23], activated BMP signaling was also observed following Myoparr knockdown, as indicated by increased levels of phosphorylated Smad1/5/8 (Figure 3a). We also noticed that Myoparr knockdown increased the expression level of Smad5 (Figure 3a,c). Taken together, these results indicated that Myoparr knockdown prevented muscle atrophy after denervation by activating BMP signaling by increasing GDF5 and Smad5 expression.
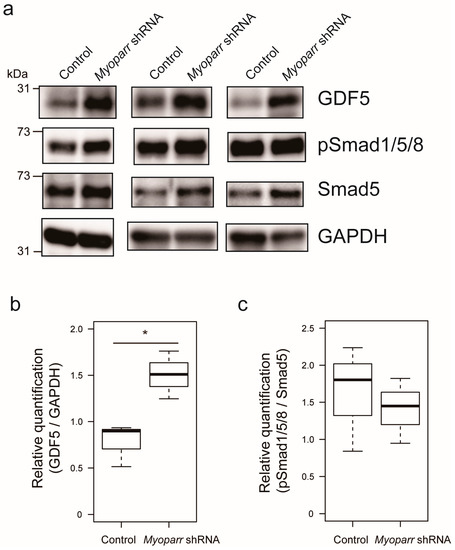
Figure 3.
Myoparr knockdown activated bone morphogenetic protein (BMP) signaling through the expression of growth/differentiation factor 5 (GDF5) in denervated TA muscles of C57BL/6J mice. (a) Western blot showing increased expression of GDF5 by Myoparr knockdown in TA muscles 3 d post denervation. The activity of BMP signaling was indicated by the levels of phosphorylated Smad1/5/8 (pSmad1/5/8). GAPDH expression served as an internal control. Three independent experiments were shown. (b) Box-and-whisker plots showed relative quantification of GDF5 expression as GDF5/GAPDH ratio (n = 3 per group). * p < 0.05, unpaired two-tailed Student’s t-test. (c) Relative quantification of the activity of BMP signaling was shown as pSmad1/5/8/Smad5 ratio (n = 3 per group).
2.3. Genes Regulated by Myoparr Knockdown Differed Between Myogenic Differentiation and Muscle Atrophy
Myoparr knockdown alters the expression of 693 genes in differentiating C2C12 cells, of which 299 are upregulated and 394 are downregulated [22]. The increased Gdf5 expression in Myoparr-depleted skeletal muscles prompted us to compare the genes regulated by Myoparr knockdown in myogenic differentiation compared to those in muscle atrophy since the expression level of Gdf5 is not changed in Myoparr-depleted C2C12 cells. We compared 423 upregulated and 425 downregulated genes in Myoparr-depleted TA muscles with 299 upregulated and 394 downregulated genes in Myoparr-depleted C2C12 cells, respectively (Figure 4a,b). Surprisingly, only 3.5% of the upregulated and 12% of the downregulated genes by Myoparr knockdown intersected between myogenic differentiation and muscle atrophy.
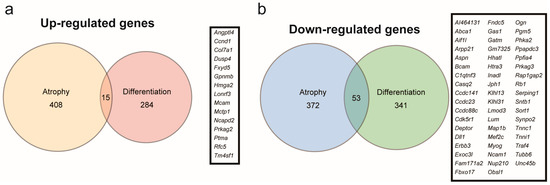
Figure 4.
Comparison of genes commonly regulated by Myoparr knockdown between myogenic differentiation and muscle atrophy. (a) The intersection of genes upregulated by Myoparr knockdown in both skeletal muscle atrophy (423 genes) and myogenic differentiation (299 genes) showed low commonality. Gene symbols of the 15 commonly upregulated genes by Myoparr knockdown in both myogenic differentiation and muscle atrophy were shown in the panel. (b) The intersection of genes commonly downregulated by Myoparr knockdown in both skeletal muscle atrophy (425 genes) and myogenic differentiation (394 genes) also showed low commonality. Gene symbols of 53 commonly downregulated genes in both myogenic differentiation and muscle atrophy were shown in the panel.
To reveal the molecular functions of genes commonly regulated by Myoparr knockdown in both myogenic differentiation and muscle atrophy, we performed functional gene ontology (GO) enrichment analysis (Table S2). Although the enrichment score was not high, the results based on the biological processes category showed that genes regulated by Myoparr knockdown in both myogenic differentiation and muscle atrophy were enriched in skeletal muscle-related processes such as myotube differentiation, regulation of muscle contraction, and skeletal muscle cell differentiation (Figure 5a). It is of interest that these genes were also enriched in skeletal muscle-related categories of cellular component such as Z disc, troponin complex, and contractile fiber (Figure 5b) and of molecular function such as protein binding, including actin binding and calcium ion binding (Figure 5c). These results indicated that although genes commonly regulated by Myoparr knockdown both in vivo and in vitro were related to skeletal muscle function, most of downstream genes of Myoparr are different between myogenic differentiation and skeletal muscle atrophy.
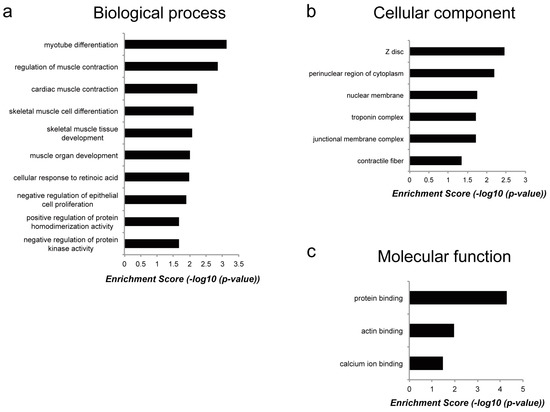
Figure 5.
Classification of the annotated downstream genes of Myoparr. Genes commonly upregulated or downregulated by Myoparr knockdown in both myogenic differentiation and muscle atrophy were grouped into different functional gene ontology (GO) categories as indicated. (a) Biological process category. (b) Cellular component category. (c) Molecular function category.
3. Discussion
Multiple biological roles of lncRNAs have been revealed in vitro [5,6]; however, the roles of lncRNAs in vivo are still not well understood. We recently identified Myoparr, a promoter-associated lncRNA, and revealed its essential role in myogenic differentiation [22]. Interestingly, increased Myoparr expression promotes skeletal muscle atrophy in denervated skeletal muscles. Myoparr activates neighboring myogenin gene expression, which is one of the inducers of muscle atrophy [29]. However, the molecules expressed downstream of Myoparr during muscle atrophy have not been fully identified. In the current work, we investigated the downstream genes of Myoparr in denervated skeletal muscles in mice. Among hundreds of genes regulated by Myoparr knockdown, we focused on 65 genes that encoded secretory proteins. While myostatin, a cytokine belonging to the transforming growth factor-β (TGF-β) superfamily, is a pivotal inducer of muscle atrophy by increasing protein catabolism [30], BMP signaling can protect skeletal muscle mass after denervation [23,31]. In addition, skeletal muscle hypertrophy induced by myostatin inhibition is also dependent on activated BMP signaling [23,31]. We narrowed the analysis of the 65 Myoparr-regulated genes to target Gdf5, since Gdf5 encodes a secretory protein belonging to BMP family and Gdf5-/- mice demonstrate aggravated muscle atrophy [23] that is accompanied with decreased BMP signaling activity. We largely observed increased GDF5 and Smad5 levels and activated BMP signaling in Myoparr-depleted skeletal muscles after denervation. It is noteworthy that the expression level of Smad5 was not increased by Myoparr knockdown in RNA-seq analysis, suggesting that Myoparr represses Smad5 expression at the post-transcriptional level. Intriguingly, Gdf5 expression in denervated muscles is independent of myogenin gene [32]. Therefore, our findings indicated that Myoparr promoted muscle atrophy in denervated muscles through the inhibition of BMP signaling by repressing Gdf5 expression in a myogenin-independent manner.
In innervated muscle, the expression of Gdf5 is repressed by the activity-regulated transcriptional co-repressors Dach2 and Hdac9/Mitr [32]. Surgical denervation reduces Dach2 and Hdac9 expression and activates BMP signaling through upregulated Gdf5 expression [23,31,32]. Notably, Dach2 and Hdac9 also repress myogenin expression in innervated muscle through the minimal promoter region [33,34]. Although it is unclear whether Dach2 and Hdac9 repress Myoparr expression in innervated muscle, it has been established that Myoparr shares the minimal promoter region with myogenin in the myogenic differentiation process [22]. Thus, Dach2 and Hdac9 are common upstream repressors of Gdf5, myogenin and Myoparr expression in innervated muscle.
Our RNA-seq analysis of genes regulated by Myoparr knockdown in denervated muscles indicated that Myoparr regulated gene expression differently between in vivo muscle atrophy and in vitro myogenic differentiation. Although several studies have previously raised the importance of different physiological functions of lncRNA in vivo and in vitro [18,19,20], we speculate that the downstream genes of Myoparr may be determined depending on the cell status and context for the following reasons: (1) Composition of cells in skeletal muscle is largely different between muscle differentiation and muscle atrophy, as demonstrated by the different target genes of myogenin protein in each situation [29,35]. Thus, the myogenin-dependent function of Myoparr likely changes in the two situations; (2) different transcription factors are used in gene expression control between muscle differentiation and muscle atrophy, as shown by the different key regulators being MyoD or Dach2 in myogenin expression in both muscle differentiation and muscle atrophy [21,33]. Myoparr binds to Ddx17, a transcriptional co-activator of MyoD, and promotes the transcriptional activity of Ddx17 during myogenic differentiation [22]. Binding partner proteins influence the function of lncRNAs [36]. Thus, the utilization of different upstream regulators would change the Myoparr-binding protein and alter the molecular function of Myoparr in each situation. Taken together, our data suggested that the molecular function of lncRNAs was to be flexibly changeable depending on the gene expression profiles or their binding-proteins in vivo and in vitro.
In conclusion, we demonstrated for the first time that the expression of GDF5 was regulated by the myogenin promoter-associated lncRNA Myoparr in denervated skeletal muscle. Recently, it has become evident that lncRNAs are novel in vivo regulators of skeletal muscle mass [37,38,39,40,41,42,43] and muscle regeneration [38,44,45,46,47], each functioning through unique mechanisms. Intriguingly, Neppl et al. described the regulation of BMP signaling by lncRNA in skeletal muscle in which Chronos inhibits muscle growth by repressing Bmp7 expression [48], indicating that several lncRNAs are pivotal negative regulators of BMP signaling in skeletal muscle. Therefore, lncRNAs that have inhibitory activity on BMP signaling such as Myoparr may be putative therapeutic targets for skeletal muscle atrophy. In addition to the role in myofibers, BMP signaling also plays an important role in the formation of blood vessels, essential components both in steady state of skeletal muscle and in the milieu of reinnervation of denervated skeletal muscle [32,49]. Collectively, examining the cell types which BMP signaling is activated by Myoparr knockdown in denervated skeletal muscle will further define the molecular roles of Myoparr in muscle atrophy.
4. Materials and Methods
4.1. Animal Experiments
All mice used were male C57BL/6J strain, purchased from the Japan SLC (Shizuoka, Japan) and housed in cages with a constant temperature (24 °C) and a 12:12 h light-dark cycle. All animal experiments were conducted under protocols approved by the Institutional Animal Care and Use Committee of Fujita Health University. Prior to electroporation, the TA muscles of 8-week-old mice were injected with 30 μg endotoxin-free plasmid DNA containing either control shRNA against LacZ or Myoparr-specific shRNA in a 30 μL saline solution using a 34-gauge needle from the ankle side to approximately 5 mm under anesthesia. The sequences of control shRNA and Myoparr shRNA are previously described [22]. Electric pulses (150 V/cm, 6 pulses, 50 ms pulses of 1-Hz frequency) were applied to the TA muscles with a tweezer-type electrode using a CUY21EDIT electroporator (Bex Co. LTD., Tokyo, Japan). A 3-mm fragment of the sciatic nerve was excised under anesthesia after electroporation. Three days after surgery, the mice were sacrificed and the TA muscles were collected, weighed, and processed for RNA and protein extraction.
4.2. RNA Isolation, Quantitative Reverse Transcription Polymerase Chain Reaction, Library Construction, Sequencing, and Data Analysis
Total RNA was extracted from the TA muscles and purified using an miRNeasy Mini kit with DNase I (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. One microgram of total RNA was used for the reverse transcription reaction using the SuperScript III First-Strand Synthesis System with random primers (Thermo Fisher Scientific, Waltham, MA, USA). The qRT–PCR was conducted using SYBR Premix Ex Taq (Takara, Shiga, Japan) according to the manufacturer’s protocol. Primers used for qRT–PCR were previously described [22].
One microgram of total RNA was used for purification of Poly(A)+ RNAs using the NEBNext Poly (A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA, USA). RNA-seq libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s protocol and sequenced with 100-bp pair-end reads using an Illumina HiSeq 1500 system (Illumina, San Diego, CA, USA) at Fujita Health University. The average number of reads of each sample was approximately 5 million. The bcl2fastq 1.8.4 software was used for base calling. The RNA-seq raw data for each sample has been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive under the Accession No. DRA007708. PRINSEQ ver. 0.20.4 software was used for quality trimming of row sequence data using the following command “-trim_qual_right 20 -min_len 30” [50]. Hisat2 ver. 2.0.5 software [51] was used for alignment of the trimmed reads to the mouse reference genome (mm10) applying the default parameters. The number of aligned reads was approximately 96% of the original reads. The aligned reads were converted and sorted to Bam files using SAMtools ver. 1.3.1 software [52] and the reads were counted with the HTSeq ver. 0.6.0 software [53] using the Mus_musculus_UCSC_mm10.gtf file and the following optional command: “--stranded=no --format=bam”. The value of the log2 fold change was calculated using DESeq2 ver. 1.12.4 software [54] and converted to a z-score using R software with the package Genefilter (https://bioconductor.org/packages/release/bioc/html/genefilter.html). Genes with z-score values >2.5 or <−2.5 were considered significant in this study. Gene ontology analysis was performed using DAVID ver. 6.8 (https://david.ncifcrf.gov/) and p-values < 0.05 were considered statistically significant.
4.3. Protein Extraction and Western Blot Analysis
Western blot analysis was performed as previously described in References [22,55]. In brief, the TA muscles were surgically isolated 3 d after denervation and frozen in liquid nitrogen. Samples were broken into pieces using a bead-homogenizer Shakeman2 (Bio Medical Science, Tokyo, Japan) and lysed in RIPA buffer consisting of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 4 μg/mL leupeptin), and phosphatase inhibitors (5 mM NaF, 5 mM β-glycerophosphate, 1 mM Na3VO4). Equal amounts of protein quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) were subjected to western blot analysis. The primary antibodies used included the GDF5 antibody A-10 (sc-373744, Santa Cruz Biotechnology, Dallas, TX, USA), the Smad5 antibody YY-6 (sc-101151, Santa Cruz Biotechnology), a phospho-Smad1(Ser463/465)/5(Ser463/465)/8(Ser426/428) antibody (#9511, Cell Signaling Technology, Danvers, MA, USA), and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (#2118, Cell Signaling Technology). The horseradish peroxidase (HRP)-linked secondary antibodies included an anti-mouse IgG (#7076, Cell Signaling Technology) and an anti-rabbit IgG (#7074, Cell Signaling Technology) as appropriate. For the detection of Smad5 and phosphorylated Smad1/5/8, Can Get Signal Immunoreaction Enhancer Solution (Toyobo, Osaka, Japan) was used. Semi-quantification analysis of western blot images was conducted with ImageJ software ver. 2.0.0-rc-69.
4.4. Statistical Analysis
Data were analyzed by using unpaired two-tailed Student’s t-test. p < 0.05 was considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/2311-553X/5/2/33/s1, Table S1: Genes upregulated or downregulated by Myoparr knockdown in denervated tibialis anterior (TA) muscle of C57BL/6J mice, Table S2: Gene ontology (GO) annotation of genes commonly upregulated or downregulated by Myoparr knockdown for myogenic differentiation and muscle atrophy.
Author Contributions
Conceptualization, K.H.; Data curation, K.H. and K.T.; Formal analysis, K.H.; Funding acquisition, K.H. and K.T.; Investigation, K.H. and M.N.; Methodology, K.H.; Project administration, K.H. and K.T.; Resources, K.H., M.N. and K.T.; Supervision, K.H. and K.T.; Validation, K.H. and M.N.; Visualization, K.H.; Writing—original draft, K.H.; Writing—review & editing, K.H., M.N. and K.T.
Funding
This work was funded in part by JSPS KAKENHI (16K08599, 17K08646 and 19H03427), Intramural Research Grant (29-4) for Neurological and Psychiatric Disorders of NCNP, and a Grant-in-Aid from the NAKATOMI Foundation.
Acknowledgments
The authors thank H. Kurahashi and H. Inagaki for support of the RNA-seq analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Ward, M.; McEwan, C.; Mills, J.D.; Janitz, M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J. Hum. Transcr. 2015, 1, 2–9. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell. Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Hirose, T.; Mishima, Y.; Tomari, Y. Elements and machinery of non-coding RNAs: toward their taxonomy. EMBO Rep. 2014, 15, 489–507. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2018, 7, 877. [Google Scholar] [CrossRef]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinformatics 2017, 18, 558–576. [Google Scholar] [CrossRef]
- Marahrens, Y.; Panning, B.; Dausman, J.; Strauss, W.; Jaenisch, R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997, 11, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Davidow, L.S.; Warshawsky, D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999, 21, 400–404. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef]
- Feil, R.; Walter, J.; Allen, N.D.; Reik, W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1994, 120, 2933–2943. [Google Scholar]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Amândio, A.R.; Necsulea, A.; Joye, E.; Mascrez, B.; Duboule, D. Hotair is dispensible for mouse development. PLoS Genet. 2016, 12, e1006232. [Google Scholar] [CrossRef]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2013, e01749. [Google Scholar] [CrossRef]
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Considerations when investigating lncRNA function in vivo. Elife 2014, 3, e03058. [Google Scholar] [CrossRef]
- Han, X.; Luo, S.; Peng, G.; Lu, J.Y.; Cui, G.; Liu, L.; Yan, P.; Yin, Y.; Liu, W.; Wang, R.; et al. Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J. Mol. Cell Biol. 2018, 10, 175–178. [Google Scholar] [CrossRef]
- Faralli, H.; Dilworth, F.J. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp. Funct. Genomics 2012, 2012, 836374. [Google Scholar] [CrossRef]
- Hitachi, K.; Nakatani, M.; Takasaki, A.; Ouchi, Y.; Uezumi, A.; Ageta, H.; Inagaki, H.; Kurahashi, H.; Tsuchida, K. Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 2019, 20, e47468. [Google Scholar] [CrossRef]
- Sartori, R.; Schirwis, E.; Blaauw, B.; Bortolanza, S.; Zhao, J.; Enzo, E.; Stantzou, A.; Mouisel, E.; Toniolo, L.; Ferry, A.; et al. BMP signaling controls muscle mass. Nat. Genet. 2013, 45, 1309–1318. [Google Scholar] [CrossRef]
- Schertzer, J.D.; Plant, D.R.; Lynch, G.S. Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 2006, 13, 795–803. [Google Scholar] [CrossRef]
- Kaspric, N.; Picard, B.; Reichstadt, M.; Tournayre, J.; Bonnet, M. ProteINSIDE to easily investigate proteomics data from ruminants: Application to mine proteome of adipose and muscle tissues in bovine foetuses. PLoS ONE 2015, 10, e0128086. [Google Scholar] [CrossRef]
- Tono-Oka, S.; Tanase, S.; Miike, T.; Tanaka, H. Transient expression of collagen type XIV during muscle development and its reappearance after denervation and degeneration. J. Histochem. Cytochem. 1996, 44, 907–918. [Google Scholar] [CrossRef]
- Brandan, E.; Fuentes, M.E.; Andrade, W. Decorin, a chondroitin/dermatan sulfate proteoglycan is under neural control in rat skeletal muscle. J. Neurosci. Res. 1992, 32, 51–59. [Google Scholar] [CrossRef]
- Svensson, A.; Norrby, M.; Libelius, R.; Tågerud, S. Secreted frizzled related protein 1 (Sfrp1) and Wnt signaling in innervated and denervated skeletal muscle. J. Mol. Histol. 2008, 39, 329–337. [Google Scholar] [CrossRef]
- Moresi, V.; Williams, A.H.; Meadows, E.; Flynn, J.M.; Potthoff, M.J.; McAnally, J.; Shelton, J.M.; Backs, J.; Klein, W.H.; Richardson, J.A.; et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 2010, 143, 35–45. [Google Scholar] [CrossRef]
- Lee, S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef]
- Winbanks, C.E.; Chen, J.L.; Qian, H.; Liu, Y.; Bernardo, B.C.; Beyer, C.; Watt, K.I.; Thomson, R.E.; Connor, T.; Turner, B.J.; et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013, 203, 345–357. [Google Scholar] [CrossRef]
- Macpherson, P.C.D.; Farshi, P.; Goldman, D. Dach2-Hdac9 signaling regulates reinnervation of muscle endplates. Development 2015, 142, 4038–4048. [Google Scholar] [CrossRef]
- Tang, H.; Goldman, D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA 2006, 103, 16977–16982. [Google Scholar] [CrossRef]
- Méjat, A.; Ramond, F.; Bassel-Duby, R.; Khochbin, S.; Olson, E.N.; Schaeffer, L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 2005, 8, 313–321. [Google Scholar] [CrossRef]
- Cao, Y.; Kumar, R.M.; Penn, B.H.; Berkes, C.A.; Kooperberg, C.; Boyer, L.A.; Young, R.A.; Tapscott, S.J. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006, 25, 502–511. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Martinet, C.; Monnier, P.; Louault, Y.; Benard, M.; Gabory, A.; Dandolo, L. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 2016, 143, 962–971. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef]
- Sun, L.; Si, M.; Liu, X.; Choi, J.M.; Wang, Y.; Thomas, S.S.; Peng, H.; Hu, Z. Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle 2018, 9, 962–974. [Google Scholar] [CrossRef]
- Jin, J.J.; Lv, W.; Xia, P.; Xu, Z.Y.; Zheng, A.D.; Wang, X.J.; Wang, S.S.; Zeng, R.; Luo, H.M.; Li, G.L.; et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA 2018, 115, E9802–E9811. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 2018, 26, 2669–2680. [Google Scholar] [CrossRef]
- Liang, T.; Zhou, B.; Shi, L.; Wang, H.; Chu, Q.; Xu, F.; Li, Y.; Chen, R.; Shen, C.; Schinckel, A.P. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2018, 32, 377–389. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef]
- Chen, X.; He, L.; Zhao, Y.; Li, Y.; Zhang, S.; Sun, K.; So, K.; Chen, F.; Zhou, L.; Lu, L.; et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017, 3, 17002. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, K.; Zhao, Y.; Zhang, S.; Wang, X.; Li, Y.; Lu, L.; Chen, X.; Chen, F.; Bao, X.; et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015, 6, 10026. [Google Scholar] [CrossRef]
- Militello, G.; Hosen, M.R.; Ponomareva, Y.; Gellert, P.; Weirick, T.; John, D.; Hindi, S.M.; Mamchaoui, K.; Mouly, V.; Döring, C.; et al. A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 2018, 10, 102–117. [Google Scholar] [CrossRef]
- Neppl, R.L.; Wu, C.L.; Walsh, K. lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 2017, 216, 3497–3507. [Google Scholar] [CrossRef]
- Moreno-Miralles, I.; Schisler, J.C.; Patterson, C. New insights into bone morphogenetic protein signaling: Focus on angiogenesis. Curr. Opin. Hematol. 2009, 16, 195–201. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Nakatani, M.; Tsuchida, K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int. J. Biochem. Cell Biol. 2014, 47, 93–103. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).