Expression Profiling Identifies the Noncoding Processed Transcript of HNRNPU with Proliferative Properties in Pancreatic Ductal Adenocarcinoma
Abstract
:1. Introduction
2. Results
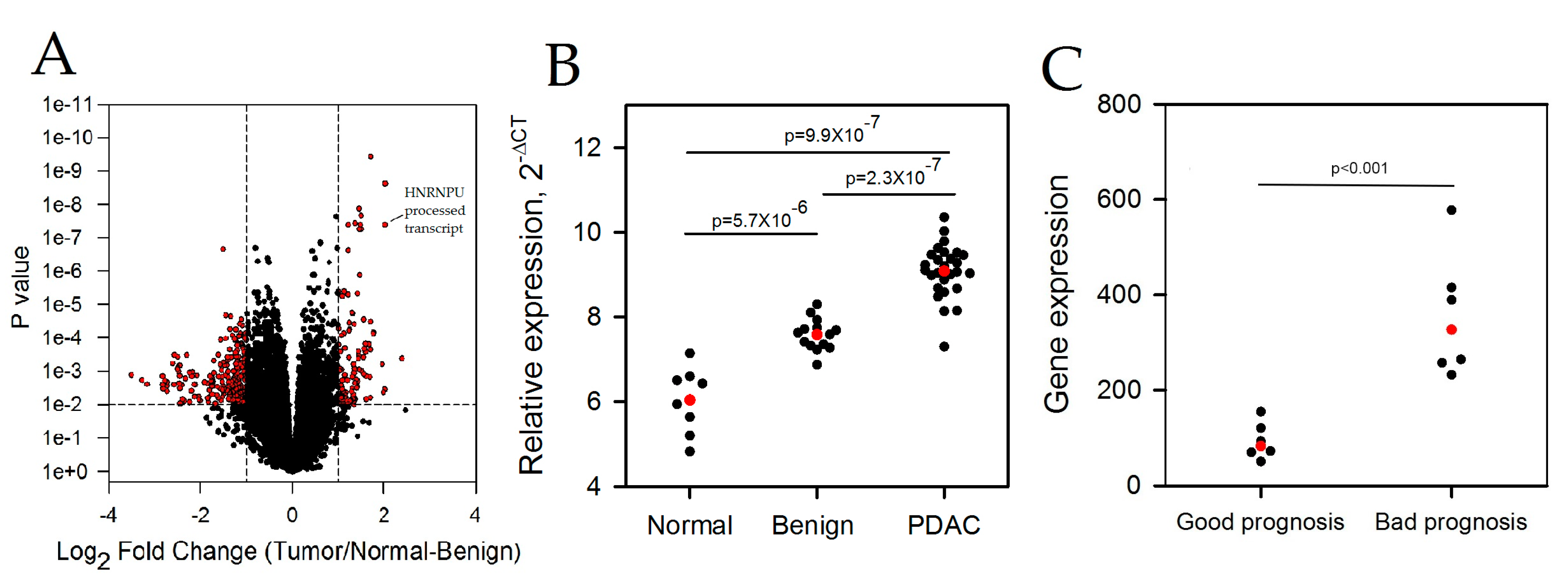
2.1. LncRNAs Are Differentially Expressed in Pancreatic Ductal Adenocarcinoma
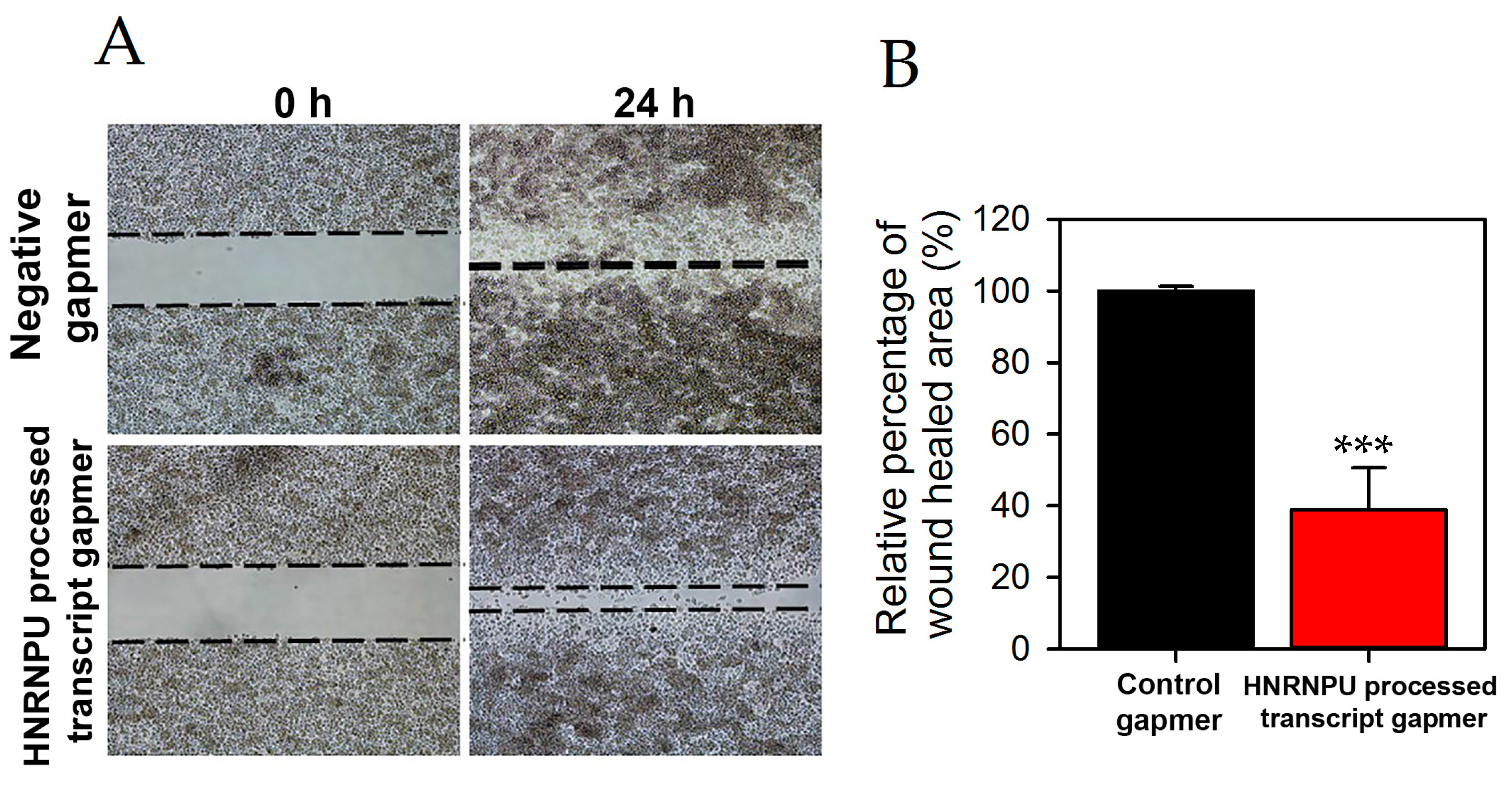
2.2. HNRNPU Processed Transcript Expression Is Increased in Pancreatic Cancer Cell Lines
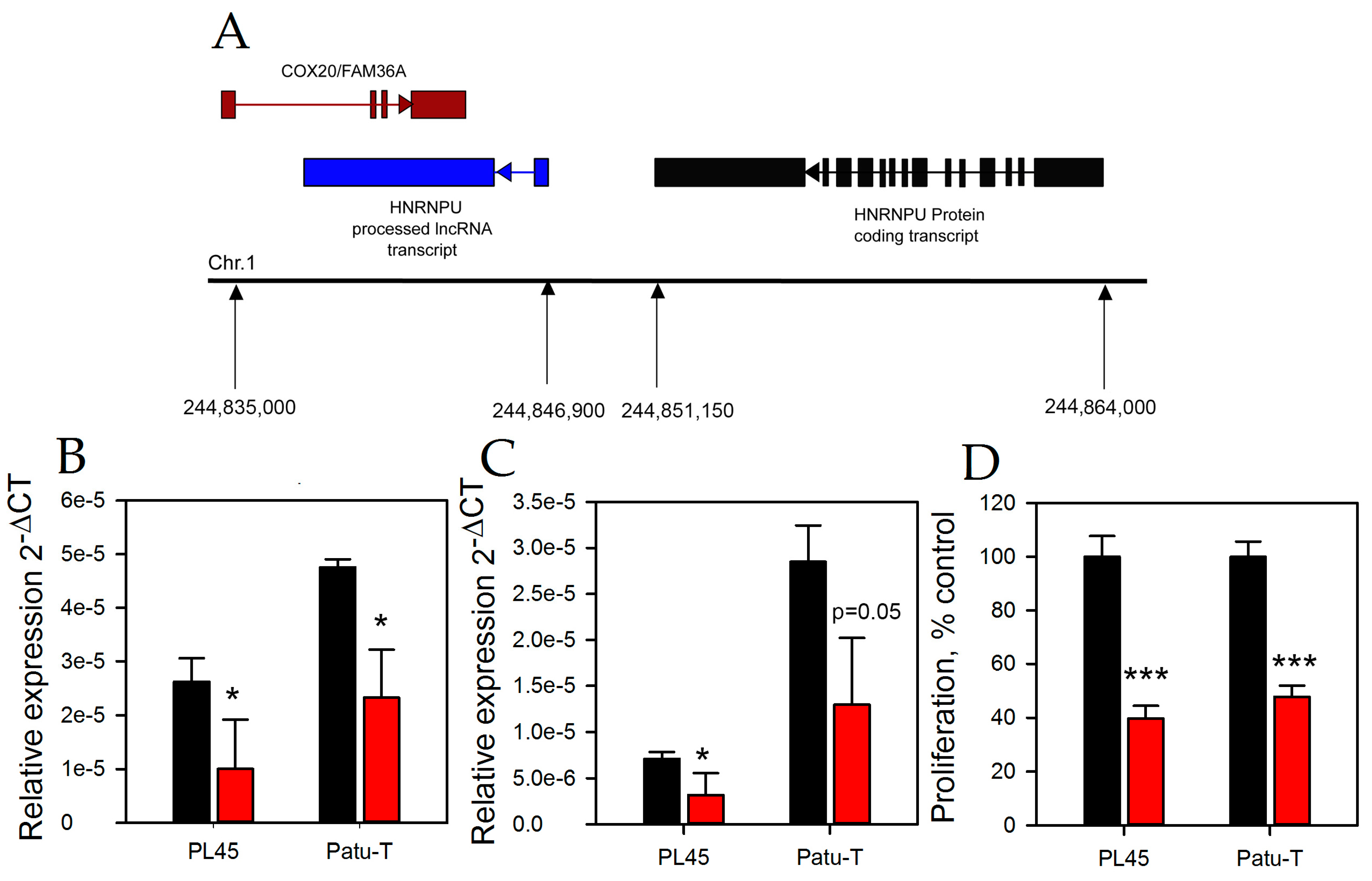
2.3. HNRNPU Processed Transcript Expression Is Regulated by HNRNPU Protein Coding Gene
3. Discussion
4. Materials and Methods
4.1. Patient Tissue Specimens
4.2. Microarray Data Processing and Analysis
4.3. Cell Culture and Treatments
4.4. qPCR
4.5. Cell Proliferation Assay
4.6. Migration and Invasion Assay
4.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Prasad, N.B.; Biankin, A.V.; Fukushima, N.; Maitra, A.; Dhara, S.; Elkahloun, A.G.; Hruban, R.H.; Goggins, M.; Leach, S.D. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005, 65, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.G.; Bhatt, D.; Taccioli, C.; Croce, C.M. Microrna expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007, 297, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression profiling identifies microrna signature in pancreatic cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.E.; Davison, T.S.; John, J.; Cannon, T.; Sipos, B.; Maghnouj, A.; Labourier, E.; Hahn, S.A. Microrna expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007, 26, 4442–4452. [Google Scholar] [CrossRef] [PubMed]
- Tahira, A.C.; Kubrusly, M.S.; Faria, M.F.; Dazzani, B.; Fonseca, R.S.; Maracaja-Coutinho, V.; Verjovski-Almeida, S.; Machado, M.C.C.; Reis, E.M. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol. Cancer 2011, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Z.; Zheng, S.; Zhou, Y.; Zhao, L.; Ye, H.; Zhao, X.; Gao, W.; Fu, Z.; Zhou, Q.; et al. Expression profile of long non-coding RNAs in pancreatic cancer and their clinical significance as biomarkers. Oncotarget 2015, 6, 35684–35698. [Google Scholar] [PubMed]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Azevedo-Pouly, A.C.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Nong, K.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014, 35, 9163–9169. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. Hotair is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, H.; Gao, Y.; Wang, Y.W.; Zhang, G.Q.; Pan, S.H.; Ji, L.; Kong, R.; Wang, G.; Jia, Y.H.; et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol. Cancer Ther. 2016, 15, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, A.; Vankelecom, H.; Van Eijsden, R.; Govaere, O.; Topal, B. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J. Exp. Clin. Cancer Res. 2012, 31, 68. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAS in cancer biology. Cancer Discov 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAS and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Mai, A.; Li, X.; Lai, Y.; Zheng, J.; Yang, Q.; Wu, J.; Nan, A.; Ye, S.; Jiang, Y. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol. Lett. 2013, 223, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, F.; Xu, Y.; Wang, B.; Zhao, Y.; Xu, W.; Shi, L.; Lu, X.; Liu, Q. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, hotair, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol. Appl. Pharmacol. 2015, 282, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ror modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. Cpc: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Beetz, N.; O'Keeffe, S.; Tapia, J.C.; Macpherson, L.; Chen, W.V.; Bassel-Duby, R.; Olson, E.N.; Maniatis, T. Hnrnp u protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. USA 2015, 112, E3020–E3029. [Google Scholar] [CrossRef] [PubMed]
- Yugami, M.; Kabe, Y.; Yamaguchi, Y.; Wada, T.; Handa, H. hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett 2007, 581, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Spraggon, L.; Dudnakova, T.; Slight, J.; Lustig-Yariv, O.; Cotterell, J.; Hastie, N.; Miles, C. hnRNP-U directly interacts with WT1 and modulates WT1 transcriptional activation. Oncogene 2007, 26, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, N.; Li, W.; Zhang, M. The function of the RNA-binding protein hnrnp in cancer metastasis. J. Cancer Res. Ther. 2013, 9, S129–S134. [Google Scholar] [PubMed]
- Chai, Y.; Liu, J.; Zhang, Z.; Liu, L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016, 5, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar]


| Gene Symbol | Fold Change | FDR | −log(p) × Fold Change |
|---|---|---|---|
| ENST00000407852 | 4.09 | 2.37 × 10−9 | 35.2 |
| TCONS_00012350 | −11.4 | 0.001301 | 32.9 |
| NCRNA00265 | 3.27 | 3.63 × 10−10 | 30.9 |
| NCRNA00201 | 4.08 | 4.03 × 10−8 | 30.1 |
| ENST00000536141 | −9.73 | 0.001841 | 26.6 |
| ENST00000543206 | −8.94 | 0.002479 | 23.3 |
| ENST00000452840 | 2.83 | 2.20 × 10−8 | 21.7 |
| TCONS_00022114 | 2.75 | 1.32 × 10−8 | 21.7 |
| ENST00000558097 | −5.94 | 0.000323 | 20.7 |
| ENST00000415104 | 2.80 | 4.03 × 10−8 | 20.7 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutaria, D.S.; Jiang, J.; Azevedo-Pouly, A.C.P.; Lee, E.J.; Lerner, M.R.; Brackett, D.J.; Vandesompele, J.; Mestdagh, P.; Schmittgen, T.D. Expression Profiling Identifies the Noncoding Processed Transcript of HNRNPU with Proliferative Properties in Pancreatic Ductal Adenocarcinoma. Non-Coding RNA 2017, 3, 24. https://doi.org/10.3390/ncrna3030024
Sutaria DS, Jiang J, Azevedo-Pouly ACP, Lee EJ, Lerner MR, Brackett DJ, Vandesompele J, Mestdagh P, Schmittgen TD. Expression Profiling Identifies the Noncoding Processed Transcript of HNRNPU with Proliferative Properties in Pancreatic Ductal Adenocarcinoma. Non-Coding RNA. 2017; 3(3):24. https://doi.org/10.3390/ncrna3030024
Chicago/Turabian StyleSutaria, Dhruvitkumar S., Jinmai Jiang, Ana Clara P. Azevedo-Pouly, Eun Joo Lee, Megan R. Lerner, Daniel J. Brackett, Jo Vandesompele, Pieter Mestdagh, and Thomas D. Schmittgen. 2017. "Expression Profiling Identifies the Noncoding Processed Transcript of HNRNPU with Proliferative Properties in Pancreatic Ductal Adenocarcinoma" Non-Coding RNA 3, no. 3: 24. https://doi.org/10.3390/ncrna3030024
APA StyleSutaria, D. S., Jiang, J., Azevedo-Pouly, A. C. P., Lee, E. J., Lerner, M. R., Brackett, D. J., Vandesompele, J., Mestdagh, P., & Schmittgen, T. D. (2017). Expression Profiling Identifies the Noncoding Processed Transcript of HNRNPU with Proliferative Properties in Pancreatic Ductal Adenocarcinoma. Non-Coding RNA, 3(3), 24. https://doi.org/10.3390/ncrna3030024






