The Chromosome 19 microRNA Cluster Facilitates Cancer Stemness in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
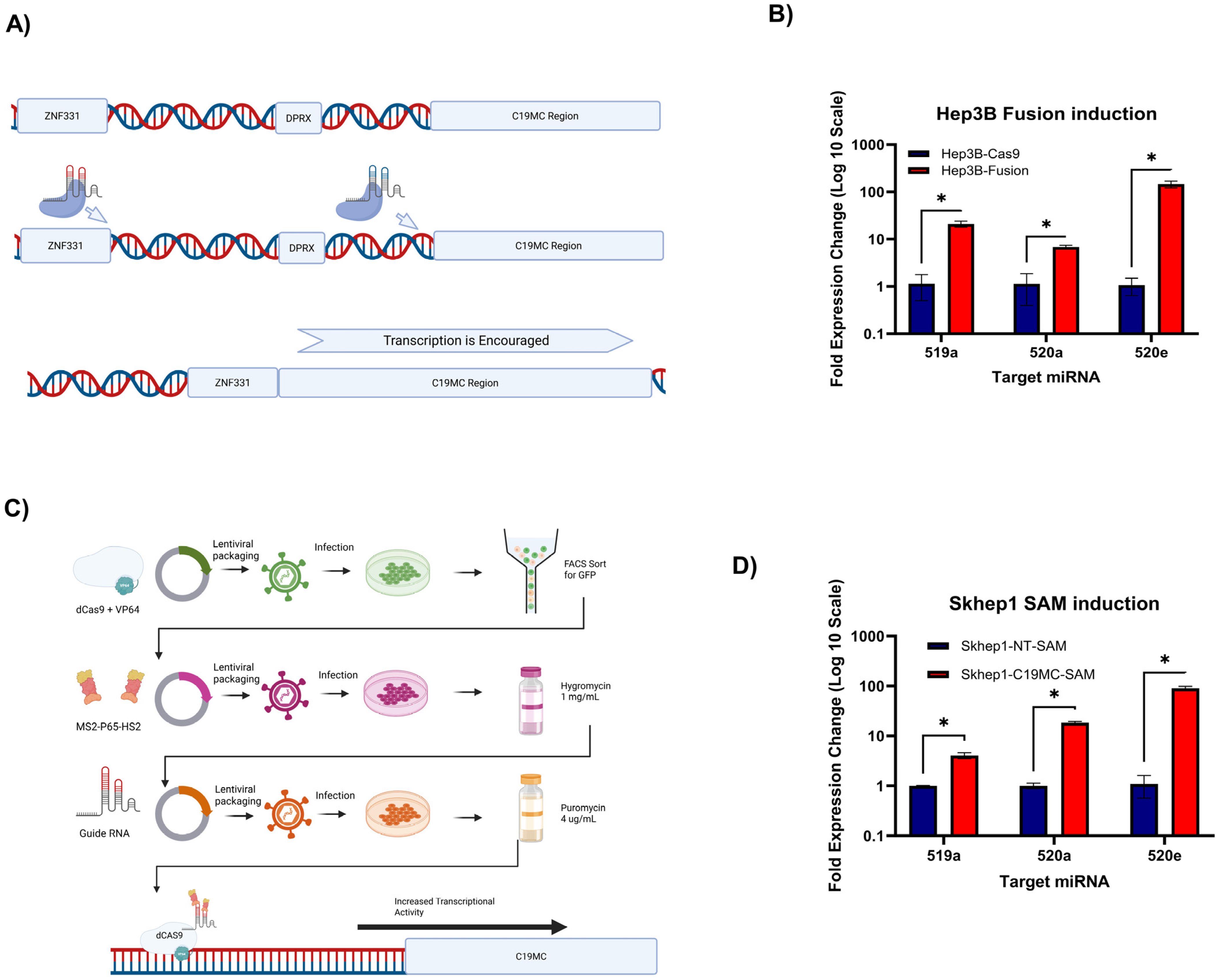
2.1. Generation of Two HCC Cell Lines That Constitutively Overexpress the Cluster-Wide C19MC miRNAs
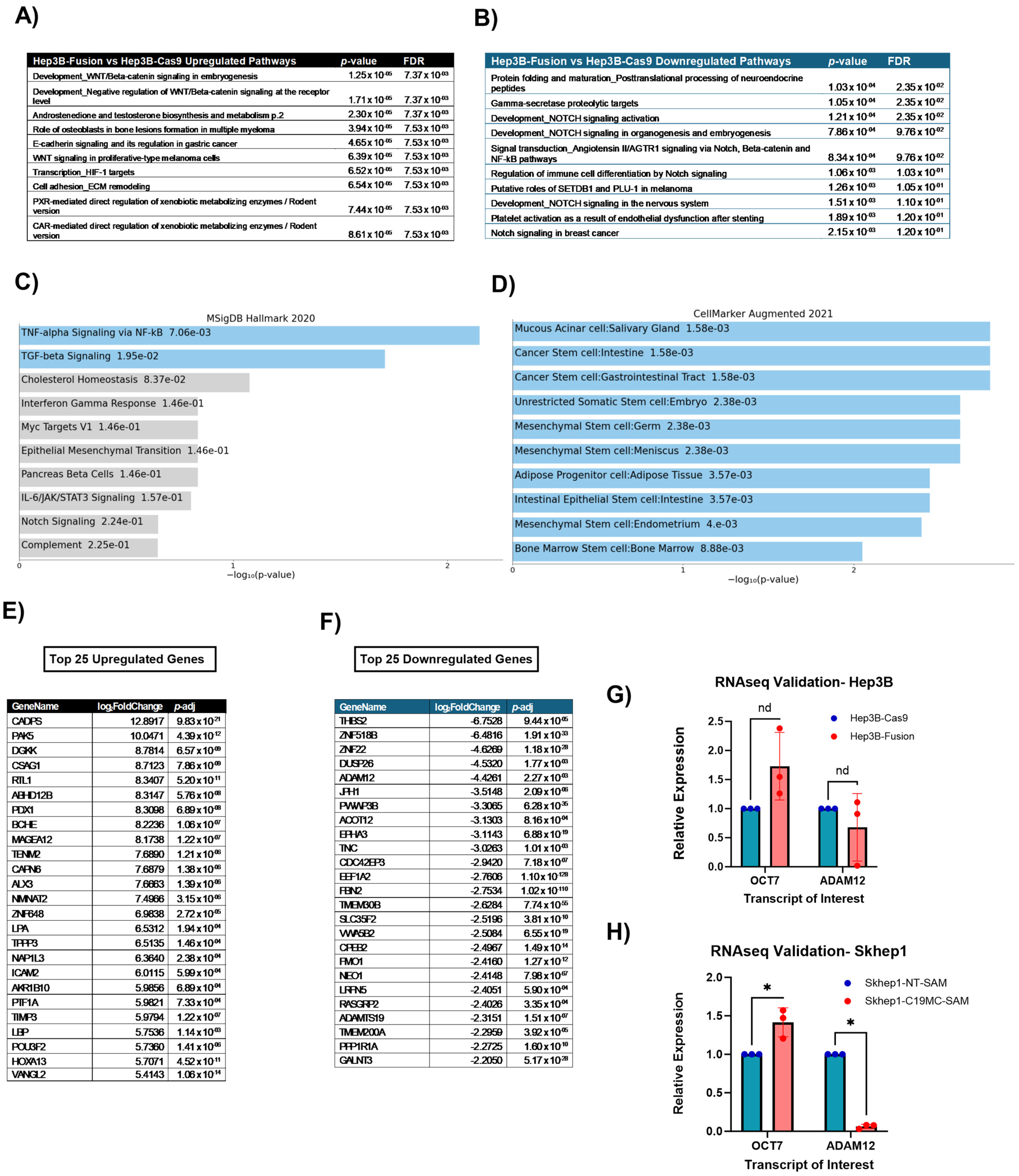
2.2. C19MC Overexpression in HCC Cell Lines Promotes Upregulation of Stemness-Related Genes and Gene Sets
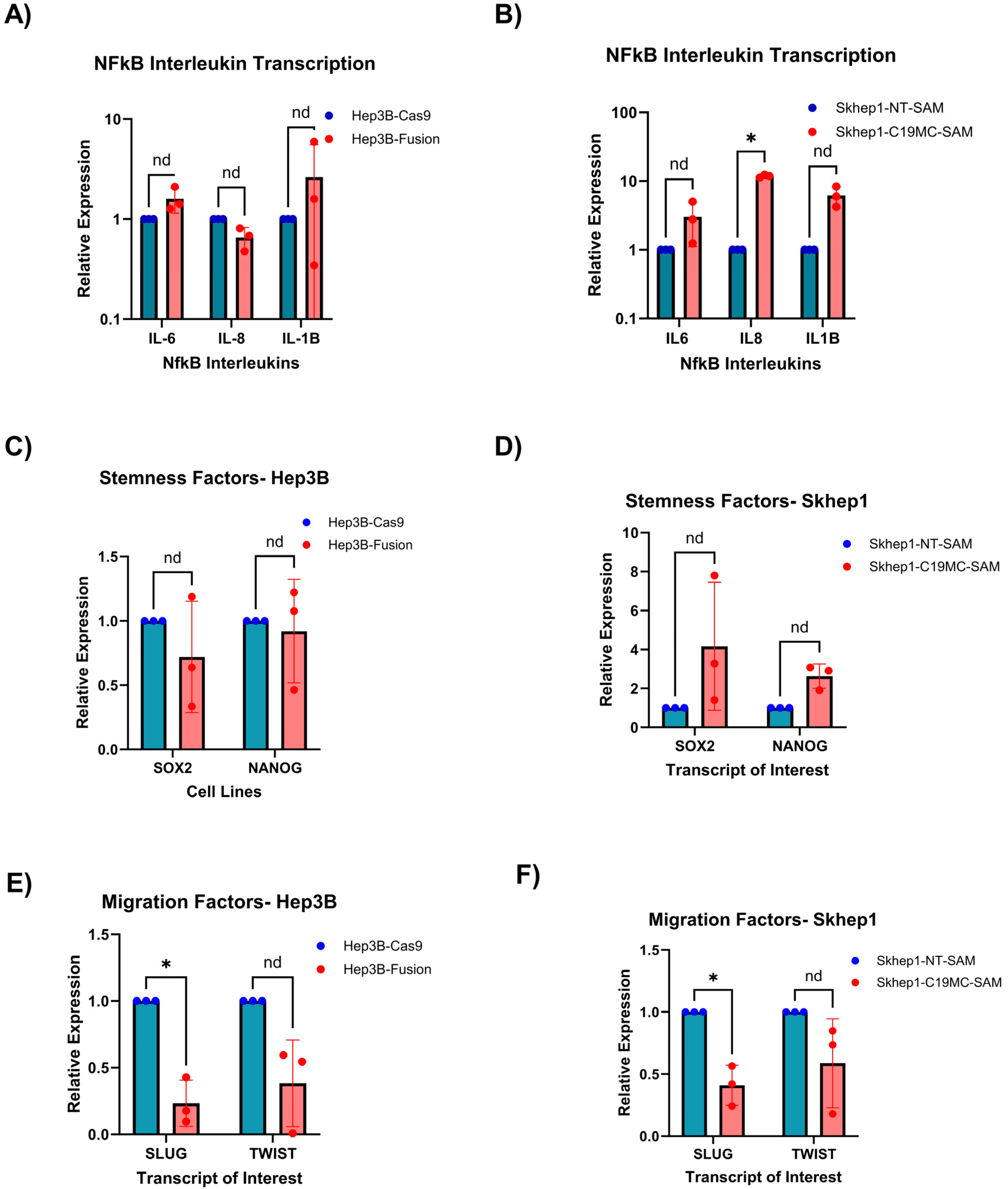
2.3. C19MC Overexpression in HCC Models Regulates the mRNA Expression of Stemness-Linked Cytokines (IL6, IL8, and IL1B) and Stemness-Related Transcription Factors
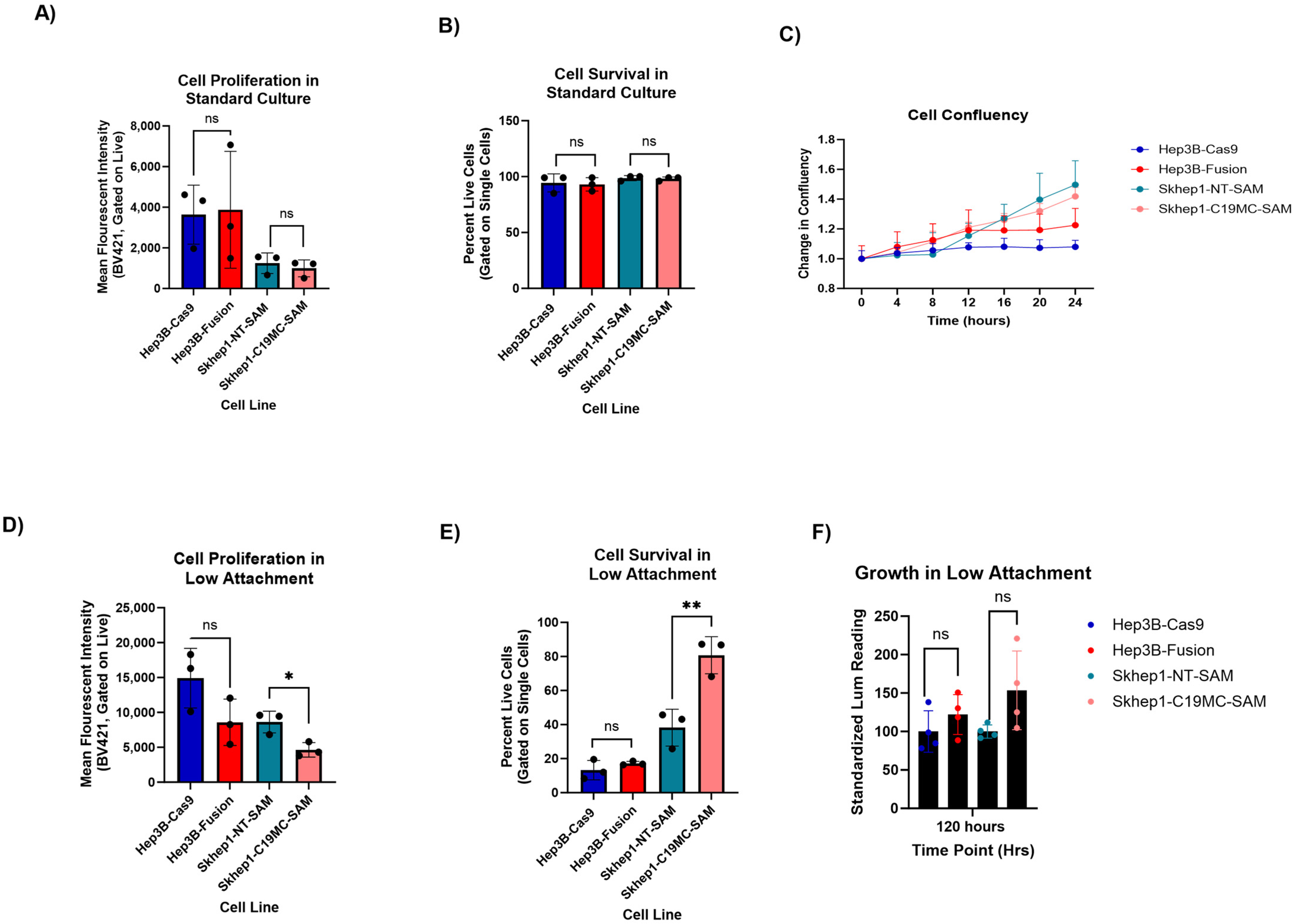
2.4. C19MC-Overexpressing HCC Models Exhibit Enhanced Cellular Fitness in Anchorage-Independent Growth Conditions
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. CRISPRa sgRNA Cloning
4.3. CRISPR Methods
4.4. Lentivirus Generation and Infection
4.5. MicroRNA Expression Analysis
4.6. RNA Sequencing and Analysis
4.7. TargetScan Enrichment and Analysis
4.8. CellTrace Violet Assay
4.9. mRNA Expression Analysis
4.10. Growth in Low Attachment (GILA) Assay
4.11. Cell Confluency Assays
4.12. Statistical Analysis
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| C19MC | Chromosome 19 microRNA Cluster |
| CSC | Cancer stem cell |
| HCC | Hepatocellular carcinoma |
References
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefèvre, A.; Coullin, P.; Moore, G.E.; Cavaillé, J. The Primate-Specific microRNA Gene Cluster (C19MC) Is Imprinted in the Placenta. Hum. Mol. Genet. 2010, 19, 3566–3582. [Google Scholar] [CrossRef]
- Lin, S.; Cheung, W.K.C.; Chen, S.; Lu, G.; Wang, Z.; Xie, D.; Li, K.; Lin, M.C.M.; Kung, H. Computational Identification and Characterization of Primate-Specific microRNAs in Human Genome. Comput. Biol. Chem. 2010, 34, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Mouillet, J.-F.; Chu, T.; Parks, W.T.; Sadovsky, E.; Knöfler, M.; Sadovsky, Y. C19MC MicroRNAs Regulate the Migration of Human Trophoblasts. Endocrinology 2014, 155, 4975–4985. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Sadovsky, E.; Sheridan, M.A.; Roberts, R.M.; Coyne, C.B.; Sadovsky, Y. Chromosome 19 microRNAs Exert Antiviral Activity Independent from Type III Interferon Signaling. Placenta 2018, 61, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Setty, B.A.; Jinesh, G.G.; Arnold, M.; Pettersson, F.; Cheng, C.-H.; Cen, L.; Yoder, S.J.; Teer, J.K.; Flores, E.R.; Reed, D.R.; et al. The Genomic Landscape of Undifferentiated Embryonal Sarcoma of the Liver Is Typified by C19MC Structural Rearrangement and Overexpression Combined with TP53 Mutation or Loss. PLoS Genet. 2020, 16, e1008642. [Google Scholar] [CrossRef]
- Beck, A.; Gabler-Pamer, L.; Alencastro Veiga Cruzeiro, G.; Lambo, S.; Englinger, B.; Shaw, M.L.; Hack, O.A.; Liu, I.; Haase, R.D.; de Biagi, C.A.O.; et al. Cellular Hierarchies of Embryonal Tumors with Multilayered Rosettes Are Shaped by Oncogenic microRNAs and Receptor–Ligand Interactions. Nat. Cancer 2025, 6, 1035–1055. [Google Scholar] [CrossRef]
- Pei, Y.-C.; Huang, G.-H.; Yao, X.-H.; Bian, X.-W.; Li, F.; Xiang, Y.; Yang, L.; Lv, S.-Q.; Liu, J. Embryonal Tumor with Multilayered Rosettes, C19MC-Altered (ETMR): A Newly Defined Pediatric Brain Tumor. Int. J. Clin. Exp. Pathol. 2019, 12, 3156–3163. [Google Scholar]
- Jinesh, G.G.; Flores, E.R.; Brohl, A.S. Chromosome 19 miRNA Cluster and CEBPB Expression Specifically Mark and Potentially Drive Triple Negative Breast Cancers. PLoS ONE 2018, 13, e0206008. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Smallin, M.T.; Mtchedlidze, N.; Godwin, I.; Napoli, M.; Hackel, N.; Phadke, M.S.; Deshpande, A.A.; Li, X.; Lockhart, J.H.; et al. C19MC miRNA-520G Induces SP100 Antiviral Gene Transcription and Inhibits Melanin Production in Skin Cutaneous Melanoma. Genes Dis. 2024, 11, 60–63. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Napoli, M.; Smallin, M.T.; Davis, A.; Ackerman, H.D.; Raulji, P.; Montey, N.; Flores, E.R.; Brohl, A.S. Mutant P53s and Chromosome 19 microRNA Cluster Overexpression Regulate Cancer Testis Antigen Expression and Cellular Transformation in Hepatocellular Carcinoma. Sci. Rep. 2021, 11, 12673. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Napoli, M.; Ackerman, H.D.; Raulji, P.M.; Montey, N.; Flores, E.R.; Brohl, A.S. Regulation of MYO18B mRNA by a Network of C19MC miRNA-520G, IFN-γ, CEBPB, P53 and bFGF in Hepatocellular Carcinoma. Sci. Rep. 2020, 10, 12371. [Google Scholar] [CrossRef]
- Zhou, Y.; Frings, O.; Branca, R.M.; Boekel, J.; le Sage, C.; Fredlund, E.; Agami, R.; Orre, L.M. microRNAs with AAGUGC Seed Motif Constitute an Integral Part of an Oncogenic Signaling Network. Oncogene 2017, 36, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Augello, C.; Vaira, V.; Caruso, L.; Destro, A.; Maggioni, M.; Park, Y.N.; Montorsi, M.; Santambrogio, R.; Roncalli, M.; Bosari, S. MicroRNA Profiling of Hepatocarcinogenesis Identifies C19MC Cluster as a Novel Prognostic Biomarker in Hepatocellular Carcinoma. Liver Int. 2012, 32, 772–782. [Google Scholar] [CrossRef]
- Rui, T.; Xu, S.; Zhang, X.; Huang, H.; Feng, S.; Zhan, S.; Xie, H.; Zhou, L.; Ling, Q.; Zheng, S. The Chromosome 19 microRNA Cluster, Regulated by Promoter Hypomethylation, Is Associated with Tumour Burden and Poor Prognosis in Patients with Hepatocellular Carcinoma. J. Cell. Physiol. 2020, 235, 6103–6112. [Google Scholar] [CrossRef]
- Augello, C.; Colombo, F.; Terrasi, A.; Trombetta, E.; Maggioni, M.; Porretti, L.; Rossi, G.; Guerneri, S.; Silipigni, R.; Bosari, S.; et al. Expression of C19MC miRNAs in HCC Associates with Stem-Cell Features and the Cancer-Testis Genes Signature. Dig. Liver Dis. 2018, 50, 583–593. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Smallin, M.T.; Mtchedlidze, N.; Napoli, M.; Lockhart, J.H.; Flores, E.R.; Brohl, A.S. C19MC Drives Nucleolar Invasion of Mitochondria and Meiotic Nuclear Division in Human Cancers. iScience 2024, 27, 111132. [Google Scholar] [CrossRef]
- Kleinman, C.L.; Gerges, N.; Papillon-Cavanagh, S.; Sin-Chan, P.; Pramatarova, A.; Quang, D.-A.K.; Adoue, V.; Busche, S.; Caron, M.; Djambazian, H.; et al. Fusion of TTYH1 with the C19MC microRNA Cluster Drives Expression of a Brain-Specific DNMT3B Isoform in the Embryonal Brain Tumor ETMR. Nat. Genet. 2014, 46, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Jinesh, G.G.; Mtchedlidze, N.; Devarapalli, V.; Adhikary, S.; Lockhart, J.H.; Napoli, M.; Godwin, I.; Reiser, M.A.; Cen, L.; Liu, X.; et al. Brohl CEBPB, C19MC, and Defective Autophagy Drive Novel Podosomal Belt to Macropinocytosis Transition, Lipid Accumulation, and HBV A-to-I RNA-Editing. Commun. Biol. 2025. [Google Scholar]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-Scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-Catenin Signaling Pathway in Cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Lv, Z.; Liu, R.-D.; Chen, X.-Q.; Wang, B.; Li, L.-F.; Guo, Y.-S.; Chen, X.-J.; Ren, X.-Q. HIF-1α Promotes the Stemness of Oesophageal Squamous Cell Carcinoma by Activating the Wnt/Β-catenin Pathway. Oncol. Rep. 2019, 42, 726–734. [Google Scholar] [CrossRef]
- Notch Signaling Pathway: Architecture, Disease, and Therapeutics. Signal Transduction and Targeted Therapy. Available online: https://www.nature.com/articles/s41392-022-00934-y (accessed on 10 July 2025).
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of microRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem. Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef]
- Bao, Z.; Ji, W.; Yang, Y.; Chen, Z.; Li, Z.; Wang, K.; Lu, T.; Yu, Y.; Xia, W.; Lu, S. PAK5 Promotes the Cell Stemness Ability by Phosphorylating SOX2 in Lung Squamous Cell Carcinomas. Exp. Cell Res. 2020, 395, 112187. [Google Scholar] [CrossRef]
- Kim, K.-P.; Han, D.W.; Kim, J.; Schöler, H.R. Biological Importance of OCT Transcription Factors in Reprogramming and Development. Exp. Mol. Med. 2021, 53, 1018–1028. [Google Scholar] [CrossRef]
- Bourd-Boittin, K.; Le Pabic, H.; Bonnier, D.; L’Helgoualc’h, A.; Théret, N. RACK1, a New ADAM12 Interacting Protein. J. Biol. Chem. 2008, 283, 26000–26009. [Google Scholar] [CrossRef]
- Enns, C.A.; Zhang, R.H.; Jue, S.; Zhang, A.-S. Hepcidin Expression Is Associated with Increased γ-Secretase–Mediated Cleavage of Neogenin in the Liver. J. Biol. Chem. 2024, 300, 107927. [Google Scholar] [CrossRef] [PubMed]
- Rico Montanari, N.; Anugwom, C.M.; Boonstra, A.; Debes, J.D. The Role of Cytokines in the Different Stages of Hepatocellular Carcinoma. Cancers 2021, 13, 4876. [Google Scholar] [CrossRef]
- Vernot, J.P. Senescence-Associated Pro-Inflammatory Cytokines and Tumor Cell Plasticity. Front. Mol. Biosci. 2020, 7, 63. [Google Scholar] [CrossRef]
- Brasier, A.R. The Nuclear Factor-κB–Interleukin-6 Signalling Pathway Mediating Vascular Inflammation. Cardiovasc. Res. 2010, 86, 211–218. [Google Scholar] [CrossRef]
- Rex, J.; Lutz, A.; Faletti, L.E.; Albrecht, U.; Thomas, M.; Bode, J.G.; Borner, C.; Sawodny, O.; Merfort, I. IL-1β and TNFα Differentially Influence NF-κB Activity and FasL-Induced Apoptosis in Primary Murine Hepatocytes During LPS-Induced Inflammation. Front. Physiol. 2019, 10, 117. [Google Scholar] [CrossRef]
- Fousek, K.; Horn, L.A.; Palena, C. Interleukin-8: A Chemokine at the Intersection of Cancer Plasticity, Angiogenesis, and Immune Suppression. Pharmacol. Ther. 2021, 219, 107692. [Google Scholar] [CrossRef]
- Lu, L.; Wang, P.; Zou, Y.; Zha, Z.; Huang, H.; Guan, M.; Wu, Y.; Liu, G. IL-1β Promotes Stemness of Tumor Cells by Activating Smad/ID1 Signaling Pathway. Int. J. Med. Sci. 2020, 17, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in Cancer Stemness: Tumor Malignancy and Therapeutic Potentials. J. Mol. Cell Biol. 2018, 12, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.R.; Yang, T.; Wang, J.; Chao, H.-P.; Tang, D.G. NANOG in Cancer Stem Cells and Tumor Development: An Update and Outstanding Questions. Stem. Cells 2015, 33, 2381–2390. [Google Scholar] [CrossRef] [PubMed]
- Mong, E.F.; Yang, Y.; Akat, K.M.; Canfield, J.; VanWye, J.; Lockhart, J.; Tsibris, J.C.M.; Schatz, F.; Lockwood, C.J.; Tuschl, T.; et al. Chromosome 19 microRNA Cluster Enhances Cell Reprogramming by Inhibiting Epithelial-to-Mesenchymal Transition. Sci. Rep. 2020, 10, 3029. [Google Scholar] [CrossRef]
- Chu, X.; Tian, W.; Ning, J.; Xiao, G.; Zhou, Y.; Wang, Z.; Zhai, Z.; Tanzhu, G.; Yang, J.; Zhou, R. Cancer Stem Cells: Advances in Knowledge and Implications for Cancer Therapy. Signal Transduct. Target. Ther. 2024, 9, 170. [Google Scholar] [CrossRef]
- Rotem, A.; Janzer, A.; Izar, B.; Ji, Z.; Doench, J.G.; Garraway, L.A.; Struhl, K. Alternative to the Soft-Agar Assay That Permits High-Throughput Drug and Genetic Screens for Cellular Transformation. Proc. Natl. Acad. Sci. USA 2015, 112, 5708–5713. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Banerjee, S.; Sarkar, F.H. Emerging Role of Notch in Stem Cells and Cancer. Cancer Lett. 2009, 279, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Zamfirescu, A.-M.; Yatsenko, A.S.; Shcherbata, H.R. Notch Signaling Sculpts the Stem Cell Niche. Front. Cell Dev. Biol. 2022, 10, 1027222. [Google Scholar] [CrossRef] [PubMed]
- Radharani, N.N.V.; Yadav, A.S.; Nimma, R.; Kumar, T.V.S.; Bulbule, A.; Chanukuppa, V.; Kumar, D.; Patnaik, S.; Rapole, S.; Kundu, G.C. Tumor-Associated Macrophage Derived IL-6 Enriches Cancer Stem Cell Population and Promotes Breast Tumor Progression via Stat-3 Pathway. Cancer Cell Int. 2022, 22, 122. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and Their Therapeutic Implications in Cancer Treatment. Stem. Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Underwood, M.T.; Devarapalli, V.; Jinesh, G.G.; Lockhart, J.H.; Napoli, M.; Mtchedlidze, N.; Flores, E.R.; Brohl, A.S. The Chromosome 19 microRNA Cluster Facilitates Cancer Stemness in Hepatocellular Carcinoma. Non-Coding RNA 2025, 11, 74. https://doi.org/10.3390/ncrna11060074
Underwood MT, Devarapalli V, Jinesh GG, Lockhart JH, Napoli M, Mtchedlidze N, Flores ER, Brohl AS. The Chromosome 19 microRNA Cluster Facilitates Cancer Stemness in Hepatocellular Carcinoma. Non-Coding RNA. 2025; 11(6):74. https://doi.org/10.3390/ncrna11060074
Chicago/Turabian StyleUnderwood, Marian T., Varsha Devarapalli, Goodwin G. Jinesh, John H. Lockhart, Marco Napoli, Nino Mtchedlidze, Elsa R. Flores, and Andrew S. Brohl. 2025. "The Chromosome 19 microRNA Cluster Facilitates Cancer Stemness in Hepatocellular Carcinoma" Non-Coding RNA 11, no. 6: 74. https://doi.org/10.3390/ncrna11060074
APA StyleUnderwood, M. T., Devarapalli, V., Jinesh, G. G., Lockhart, J. H., Napoli, M., Mtchedlidze, N., Flores, E. R., & Brohl, A. S. (2025). The Chromosome 19 microRNA Cluster Facilitates Cancer Stemness in Hepatocellular Carcinoma. Non-Coding RNA, 11(6), 74. https://doi.org/10.3390/ncrna11060074





