Exploring microRNAs, One Cell at a Time
Abstract
1. Introduction
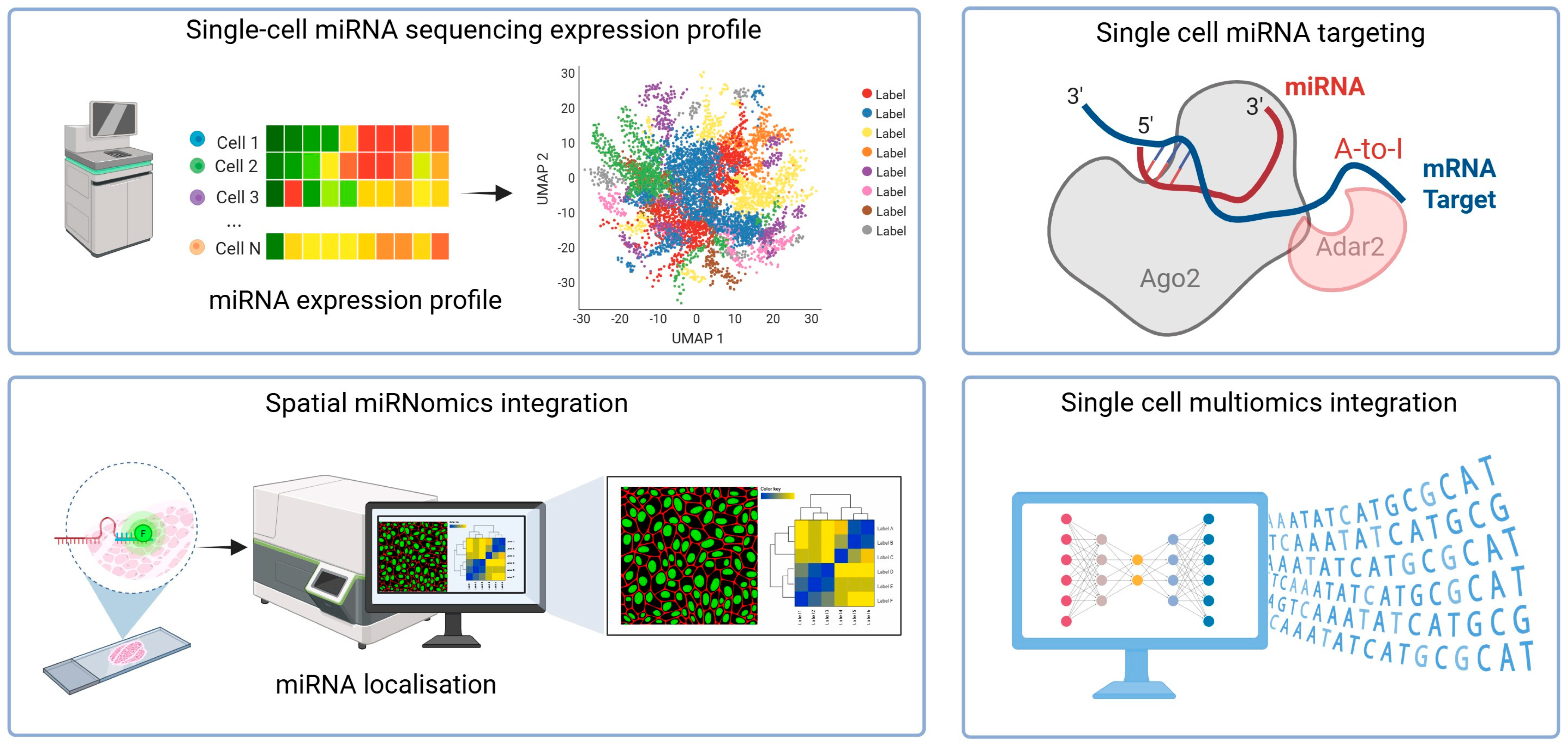
2. Single-Cell microRNA Sequencing
3. Spatial microRNA Detection
4. MicroRNA Targeting at the Single-Cell Level
5. Bioinformatics Approaches
6. Conclusions and Future Directions
6.1. Multimodal Analysis
6.2. Clinical Implications of Spatial miRNomics
6.3. Sequencing Sensitivity and Throughput
6.4. Evolution of Single-Cell microRNA-mRNA Co-Sequencing Techniques
6.5. Data and Code Sharing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kim, H.; Lee, Y.Y.; Kim, V.N. The biogenesis and regulation of animal microRNAs. Nat. Rev. Mol. Cell Biol. 2025, 26, 276–296. [Google Scholar] [CrossRef]
- Lokhande, H.A. Bioinformatics Analysis of miRNA Sequencing Data. Methods Mol. Biol. 2023, 2595, 225–237. [Google Scholar]
- Benesova, S.; Kubista, M.; Valihrach, L. Small RNA-Sequencing: Approaches and Considerations for miRNA Analysis. Diagnostics 2021, 11, 964. [Google Scholar] [CrossRef]
- Faridani, O.R.; Abdullayev, I.; Hagemann-Jensen, M.; Schell, J.P.; Lanner, F.; Sandberg, R. Single-cell sequencing of the small-RNA transcriptome. Nat. Biotechnol. 2016, 34, 1264–1266. [Google Scholar] [CrossRef] [PubMed]
- Giraldez, M.D.; Spengler, R.M.; Etheridge, A.; Godoy, P.M.; Barczak, A.J.; Srinivasan, S.; De Hoff, P.L.; Tanriverdi, K.; Courtright, A.; Lu, S.; et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018, 36, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Dard-Dascot, C.; Naquin, D.; D’aUbenton-Carafa, Y.; Alix, K.; Thermes, C.; van Dijk, E. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genom. 2018, 19, 118. [Google Scholar] [CrossRef]
- Hücker, S.M.; Fehlmann, T.; Werno, C.; Weidele, K.; Lüke, F.; Schlenska-Lange, A.; Klein, C.A.; Keller, A.; Kirsch, S. Single-cell microRNA sequencing method comparison and application to cell lines and circulating lung tumor cells. Nat. Commun. 2021, 12, 4316. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zheng, J.; Chen, Z.; Liu, Y.; Dura, B.; Kwak, M.; Xavier-Ferrucio, J.; Lu, Y.-C.; Zhang, M.; Roden, C.; et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 2019, 10, 95. [Google Scholar] [CrossRef]
- Li, J.; Tian, J.; Cai, T. Integrated analysis of miRNAs and mRNAs in thousands of single cells. Sci. Rep. 2025, 15, 1636. [Google Scholar] [CrossRef]
- Bai, Z.; Zhang, D.; Gao, Y.; Tao, B.; Zhang, D.; Bao, S.; Enninful, A.; Wang, Y.; Li, H.; Su, G.; et al. Spatially exploring RNA biology in archival formalin-fixed paraffin-embedded tissues. Cell 2024, 187, 6760–6779 e24. [Google Scholar] [CrossRef]
- Sekar, V.; Mármol-Sánchez, E.; Kalogeropoulos, P.; Stanicek, L.; Sagredo, E.A.; Widmark, A.; Doukoumopoulos, E.; Bonath, F.; Biryukova, I.; Friedländer, M.R. Detection of transcriptome-wide microRNA-target interactions in single cells with agoTRIBE. Nat. Biotechnol. 2024, 42, 1296–1302. [Google Scholar] [CrossRef]
- Hagemann-Jensen, M.; Abdullayev, I.; Sandberg, R.; Faridani, O.R. Small-seq for single-cell small-RNA sequencing. Nat. Protoc. 2018, 13, 2407–2424. [Google Scholar] [CrossRef] [PubMed]
- Shore, S.; Henderson, J.M.; Lebedev, A.; Salcedo, M.P.; Zon, G.; McCaffrey, A.P.; Paul, N.; Hogrefe, R.I. Small RNA Library Preparation Method for Next-Generation Sequencing Using Chemical Modifications to Prevent Adapter Dimer Formation. PLoS ONE 2016, 11, e0167009. [Google Scholar] [CrossRef] [PubMed]
- Robles-Remacho, A.; Nilsson, M. Spatial miRNomics: Towards the integration of microRNAs in spatial biology. Nat. Rev. Genet. 2025, 26, 291–292. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.; Rishik, S.; Hirsch, P.; Keller, V.; Fehlmann, T.; Kern, F.; Keller, A. SingmiR: A single-cell miRNA alignment and analysis tool. Nucleic Acids Res. 2024, 52, W374–W380. [Google Scholar] [CrossRef]
- Herbst, E.; Mandel-Gutfreund, Y.; Yakhini, Z.; Biran, H. Inferring single-cell and spatial microRNA activity from transcriptomics data. Commun. Biol. 2025, 8, 87. [Google Scholar] [CrossRef]
- Gondal, M.N.; Farooqi, H.M.U. Single-Cell Transcriptomic Approaches for Decoding Non-Coding RNA Mechanisms in Colorectal Cancer. Noncoding RNA 2025, 11, 24. [Google Scholar] [CrossRef]
- Stanojevic, S.; Li, Y.; Ristivojevic, A.; Garmire, L.X. Computational Methods for Single-cell Multi-omics Integration and Alignment. Genom. Proteom. Bioinform. 2022, 20, 836–849. [Google Scholar] [CrossRef]
- Du, Y.; Ding, X.; Ye, Y. The spatial multi-omics revolution in cancer therapy: Precision redefined. Cell Rep. Med. 2024, 5, 101740. [Google Scholar] [CrossRef]
- Hücker, S.M.; Kirsch, S. Single Cell Micro RNA Sequencing Library Preparation. Methods Mol. Biol. 2024, 2752, 189–199. [Google Scholar]
- Zhang, J.; Yan, S.; Chang, L.; Guo, W.; Wang, Y.; Wang, Y.; Zhang, P.; Chen, H.-Y.; Huang, S. Direct microRNA Sequencing Using Nanopore-Induced Phase-Shift Sequencing. iScience 2020, 23, 100916. [Google Scholar] [CrossRef]
- Maguire, S.; Lohman, G.J.S.; Guan, S. A low-bias and sensitive small RNA library preparation method using randomized splint ligation. Nucleic Acids Res. 2020, 48, e80. [Google Scholar] [CrossRef]
- Velut, L.; Fancello, L.; Cherradi, N.; Guyon, L. Single-cell microRNA-mRNA co-sequencing techniques convey large potential for understanding microRNA regulations but require careful and systemic approaches. Nat. Commun. 2025, 16, 5255. [Google Scholar] [CrossRef]
- Li, M.; Ang, K.S.; Teo, B.; Rom, U.; Nguyen, M.N.; Maurer-Stroh, S.; Chen, J. Rediscovering publicly available single-cell data with the DISCO platform. Nucleic Acids Res. 2025, 53, D932–D938. [Google Scholar] [CrossRef]
| Method | Summary | Advantages | Limitations | Ref |
|---|---|---|---|---|
| Sandberg Protocol I (SB) | A small RNA sequencing protocol for single cells that captures miRNAs using sequential 3′ and 5′ adapter ligation. |
|
| [7] |
| Sandberg Protocol II CleanTag (SBN_CL) | An “optimised” version of SB that incorporates chemically modified CleanTag adapters to suppress the formation of adapter dimers. |
|
| [7] |
| Half-Cell Genomics Approach | A co-sequencing method where a single cell’s lysate is split into two equal frac-tions, enabling the simulta-neous profiling of both miRNAs and mRNAs from the same cell. |
|
| [8] |
| PSCSR-seq V2 | A parallel, barcoded sin-gle-cell coprofiling method that integrates a SMART-seq reaction into the PSCSR small RNA workflow, ena-bling high-sensitivity se-quencing of miRNAs along-side rich mRNA information from thousands of individ-ual cells. |
|
| [9] |
| Patho-DBiT | A spatial transcriptomics platform that conducts whole transcriptome se-quencing on archival for-malin-fixed paraf-fin-embedded tissues using in situ polyadenylation for diverse RNA capture and microfluidic barcoding for spatial mapping. |
|
| [10] |
| agoTRIBE | A method for detecting miRNA–target interactions in single cells involves fusing Argonaute2 with a hyper-active ADAR2 RNA-editing domain. This fusion allows endogenous miRNAs to edit target mRNAs, producing A-to-I (read as A-to-G) marks that can be identified through single-cell RNA sequencing. |
|
| [11] |
| Method | Summary | Advantages | Limitations | Ref |
|---|---|---|---|---|
| SingmiR | A user-friendly web server with an intuitive interface provides a comprehensive bioinformatics pipeline for single-cell miRNA-seq data, encompassing everything from raw read pre-processing and miRNA quantification to di-mension reduction and dif-ferential expression analysis. |
|
| [15] |
| miTEA-HiRes | A statistical method inferring miRNA activity from sin-gle-cell and spatial tran-scriptomics data based on validated target expression patterns. It enables the creation of spatial activity maps and the assessment of overall activity, as well as differential analysis in single-cell data. |
|
| [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreutz, J.; Mitić, T.; Caporali, A. Exploring microRNAs, One Cell at a Time. Non-Coding RNA 2025, 11, 73. https://doi.org/10.3390/ncrna11060073
Kreutz J, Mitić T, Caporali A. Exploring microRNAs, One Cell at a Time. Non-Coding RNA. 2025; 11(6):73. https://doi.org/10.3390/ncrna11060073
Chicago/Turabian StyleKreutz, Jessica, Tijana Mitić, and Andrea Caporali. 2025. "Exploring microRNAs, One Cell at a Time" Non-Coding RNA 11, no. 6: 73. https://doi.org/10.3390/ncrna11060073
APA StyleKreutz, J., Mitić, T., & Caporali, A. (2025). Exploring microRNAs, One Cell at a Time. Non-Coding RNA, 11(6), 73. https://doi.org/10.3390/ncrna11060073







