Abstract
Long noncoding RNAs (lncRNAs) are transcripts generated by polymerase II, therefore subject to 5′ capping and 3′ polyadenylation, categorized as such when they are at least 200 nt in size and lack coding function. The lncRNAs were initially interpreted as spurious transcription products, but over the last two decades an increasing amount of evidence has accumulated for regulatory functions. They are found in all taxonomic groups, including bacteria, archaea, fungi, animals and plants. In fungi, global analyses anticipate their presence in higher numbers than initially expected considering the simplicity of these organisms. Except for the numerous studies performed in budding and fission yeast, relatively few lncRNAs have been investigated in sufficient detail in the rest of the fungi, but their number has increased steadily in recent years. The lncRNAs can be transcribed from intergenic regions or coincide totally or partially with protein-coding genes, in which case they are most frequently antisense transcripts. Their regulatory functions can be performed by a wide variety of mechanisms, both in cis on neighboring genes and in trans on distant genes or on proteins. Among the most frequent mechanisms are interference on the transcription of neighboring genes and generation of epigenetic modifications in the environment of target genes. Here, we review the most representative cases of global analyses of the presence of lncRNAs in fungal transcriptomes and describe the lncRNAs that have received more detailed attention.
1. Introduction
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts that are larger than 200 nt in size and, unlike messenger RNA (mRNA), do not possess significant or recognizably functional reading frames, and therefore are not expected to encode proteins [1]. Initially they were considered functionless transcriptional products or mere “genetic noise”, but today it is known that many lncRNAs play important roles in regulating the expression of other genes through very diverse mechanisms, covering epigenetic, transcriptional and post-transcriptional levels [2,3]. The interaction of lncRNAs with their regulatory targets does not conform to easily predictable mechanisms. In many cases, these are non-canonical interactions, such as the formation of a triple helix with a double-strand DNA. The diversity of mechanisms makes them difficult to study, despite efforts to understand and classify them [4]. The synthesis of lncRNAs is carried out by the same process as that of mRNAs. That is, they are transcribed by RNA polymerase II and can undergo maturation steps at the level of intron splicing, and additions of 5′ cap and poly-A tail [5].
There are different types of lncRNAs. From the point of view of their location in the genome or their relationship with coding genes, they can be intergenic, or associated with coding genes, in which case they can be transcribed antisense to the coding sequence, be located in an intron, or even be variants of the coding strand [6]. In overall, lncRNAs may exert their action by interacting with other molecules in the cell [7], which may be DNA, other mRNAs, miRNAs, or proteins, ranging from transcription factors, chromatin, or proteins mediating different processes, and there are computational tools to predict many of these interactions [8]. When they are antisense, they can form double-stranded RNAs with their complementary RNAs, which can affect their stability and availability for translation [9,10,11,12]. In higher organisms, their expression is usually highly specific to the level of tissue, developmental stage or physiological conditions, suggesting specialized functions [13,14,15].
Discovery of lncRNAs arose from the development of global transcriptomic techniques, first with cDNA microarrays, followed by cDNA tiling arrays, and fostered in the last decade by the enormous development of high-throughput sequencing technologies, such as RNA-seq [5]. At present, the large amount of accumulated information makes lncRNAs a type of regulatory elements with a wide variety of functions and mechanisms that are increasingly better characterized, and for which there are well-established study procedures [16] and databases that keep growing, including many involved in human diseases [17]. Initiatives such as ENCODE [18] or NONCODE [19] revealed the existence of thousands of previously unannotated non-coding transcripts in the human genome or in other organisms. Their presence, however, is widespread in all species investigated. This review summarizes the current state of knowledge on lncRNAs in fungi, about which new information is currently accumulating exponentially.
2. Identification of Antisense RNAs and lncRNAs in Fungi
Global transcriptomic analyses, and more recently those based on RNA-seq technologies, have revolutionized the identification and study of RNAs of unknown functions and in all types of organisms, including fungi [20], with long non-coding RNAs (lncRNAs) as an outstanding class. Large-scale detection and quantification of any type of transcripts have revealed the wide diversity of lncRNAs involved in gene regulation of a wide range of processes [16]. The data obtained in fungi has expanded the knowledge about their transcriptional complexity and the evolution of their genomes and has allowed us to glimpse that lncRNAs play more important roles than might have been expected considering the apparent simplicity of these organisms.
Recent works are often focused on the identification of lncRNAs, as a functionally open group with similar transcriptional characteristics, but previous studies focused on non-coding RNAs, with special emphasis on antisense RNAs because of their obvious regulatory characteristics [21]. For the most part, the latter are in fact a subclass of lncRNAs.
2.1. Saccharomyces cerevisiae
The existence of transcripts that do not match annotated genes, or that match but are transcribed from the complementary strand (antisense transcripts), has been known in S. cerevisiae for more than two decades. Before RNA-seq techniques became available, the identification of non-coding transcripts of unknown functions, including the first lncRNAs, was initiated in S. cerevisae using tiling array technology [22,23]. This technique, consisting in the use of overlapping probes (tiles) that cover the entire genome (or specific regions of interest) in a uniform manner, without bias towards annotated genes, allows the detection of any type of transcripts, including non-coding and antisense transcripts (Figure 1).
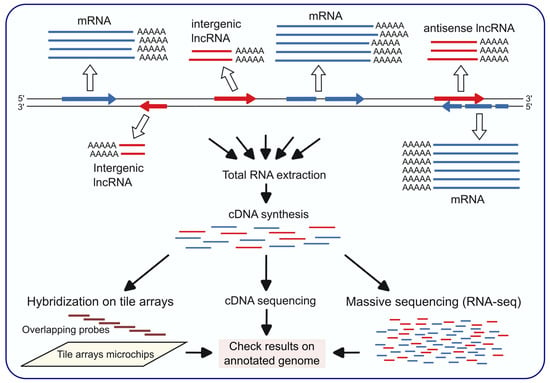
Figure 1.
Procedures most frequently used for the detection of lncRNAs. Open and black arrows represent PolII transcription and experimental steps, respectively. Protein-coding sequences are indicated in blue. The two major classes of lncRNAs, antisense and intergenic, are represented in red.
In addition to genome tiling array, cDNA sequencing and RNA-seq analyses (Figure 1) of S. cerevisiae transcripts uncovered the existence of many non-coding RNAs [24]. The different techniques are complementary and give rise to different sets of lncRNAs, although they may overlap. For example, in some of the first studies the number of intergenic lncRNAs identified by cDNA analysis was 667 [25], by tiling arrays 234 [23], and by RNA-seq 487 [26]. Expression levels are highly variable, and in many cases are similar to those of coding genes. Overall, the transcription of non-coding RNAs is more extensive than expected, as indicated by the approximately 1000 intergenic or antisense transcripts detected by full-length cDNA analysis [25], subsequently corroborated by the identification of 1103 antisense transcripts by strand-specific RNA-seq analyses, many of them with differential expression depending on the growth conditions and conserved in other close species [27].
Not all antisense transcripts lack coding functions. The S. cerevisiae MDF1 gene, a negative regulator of the mating pathway that interacts with MATa2, completely overlaps with the antisense gene ADF1, which encodes a protein that blocks MDF1 transcription [28]. This is expected to be a rare case, due to coding constraints imposed by the overlap of the two sequences. On the other hand, the presence of an antisense RNA does not necessarily imply regulation of the involved protein-coding gene. In a study in which the transcription of antisense RNAs from 162 coding genes in S. cerevisiae was repressed and the impact on the amounts of proteins from their target genes was analyzed under four different culture conditions, a significant impact on protein levels in at least one condition was observed in only a quarter of them [29]. The effects were less likely for genes that showed high expression and more likely when the antisense RNA overlapped with the transcription start sites (TSSs).
The finding of lncRNAs in S. cerevisiae was promoted by global transcriptomics analysis of mutants affected in RNA degradation mechanisms. The reason for this is that, although non-coding lncRNAs undergo a maturation process at their 5′ and 3′ ends like that of coding mRNAs, many are not exported to the cytoplasm and remain in the nucleus performing short-term regulatory functions, until they are degraded by the exosome, an enzyme complex involved in RNA processing and degradation [30,31]. On the other hand, a population of more stable lncRNAs that are not rapidly degraded, described as Stable Unannotated Transcripts (SUTs), is also detected [32]. In both cases, they are frequently transcribed from nucleosome-free regions associated with the promoters of other genes or with their 3′ regions, from which bidirectional transcription is often observed [32,33]. The occurrence of unstable RNAs, initially called Cryptic Unstable Transcripts (CUTs), was evidenced in mutants affected in exosome function, where they accumulate abnormally [34]. The reason for their different behavior is that, unlike the reproducibility of the polyadenylation sites of the mRNAs to be translated, these lncRNAs have greater heterogeneity at their 3′ ends. In S. cerevisiae, this is due to their premature transcriptional termination by the Nrd1 complex, which interacts with the PolII complex and marks it for future exosomal degradation [35,36]. This mechanism plays an important role in the control of lncRNAs, as indicated by the 1526 transcripts that increase in the absence of Nrd1, which led to them being named NUTs, from Nrd1-unterminated transcripts [37].
As a representative example, transcriptome comparison between wild type and a mutant lacking Rrp1p, an essential component of the nuclear exosome, allowed the identification of 98 non-coding transcripts of 350–800 nt, half of them corresponding to promoter regions and the rest to intergenic or antisense transcripts [38]. On the other hand, comparison with the transcriptome of the mutant lacking Rrp6p, an essential component of the RNA processing exosome [39], revealed a strong increase in polyadenylated RNAs of sizes around 250–500 nt, largely associated with promoters [40]. Because of their length, they are not expected to be full-length mRNAs and they could regulate the transcription of downstream RNAs.
High-resolution oligonucleotide arrays were used to study the role of the nuclear exosome component Rrp6 in the transcriptome of S. cerevisiae involved in the vegetative and sexual phases of its life cycle [41]. The data allowed the identification of 1452 differentially expressed non-coding RNAs specific to meiosis, named as unannotated meiotic transcripts (MUTs). Rrp6 keeps MUT levels low during the budding stage, but the degradation of Rrp6 at the onset of meiosis leads to the accumulation of MUT in successive waves. As a result, diploid cells lacking Rrp6 cannot undergo premeiotic DNA replication and subsequent meiotic development. Therefore, Rpr6 plays a critical role in the asexual-sexual transition and the initiation of meiosis through differential ncRNA degradation depending on its activity levels.
The involvement of epigenetic regulation in the mechanisms of action of 566 antisense lncRNAs that increase in quantity in the absence of Rrp6 was also investigated using high-density tiling arrays [42]. For this goal, the regulation of the target genes was analyzed in mutants for the H3K4 histone methyl transferase Set1 or the histone deacetylases Hda1 and Rpd3. For 469 of the antisense lncRNAs, their increase in the Rrp6 mutant did not affect the transcription of their sense genes. Among the rest, for which the higher lncRNA level resulted in a repressing effect on the target gene, two types were found which differed in the silencing mechanism. For 69, silencing of the sense gene involved histone modification activities, as revealed by the effects of modifying enzymes, and for 28, silencing occurred through another mechanism, possibly transcription interference. The repressive effect of antisense transcription on expression of the sense genes was related to the efficiency of early termination of the antisense RNA and its polyadenylation. The data suggested that a subset of antisense lncRNAs, or their transcription, can recruit histone-modifying proteins to specific targets to control their expression.
A quality control checkpoint for aberrant transcripts, known as nonsense-mediated decay (NMD) pathway, is involved in the degradation of mRNAs with a premature termination codon, an upstream open reading frame, or an abnormally long 3′ UTR [43]. The transcripts recognized by the NMD machinery are degraded by exonuclease Xrn1 in the cytoplasm. In S. cerevisiae an important proportion of lncRNAs are targeted by NMD and degraded by Xrn1. These lncRNAs are known as XUTs (Xrn1-sensitive unstable transcripts), as their levels increase in the absence of Xrn1 activity [44]. The RNA-seq study with the mutant revealed 1658 XUTs, of which two-thirds are antisense to coding genes. XUTs are targeted to Xrn1 through the translation-dependent Nonsense-Mediated mRNA Decay (NMD) pathway [45], as indicated by their accumulation in mutants lacking Upf1, an RNA helicase essential for NMD, and Rrp6, a nuclear exosome catalytic subunit [46]. This yeast contains seven other cytoplasmic helicases, two of which have also been shown to be actively involved in the control of XUT levels [47].
Techniques based on the use of tile arrays or cDNA sequencing preceded those based on massive RNA sequencing. The latter, due to their affordability and high resolution, especially strand-specific sequencing, has gradually replaced other techniques, and has facilitated its extension to a growing number of fungi. Table 1 shows the results of the identification of antisense and intergenic lncRNAs in some representative cases of S. cerevisiae and the most informative ones of those presented in the following sections.

Table 1.
Examples of global identifications of intergenic and antisense lncRNAs in wild-type fungi.
2.2. Other Yeasts
The content of antisense lncRNAs has also been analyzed in other species of budding yeasts, where they have been found in smaller numbers. Thus, while 2230 were detected in S. cerevisiae, their number was 810 in Saccharomyces mikatae, 525 in Saccharomyces kudriavzevii, and 431 in Saccharomyces uvarum, with very similar size distributions [61]. The lower number in other Saccharomyces species can be attributed to the partial loss of the RNAi machinery, totally absent in S. cerevisiae, since in a very closely related budding yeast species that retains fully active this machinery, Naumovozyma castellii, the number of antisense lncRNAs identified is only 177, and their average sizes are smaller. Although it has been proposed that the presence of cytoplasmic RNAi in N. castellii affects the antisense lncRNA transcriptome, in this species too, its levels are mainly controlled by the Xrn1 degradation system [62].
Another yeast for which lncRNA analyses have been carried out is Schizosaccharomyces pombe. A detailed study achieved in this yeast, combining RNA-seq and high-density tiling array techniques, and comparing different growth conditions, such as minimal or rich media, thermal, and oxidative stress, and different phases of meiosis, allowed for a highly detailed characterization of its transcriptome [48]. This work revealed the existence of transcripts for numerous previously unidentified genes, including 427 for lncRNAs, whose average sizes and expression levels were lower than those of coding genes. A high degree of bidirectional transcription throughout the genome was found, although in most cases transcripts from one of the strands predominate. Comparison of the data with those obtained with a rrp6 mutant, affected in nuclear exosome function, showed that 36 of the new transcripts accumulate in greater quantities in the mutant, indicating that they are normally degraded in the nucleus. Later, a different protocol for the analysis of high-resolution tiling microarrays allowed the identification of 510 lncRNAs differentially expressed under oxidative stress [63].
A strand-specific RNA-seq study of the transcriptomes of S. pombe and three other related fission yeast species included a comprehensive survey of putative noncoding RNAs [49]. As a result, 1097 putative noncoding transcripts were found, of which 213 might be alternative UTRs due to their overlap with annotated UTRs on the same strand. Of the remaining 884 ncRNAs, 338 were intergenic and 546 antisense, most of them presumably lncRNAs. Of the 338 intergenic ones, 138 were conserved in location and 26 were conserved in sequence in at least another related species, suggesting the existence of potentially biologically important non-coding RNAs. Of the 546 antisense transcripts, 328 were conserved in two or species, also suggesting that they are functionally relevant.
A more exhaustive analysis of potential lncRNAs was carried out in S. pombe by comparing RNA-seq data from 12 mutants of nuclear and cytoplasmic RNA processing systems, and nine different physiological conditions, quiescent or stationary phase cells at different incubation times and several stages of meiotic differentiation development [64]. As a result, 5775 potential lncRNAs were identified, most of which had low expression levels but were induced in some of the conditions studied. They have been classified in three classes depending on the predominant system involved in their degradation, CUTs, degraded by the exosome, XUTs, degraded by Exo2/Xrn1, and DUTs (from Dicer-sensitive Unstable Transcripts), degraded by the RNAi machinery. The analysis was completed with their relationship with neighboring mRNAs and nucleosome positioning patterns, observing that many lncRNAs originate from nucleosome-depleted sites, often with bidirectional transcription. The three types of lncRNA degradation mechanisms have overlapping functions, as they can degrade the same lncRNAs, albeit with different affinities. Together they form a sophisticated RNA surveillance network that is partly responsible for the proper functioning of gene regulation in the cell to optimize its viability at different stages of its life cycle or under different growth conditions.
Other studies in S. pombe have also investigated different aspects of lncRNA accumulation. The histone chaperone Spt6 participates in the regulation of the expression of numerous genes by interacting with histones and RNA polymerase II. A combined approach in a spt6 mutant, both at the transcriptome and chromatin structure levels, showed an increase in antisense RNAs in 70% of genes, probably due to alterations in histone modifications [65]. An RNA-seq variant, NET-RNA-seq, consisting in specific sequencing of nascent RNAPII RNAs, was applied to investigate antisense transcription in S. pombe and its connection with Exo2/Xrn1 through the effect of the exo2 mutation [66]. This method, that detects only elongating transcripts, found antisense transcription in 3455 protein-coding genes (68% of total), mostly XUTS, thus increasing the catalog of antisense RNAs known to date in this yeast. A study on the role of histone deacetylation in the regulation of meiosis genes included a global analysis of CUTs, corresponding to those whose levels rise at least twice in a mutant lacking Rrp6, allowing the identification of some 2500 [67]. Noncoding RNA sets depend on the methodologies used. Improved genome annotation redefining mRNA borders allowed to detect 487 novel ncRNAs [68].
There are fewer examples of global lncRNA studies in other yeasts. An RNA-seq study of the methylotrophic species Pichia pastoris to investigate the effect of stress caused by overexpression of heterologous phospholipase A2 and exposure to methanol led to the identification of 208 lncRNAs, of which 168 were antisense, 36 intergenic, and 4 intronic [50]. As found in other yeasts, their average sizes and expression levels were lower than those of coding genes. Of the 208 lncRNAs, 18 showed differential changes in their expression in the investigated conditions, and three were chosen for a more detailed study, described in Section 4.3.
2.3. Dimorphic Fungi
Dimorphic fungi are characterized for their ability to alternate between yeast and filamentous forms in their life cycles. The ability for this developmental switch provides them enormous adaptability, allowing them to thrive in very different environments. This morphological plasticity, often linked to pathogenicity, makes them organisms with a great capacity to adapt to adverse situations or respond to environmental signals.
In its process of maize infection, the basiodimycete U. maydis changes from haploid yeast to dikaryon hyphae, which form thick-walled diploid teliospores that are used for dispersal [69]. In a sequencing study of expressed sequence tags (ESTs) libraries from different developmental stages, transcripts for 4675 genes were found, of which antisense transcripts were detected for 210 of them, most with possible ORFs but no real recognizable coding function [51]. Subsequent analysis of a cDNA library of dikaryotic cells brought the number of antisense transcripts to 247 and extended the analysis to the identification of more than a hundred non-coding RNAs, defined by the absence of ORFs with database matches [70]. Some of them could be associated with pathogenesis, according to their expression patterns (see Section 4.4.1).
Subsequently, a comparative study between U. maydis and two other species of the smut fungi group, Ustilago hordeum and Sporisorium reilianum, revealed a high number of lncRNAs, many shared and others species-specific [52]. Most of them were antisense or intergenic transcripts, with total numbers for the first class of 2624 in U. maydis, 1606 in U. hordeum, and 1949 in S. reilianum, about half conserved in all three species, indicating relevant regulatory functions. On the other hand, 2414, 1206 and 1776 intergenic transcripts were found, respectively, in the same species. Some of them could be protein-coding transcripts that escaped the annotation processes, but presumably the vast majority are true lncRNAs. The high numbers of lncRNAs found in these fungi reveal a high regulatory complexity in which lncRNAs play an important role.
In an RNA-seq analysis specifically aimed at identifying lncRNAs in the rice pathogen Ustilaginoidea virens, samples from five stages of infection were analyzed, accompanied by microscopic follow-up [53]. As a result, 1724 lncRNAs were found, of which 1084 corresponded to intergenic regions and 566 to antisense transcripts. Of the latter, some were antisense of transport genes, and one of them was investigated in detail (described in Section 4.4.4).
Due to its importance as a human pathogen, several studies focused on the role of lncRNAs in the pathogenic activity in Candida species. A comparative analysis of numerous transcriptomic datasets from five pathogens of this genus: Candida albicans, Candida tropicalis, Candida parapsilosis, Candida auris and Candida glabrata identified hundreds of lncRNAs, located in both intergenic regions and protein-coding genes [71]. Although many show low sequence conservation between species, some lncRNAs are syntenic and enriched in common motifs. Coexpression between certain lncRNAs and protein-coding genes was also detected, suggesting functional interactions. They also identified differentially expressed lncRNAs during infection of human epithelial cells in four of the species studied. The findings highlight the relevance of lncRNAs as potential regulators of virulence and adaptation in the genus Candida.
2.4. Filamentous Fungi
Filamentous fungi constitute the most extensive and heterogeneous group within fungi. Their multicellular growth form and the formation of hyphae, mycelia, and reproductive structures, the latter normally involved in the formation of asexual or sexual spores for dispersal, make many of them models of great interest for studies of development and differentiation. In many cases, they are pathogenic organisms, but they are also capable of living freely, for which they have developed highly versatile metabolisms. In some cases, they stand out for their ability to degrade biopolymers, such as cellulose or chitin, or to produce secondary metabolites with different biological properties, such as antibiotics or mycotoxins.
The ascomycete Neurospora crassa is an important model in molecular biology of filamentous fungi [72], with relevant contributions in the areas of photobiology and circadian rhythms, and with extensively developed genetic and molecular tools [73]. In this organism, which is non-pathogenic and does not produce mycotoxins, different illumination and temperature conditions were studied by RNA-seq and 939 lncRNAs were identified, evenly distributed across its seven chromosomes [54]. Of these, 477 were antisense with varying degrees of overlap with coding genes. Eleven lncRNAs were stimulated by light, opening up new fields in the photobiology of this fungus. In a subsequent study combining ChiP-seq, RNA-seq, and polysome fractionation, the number of lncRNAs rose to 1478 intergenic lncRNAs and 1056 antisense lncRNAs, most of which were shorter, typically without introns, and with lower expression levels than the average for coding genes [74]. This study highlights the importance of non-coding transcription in N. crassa, with ca. 20% of RNA polymerase II transcripts being lncRNAs.
Aspergillus flavus is a phytopathogenic fungus and opportunistic human pathogen of great interest due to its ability to produce mycotoxins, among which aflatoxin stands out for its health risks [75]. Several surveys have been carried out with this fungus. In an analysis of EST sequences by microarray technology, 352 antisense transcripts were detected [76]. Temperature is a determining environmental factor for aflatoxin production in infected maize. Temperature regulation was detected in 32 of these transcripts, some of them related to secondary metabolism and aflatoxin metabolism. A later RNA-seq analysis in mycelia and sclerotia in the same fungus showed that 30% of all transcripts were unknown, the vast majority, approximately one thousand, corresponding to antisense transcripts presumably involved in post-transcriptional regulation of coding genes [55]. The new ncRNA transcripts identified have globally similar expression levels to those of the coding genes, and 62% of these have lengths greater than 500 nt, so the vast majority must be lncRNAs.
Another RNA-seq study in A. flavus under different stress conditions, such as changes in water activity, CO2 concentration and temperature, identified 472 putative lncRNAs [77]. Many of them showed differential expressions to varying degrees under stress, suggesting key roles in aflatoxin biosynthesis, respiration, cell survival, and metabolism. In addition, some sense lncRNAs, downregulated by temperature, osmotic stress and CO2, could indirectly modulate proline metabolism. Subcellular localization analysis indicated that some regulated lncRNAs are concentrated in the nucleus, whereas others accumulate mainly in the cytoplasm. Possible regulatory targets based on chromosomal locations, correlation with the expression patterns of other genes, e.g., those for aflatoxin biosynthesis, and predicted interactions with milRNAs were suggested, laying the groundwork for further studies on specific regulatory mechanisms.
Numbers of lncRNAs were found to be very high in some fungi. Transcriptome analysis of Cordyceps militaris, an insect pathogenic fungus, identified 4140 putative lncRNAs, many of them manifesting changes in their transcript levels in the transition from mycelium to fruiting bodies [78]. As found in other fungi, on average, their sizes and numbers of exons were smaller than those of coding mRNAs, and a very high proportion of them are located at the 5′ region of neighboring genes, in many cases probably their regulatory targets. Deletion of the xrn1 gene in C. militaris, with a central role in the NMD pathway [79] and affecting lncRNAs, reduced virulence on insects and slowed growth, strongly suggesting the involvement of lncRNAs in the pathogenesis and other aspects of the biology of this fungus.
A high number of lncRNAs was also found in the rice pathogen Magnaporthe oryzae. Transcriptomic analysis at 6 different stages of the infection process identified 2601 lncRNAs, including 1286 antisense and 980 intergenic lncRNAs [56]. A high proportion, 755, showed differential expression at different stages of infection and 560 were found specifically in pathogenesis. Many of them are neighbors of genes with pathogenesis-related functions or are unlinked but show complementarity with their sequences. It is concluded that regulation by lncRNAs in M. oryzae plays an important role in the regulation of genes required for pathogenesis.
An analysis aimed at completing the sequence and annotation of the genome of Botrytis cinerea, a pathogen of grapes, strawberry or tomato, and many other commercial plants [80] included a detailed preliminary analysis of the first of its 18 chromosomes, in which 30 antisense transcripts were found that had undergone intron splicing and that at least partially matched the coding sequence of the gene in which they are located [81]. A thorough survey of B. cinerea lncRNAs was recently carried out during different stages of infection on tomato, from inoculation to 48 h of growth in the plant [57]. As a result, 1837 lncRNAs were identified compared to a total of 18063 annotated coding genes, of which transcripts were detected for 14236. More than 40% of the lncRNAs possessed introns, but their average number, as well as the average transcript size, were lower than those of the coding genes. Of the 1837 lncRNAs, 743 were antisense, of which 55 were induced in the late stages of infection, in parallel to the overlapping coding genes. Interestingly, alternative splicing was observed in 123 of the lncRNAs, adding another level of complexity to their possible regulatory functions.
In the cotton pathogen Verticillum dahliae, the effect of nutrient starvation or growth on pectin as pathogenesis-associated conditions was investigated by RNA-seq in the wild-type strain and in five virulence gene mutants. As a result, 2965 putative lncRNAs were identified, distributed in similar numbers of intergenic and antisense lncRNAs [58]. Many of them were upregulated under starvation conditions or in the presence of pectin in different ways in the five mutants, with the highest numbers in nitrogen-deprived medium, indicating an association with pathogenesis. However, in another study with the same V. dhaliae strain, only 352 novel lncRNAs were detected [82]. In this case, RNA-seq was performed at three stages of infection on cotton, and 47 of the lncRNAs significantly changed their transcript levels during pathogenesis and parallel changes were detected in neighboring genes, presumably regulatory targets. In both studies, the involvement of some lncRNAs on pathogenesis was confirmed by the effects of their overexpression, as described in Section 4.5.10.
In an RNA-seq analysis of Pyricularia oryzae, a phytopathogen causing rice blast, 3374 previously unidentified genes were found, mostly putative lncRNAs, with a predominance of intergenic transcripts with parallel expression to neighboring genes [59]. Many of them changed their expression patterns in hyphae or conidia, suggesting that they may be involved in the regulation of development, a hypothesis that was confirmed for at least one lncRNA investigated in more detail (Section 4.5.8). As a last example on pathogenesis studies, genomic features of lncRNAs from the wheat pathogen Zymoseptoria tritici were identified by analysis of their expression during the infection cycle. Their characteristics and their distribution along the chromosomes were described as well as their differential expression during plant infection [83].
The species of the genus Fusarium are widespread plant pathogens, producing important losses in agriculture. Fusarium graminearum transcriptome data were obtained from five successive sexual stages and from hyphae to search for differentially expressed mRNAs and lncRNAs [60]. Among the 2574 noncoding transcripts, 547 were differentially expressed at least in one of the developmental stages. Of them, 280 were antisense lncRNAs that overlap at least 100 bp with an mRNA, 237 were long intergenic ncRNA (lincRNA). The expression of some lncRNAs were validated by amplification by PCR of their cDNA. Deletion of xrn1 resulted in a defective sexual phenotype, although the mutant produced normal asexual conidia. Transcriptomics of wild type and Δxrn1 mutant from asci and vegetative stages were compared to search for lncRNAs involved in development of sexual fruiting body. Expression of 25 lncRNAs were upregulated in the Δxrn1 mutant in hyphae and increased in asci (meiotic stage).
2.5. Other Global Studies
The cases described above are representative of the works on fungi that are proliferating in increasing numbers based on computational analysis of transcriptomes. These are particularly powerful in the case of those that are strand-specific, as they allow the identification of antisense RNAs. Other examples on global lncRNA studies in fungi may derive from the study of the transcriptome in a single condition, as the analyses carried out in the human pathogens Paracoccidioides brasiliensis [84] and Cryptococcus neoformans [85]. However, the analyses usually compare different biological conditions and are aimed at associating lncRNAs with specific physiological processes, as those involved in regulatory effectors in the soy pathogen Phytophthora sojae [86], stress in the entomopathogen Metharizium robertsii [87], virulence decline in the aphid pathogen Conidiobolus obscurus [88], pathogenesis in the development in the pome fruit pathogen Penicillium expansum [89], heat-shock in the thermophilic fungus Thermothelomyces thermophilus [90], nucleus–mitochondria interaction in the edible fungus Lentinula edodes [91], growth rate in the biotechnological yeast Komagataella phaffii [92], or sexual and asexual development in the potato pathogen Phytophthora infestans [93].
3. Mechanisms of Action of Fungal lncRNAs
In recent years, fungal lncRNAs have been reviewed, including their proposed mechanisms of action. The reviews are either general [94,95] or specifically focused on yeasts [96,97,98] or antisense lncRNAs [99]. The cases of lncRNAs studied in more detail allow us to sketch a general scenario of the different regulatory mechanisms in which they may be involved. This section may serve as a guide for a better understanding of possible mechanisms for the specific lncRNAs described in Section 4, especially for those in which there is sufficient experimental support.
As a first criterion for classification, there are cis- and trans-acting lncRNAs. Cis-acting lncRNAs affect immediately close regions of the genome, and they can be in sense or in anti-sense orientation with respect to the region they regulate. The regulatory effects of some lncRNAs are a consequence of their own transcription, being able to negatively affect the transcription of genes in the opposite or same strand. This ‘Transcriptional Interference’ (Figure 2A) can be caused by transcriptional machinery collision, promoter occlusion, or interferences with the initiation or elongation of overlapping or nearby genes (e.g., RME2 on IME4 in S. cerevisiae, ref. [100]). In other cases, a ‘Transcription Repression’ (Figure 2B) may occur via histone modification associated with the ability of lncRNA to modify complexes such as histone deacetylases and histone methyltransferases (HMTs), altering chromatin accessibility and reducing transcriptional activity. An example is provided by PHO84 antisense on PHO84 in S. cerevisiae [101,102]. However, not all lncRNAs act as inhibitors, and their action may result in ‘Transcriptional Enhancement’ (Figure 2C), as occurs with PHO5 antisense on PHO5 in the same yeast [102,103]. In these cases, lncRNAs facilitate activation of neighboring genes by promoting chromatin accessibility, typically recruiting histone acetyltransferases (HATs) or transcription factors. The transcription of lncRNAs can strongly alter the nucleosome environment resulting in ‘Nucleosome Deposition’ (Figure 2D) over adjacent promoter regions, thereby repressing gene expression, as described for SRG1 on SER3 in S. cerevisiae [104,105,106]. This mechanism is prominent in tightly packed genomes and can serve as a rapid response to developmental or environmental signals. Other lncRNAs act as chromatin stabilizers by maintaining either active or repressed chromatin states. This ‘Chromatin Stabilization’ (Figure 2E) mechanism can involve interactions with chromatin readers or remodelers that prevent the spread of heterochromatin or protect euchromatin from silencing. A good example is the action of Ty1 antisense on Ty1 transposon in S. cerevisiae [107]. Such stabilization is frequently observed in pericentromeric or sub-telomeric regions, where the chromatin landscape is dynamic.
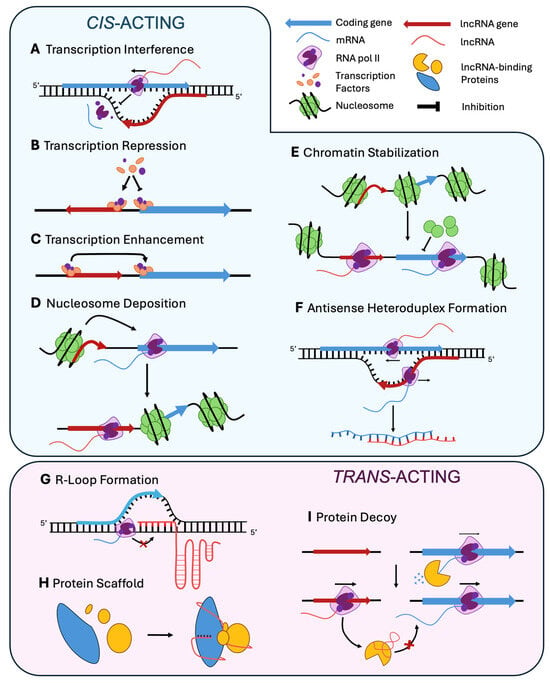
Figure 2.
Representative examples of the mechanism of action of lncRNAs in fungi. (A–F) LncRNAs acting in cis. (G–I) LncRNAs acting in trans. Arrows crossed out in red indicate inhibitory effects.
LncRNAs can interact with other functional molecules, either RNA, DNA or proteins. LncRNA-mRNA interaction involves ‘Antisense Heteroduplex Formation’ (Figure 2F), as found with as-ssm1 on ssm1 in U. maydis [108]. In this mechanism, antisense lncRNAs base-pair with complementary mRNAs to alter their stability or translation, protect them from degradation or induce granules formation. Interaction with DNA or proteins can expand the location of action of lncRNA, which shows a wide spectrum of effects. A characteristic lncRNA-DNA binding mechanism is the ‘R-loop Formation’ (Figure 2G), in which lncRNAs directly interact with genomic DNA through sequence complementarity or structural motifs. These interactions can promote the formation of lncRNA: DNA hybrids creating R-loops, which influence local chromatin accessibility, transcription, or recombination. As a paradigmatic case, lncRNAs derived from telomeric regions, such as TERRA of S. cerevisiae [109], bind DNA to modulate telomere length or genome stability. Such mechanisms may also be behind the long-range regulation of target genes by enhancer-associated lncRNAs that fold into tertiary structures capable of bridging distal genomic elements. LncRNAs may also work as ‘Protein Scaffold’ (Figure 2H) that facilitate the assembly of protein complexes by bringing together multiple components with distinct binding domains. A representative example is provided by the action of meiRNA at sme2 locus in S. pombe [110,111]. This mechanism is particularly effective in the coordination of chromatin modifiers, splicing factors, or signaling proteins. Such scaffold functions are highly sensitive to lncRNA structure, expression levels, and cellular localization, often allowing for fine-tuned regulatory control across multiple processes. Other lncRNAs operate as ‘Protein Decoy’ (Figure 2I), binding to regulatory proteins and preventing them from interacting with functional targets, as proposed for HAX1 on the Xyr1 activator in Trichoderma reesei [112]. This competitive inhibition mechanism can modulate diverse regulators, as transcription factors, kinases, or RNA-binding proteins.
4. Functions of Specific lncRNAs Investigated in Fungi
The study of long non-coding RNAs (lncRNAs) in fungi has revealed important roles for these transcripts in gene regulation, cell development, and response to environmental conditions. This section is a recompilation of the studies on specific fungal lncRNAs. The diversity of the cases studied provides insight into the heterogeneity of the processes and mechanisms of action in which they are involved, which are summarized in Figure 1. Special attention is paid to the yeasts S. cerevisiae and S. pombe, which stand out for the magnitude of research carried out.
4.1. Saccharomyces cerevisiae
The finding of pervasive transcription as a widespread phenomenon in S. cerevisiae suggested a high transcriptional complexity in this simple eukaryote [113,114], indicating the existence of additional levels of regulation beyond those previously known [115]. The functions of numerous lncRNAs have been investigated in more detail in this yeast. They are listed in Table 2.

Table 2.
Specific lncRNAs investigated in S. cerevisiae.
4.1.1. Metabolism and Cellular Functions
A classic example in budding yeast, and one of the best known at the mechanism level, is the lncRNA SRG1, which is transcribed upstream of SER3, a structural gene of the serine biosynthetic pathway, and represses its transcription [106]. This repression is regulated by serine via the activator Cha4 and coactivators SAGA and SWI/SNF. Additionally, SRG1 transcription directs nucleosome deposition over the SER3 promoter, blocking transcription factor access [104,105]. This process depends on elongation factors Spt6 and Spt16, which maintain nucleosome positioning without affecting SRG1 transcription levels. Furthermore, Spt2, an RNA polymerase II-associated factor, facilitates nucleosome reassembly behind transcribing RNAP II, reinforcing repression [143].
Two genes involved in nucleotide metabolism, URA2 in pyrimidine synthesis and IMD2 in inosine synthesis, share a similar regulatory mechanism mediated by TSS selection [116,117]. Both genes are only induced when there is a shortage of a base involved in their pathway, uracil in the first case and guanine in the second. Regulation is carried out by the use under repressive conditions of an upstream TSS, which results in the synthesis of a SUT that is rapidly degraded. Under induction conditions, a TSS closer to the start codon is used instead, which gives rise to a stable and functional mRNA. In the case of IMD2, SUT transcripts (called here “attenuated transcripts”) start with guanine, which facilitates their preferential transcription in the presence of abundant GTP.
One of the lncRNAs identified in the transcriptomic analysis of the mutant lacking the exosome component Rrp1p was HRA1 (from hidden in reading-frame antisense), antisense of the DRS2 gene, which encodes a protein associated with the Golgi apparatus [38]. However, a role in the maturation of 18S rRNA was attributed to this gene because of its mutant phenotype. The authors suspect that this second phenotype is due to the loss of HRA1 and not to the loss of the DRS2 gene itself, providing an example of possible multifunctionality of the same DNA sequence.
A well-studied example of regulation by lncRNA in S. cerevisiae is the GAL1-10 gene regulon [118], which responds to sugar availability, and which exhibits unusual Set1-dependent methylation at GAL10 (H3K4me3) under repressive conditions [119]. This repression is associated with the transcription of the GAL10-ncRNA, transcribed in the opposite direction to GAL10 at a very low rate, approximately once every 50 min. Its transcription depends on the binding of the Reb1 protein to specific sites and is rapidly degraded by the TRAMP and exosome complexes, keeping it at very low levels (∼1 molecule/14 cells). H3 methylation delays the recruitment of RNA polymerase II (RNAPII) and TBP on GAL1 promoter. In addition, transcription of GAL10-ncRNA recruits Set2 and induces histone deacetylation through the Rpd3S complex, modifying chromatin structure. Other genes of the GAL regulon are also associated with lncRNAs, whose levels have been shown to be regulated by 5′ decapping, which results in their destabilization [144]. The GAL genes, as well as other inducible genes in S. cerevisiae, have evolved the use of regulatory lncRNAs to maintain their inactive state through an epigenetic mechanism, which involves histone modifications. Their regulation by decapping is evidenced by mutants of the enzyme that carries out this process, DCP2, which show increased expression levels of the GAL1, GAL2, GAL4, and GAL10 genes.
Two lncRNAs involved in the regulation of phosphorus utilization have been analyzed. In the absence of Rrp6, the expression of the PHO84 gene, which encodes a phosphate transporter, is repressed. This led to the detection of a regulation of PHO84 by two antisense transcripts, whose levels remain higher in the absence of Rrp6 and repress PHO84 transcription [103]. Under these conditions, there is a recruitment of the histone deacetylase Hda1 to the PHO84 region, indicating that regulation by these antisense RNAs takes place at the epigenetic level, promoting deacetylation of histones by the Hda1/2/3 complex. On the other hand, the PHO5 gene, which encodes an acid phosphatase that is excreted into the medium to hydrolyze organic phosphates, is regulated by another 2.4 kb antisense RNA, which is transcribed from the PHO5 terminator and covers the entire gene [101]. In the wild-type strain, antisense transcript levels are low in the absence of P and higher in its presence or in mutants lacking Rrp6, and PHO5 mRNA levels show the opposite pattern. However, loss of this transcript in a mutant lacking the 3′ region of PHO5 results in a delay in chromatin remodeling, and subsequent recruitment of polymerase II. Thus, transcription of this antisense transcript promotes PHO5 activation, possibly facilitating proper nucleosome positioning. A search for new factors participating in antisense RNA-mediated transcription interference led to the identification of the HIR histone chaperone complex, involved in histone deposition on DNA in replication-independent chromatin assembly [145]. Antisense RNA repression of genes whose expression is mediated by the SAGA complex requires HIR activity, which was also demonstrated for the PHO84 and PHO5 genes [102]. Knowledge of chromatin dynamics in the PHO5 promoter, in parallel with the coregulated PHO84 promoter among others, constitutes a paradigmatic model of epigenetic regulation of eukaryotic promoters [146].
A very different regulatory system has been described for the ASP3 gene, which encodes an asparaginase that is activated only in the absence of nitrogen in the medium. An important role in its activation is played by an RNA transcribed from the central region of the gene in the sense orientation [120]. This unusual transcription occurs irrespective of the nitrogen availability and is independent of the ASP3 GATA activators Gat1p and Gln3p, and of polymerase II itself, but results in an appropriate level of trimethylation of histone H3 at lysine 4 (H3K4me3) at the ASP3 promoter in a manner that facilitates gene activation in the absence of nitrogen. This is therefore a case of positive epigenetic regulation by an internal sense transcript.
Target of Rapamycin (TOR) is a protein kinase that acts as a central nutrient and energy sensor, regulating cell growth and metabolism in response to environmental signals [147]. Its regulatory targets include amino acid transporters, and different lncRNAs found near transporter genes are assumed to be involved in their regulation. This is the case of the lncRNA XUT_2F-154 (TBRT), antisense to the transporter gene TAT1, which regulates this gene and the linked transporter gene BAP2. The expression of all three is TOR-dependent, and when TOR activity is inhibited, TBRT levels fall and BAP2 and TAT1 mRNAs increase, and the same result is obtained by blocking TBRT transcription [121]. It is concluded that TOR keeps the expression of these two transporters under control by inhibiting them through TBRT, which takes place at the epigenetic level, establishing a state of repression in chromatin structure.
The ATP16 gene encodes a subunit of mitochondrial ATP synthetase, which plays a key role in energy generation by coupling proton translocation to ATP synthesis. Upstream of ATP16 there is a lncRNA gene called CUT60, whose transcription facilitates ATP16 transcription [122]. For this purpose, it is necessary that CUT60 terminates transcription at the appropriate site to facilitate accessibility of the transcription machinery to the ATP16 promoter, since if the termination signal is removed and transcription of CUT60 is prolonged, ATP16 expression is greatly reduced, resulting in loss of mitochondrial function and a petite phenotype.
In an investigation of biotechnological interest due to the use of S. cerevisiae for the production of recombinant enzymes, the effect of the deletion of 208 intergenic lncRNAs, chosen on the basis of being at least 200 bases from the start codon of the nearest gene, on the secretion of α-amylase from an engineered plasmid was analyzed [123]. As a result, 21 lncRNAs were found to influence production, and of these, the deletion of SUT067, SUT433, and CUT782 allowed for the greatest increases. The three deletion strains showed enhanced energy metabolism and translational activity, allowing for a greater supply of ATP and protein synthesis. This study shows the applied interest that lncRNA-mediated regulatory mechanisms may have.
Another extensive study investigated the phenotypic consequences of 372 mutants with deletions for ncRNAs, which included 50 CUTs and 93 SUTs [124]. Their phenotypes were tested under 23 different environmental conditions, mainly changes in carbon sources and stress conditions. As a result, 29% of SUTs and 22% of CUTs exhibited phenotypic alterations. Of these, 19 were selected, together with a snoRNA (small nucleolar RNA), to analyze the consequences of their deletions on the transcriptome. Six of them showed changes in the expression of hundreds of genes, while others showed more limited effects. Of the first, the deletions of SUT125, SUT126, SUT035, and SUT532 showed the most severe changes in the greatest number of conditions and were investigated in more detail. The number and diversity of the affected genes indicated an action of these ncRNAs on transcription factors, leading to cascading effects.
4.1.2. Cell Cycle and Stress Responses
One strategy to identify lncRNAs potentially related to cell cycle control is to detect those that oscillate throughout the cycle. This approach revealed that 80 of 523 antisense transcripts undergo cyclical changes in accordance with the cell cycle or are antisense to genes that do so [125]. On the other hand, of 135 intergenic RNAs, 11 show cyclical changes in their levels, giving a total of 91 non-coding RNAs potentially associated with cell cycle regulation. These include antisense RNAs for well-known genes such as FAR1, CTF4, and TAF2. FAR1 plays a key role in the G1/S transition, and it is expressed at the M/G1 transition and inactivated in the late G1 phase. Its antisense RNA shows the opposite pattern, as it appears when FAR1 mRNA disappears. CTF4 is required for the maintenance of chromatin structure to overcome the S-phase checkpoint, and its mRNA peaks at the G1/S and G2/M transitions, whereas its antisense RNA peaks at the G1/S and G2/M transitions. Other genes are not directly related to the cell cycle, but their regulation is connected to it. This is the case of TAF2, which assists TFIID in the initiation of transcription, and indirectly participates in cell cycle regulation by ensuring the expression of genes necessary for cell growth and division. The TAF2 gene shows peaks of expression in late M phase and early G1, while its antisense RNA peaks at the end of G1 and throughout S phase.
A relevant case of cell cycle gene regulated by lncRNA is CDC28, which encodes cyclin-dependent kinase 1. One of the transcription factors regulating this gene is Hog1, the protein kinase that executes the response of the osmotic stress signal transduction chain. This response consists of the activation of transcription of hundreds of stress-responsive genes to adapt to the new conditions [148]. An analysis by whole-genome tiling arrays revealed that Hog1 also activates approximately one hundred lncRNAs, one of them an antisense lncRNA of the cdc28 gene, also regulated by Hog1 [127]. The CDC28 kinase plays a key role in the control of the cell cycle, which is temporarily stopped in response to stress pending its resumption once adaptation has occurred. In this case, the transcription of this lncRNA exerts a positive effect on CDC28 transcription, an action it exerts by facilitating the correct positioning of Hog1 and the Chromatin Remodeling Complex to facilitate the initiation of CDC28 transcription and thus the resumption of the cell cycle.
When S. cerevisiae cells are under severe nutrient stress, they enter a resting stage known as quiescence. In a global analysis of antisense RNAs, comparing the wild-type strain and an NMD upfl1 mutant, it was observed that many genes expressed in quiescent cells show very low expression levels in the exponential phase, and that this repression is often associated with the presence of antisense RNAs, very often also covering the promoters. This cause-effect relationship was confirmed for the PET10, CLD1, MOH1, and SSH3 genes by prematurely interrupting the expression of antisense RNAs [126].
Ethanol stress is an important environmental scenario in S. cerevisiae. Among the hundreds of lncRNAs in S. cerevisiae, those that change their levels under high ethanol concentrations have been identified, and considerable differences have been observed between strains with different tolerance [149,150]. The ethanol stress response is complex, affecting numerous aspects of cell metabolism and function. In a strain with low ethanol tolerance, the lncRNA lnc9136 has been studied by computational analysis and the effect of its deletion [128]. The results led to the conclusion that lnc9136 acts by preventing ethanol-induced mitosis arrest; this is done by facilitating the binding of the cell cycle regulatory proteins Swe1p and Clb1/2 to the proteins Gin4p and Hsl1p. On the other hand, computational predictions of lncRNA lnc10883 in a highly ethanol-tolerant strain indicated that it bypasses DNA damage and the mitotic spindle checkpoints to allow cell cycle progression through its interaction with Mec1p, involved in detecting DNA damage, or Bub1, involved in checking the correct attachment of chromosomes to the mitotic spindle. Analysis of RNA and protein levels under stress conditions with respect to control cells led to the conclusion that lnc9136 also acts on the general level of translation in the cell [151], which is attributed to its action on the levels of 17S rRNA and Rrp1, a nucleolar protein involved in the processing of rRNAs [152]. On the other hand, the same experimental approach suggested that lnc10027, from a different strain, inhibits the formation of processing body and stimulates translation activity, linking ethanol stress to these cellular processes.
4.1.3. Sexual Cycle and Development
At least two lncRNAs play a relevant role in the control of meiosis entry in S. cerevisiae [129,131]. The gene IME1 (Inducer of Meiosis 1) is downregulated by the lncRNA IRT1 (IME1 Regulating Transcript 1). IRT1 is transcribed upstream towards IME1, and its transcription inhibits IME1 expression through competition with the IME1 promoter. In addition, IRT1 acts by remodeling chromatin, as its transcriptional elongation recruits histone deacetylases that generate a closed chromatin state, such as Rpd3, blocking the binding of IME1 activators. Moreover, its transcription induces repressive marks, such as H3K36me3, that maintain the region in a non-permissive state. Expression of IRT1, as that of IME1, is regulated by nutrient availability through the PKA and TORC1 activities [130]. Under conditions of sufficient nutrients, IRT1 is repressed, but this repression is lost in the absence of PKA and TORC1. In turn, IRT1 is controlled by another lncRNA located upstream, IRT2, transcribed in the same orientation. In haploid cells, IRT2 has a low level of expression, which facilitates acetylation in histones H3 in the IRT1 promoter region, and therefore its transcription, resulting in low expression of IME1 [132]. However, in diploid cells, IRT2 is actively expressed, preventing such acetylation and leading to low levels of IRT1 transcription and therefore the derepression of IME1, enabling entry into meiosis.
Another gene required to initiate meiosis, IME4, is also regulated by an antisense RNA [100]. In haploid cells, IME4 antisense RNA is produced, and in diploid cells, IME4 mRNA is produced instead. The system appears to be regulated by interference between the transcription of both transcripts. In diploid cells, the heterodimer formed by transcription factors a1 and α2, encoded by the MATa and MATα loci of each sex, prevents transcription of the antisense IME4 gene by binding to a site near its 5′ region, thus facilitating expression of the sense IME4 and the ability of the cells to enter meiosis under the right environmental circumstances. Another case of regulation by antisense RNA of a gene activated by the a1/α2 heterodimer is ZIP2, involved in the process of recombination between homologous chromosomes in meiosis. This gene is negatively regulated by the lncRNA RME3 [133]. In diploid cells, this antisense transcript is blocked by a1/α2, thus facilitating the transcription of ZIP2. These same authors named RME1 and RME2, from Regulator of Meiosis, the antisense lncRNAs that inhibit IME1 and IME4 in haploid cells.
The expression of the FLO11 gene produces a morphological alteration in yeast growth, leading to the formation of pseudohyphal filaments [153]. This gene has two epigenetically controlled states, induced or repressed. In the same clone of cells incubated under the same conditions, the cells show one or the other state, giving rise to a variegated phenotype. The alternation between the two states is controlled by the simultaneous action of two lncRNAs, PWR1 and ICR1, which are transcribed in opposite and overlapping directions in the upstream region of the FLO11 gene [134]. Transcription of ICR1 results in chromatin compaction of the FLO11 region, preventing its expression, whereas transcription of PWR1 prevents ICR1 transcription and thus promotes FLO11 expression. Consequently, FLO11 expression depends on whether PWR1 or ICR1 is expressed predominantly, which in turn depends on competition between an activator of PWR1 transcription, Flo8, or a repressor, Sfl1. In some cells the balance is inclined towards PWR1 and FLO11 is inhibited (yeast-like phenotype), and in others it is inclined towards ICR1 and FLO11 is activated (filamentous growth), giving rise to the variegated appearance of the colony.
The cell wall is an essential structure for fungal survival and development, and its correct maintenance is subject to a fine control of the numerous genes involved, in which different lncRNAs participate in S. cerevisiae. In addition to FLO11, involved in this process, lncRNAs have been found controlling the ECM3 [136], PIR3 [137], SPS100 [138] and TIR1 [44] genes, regulated respectively by the lncRNAs EUC1 and SUT228, transcribed from the same strand upstream of their target genes, and the antisense lncRNAs SUT169 and TIR1axut [137]. In some cases the regulation is positive and in others negative. Many more cases are expected, since of 201 cell wall-related genes, overlap with antisense RNA is detected in 88 and overlap of their promoters with sense RNA in 15, most of which are likely to be functional regulatory lncRNAs.
The HO gene encodes an endonuclease involved in the switching of the mating type by a gene conversion mechanism [154]. Regulation of the HO gene is coordinated with the cell cycle, as it is always repressed except at the end of G1 phase, at which time it is induced following sequential binding of the SBF complex [155]. Cells can be stalled in G1 phase by nutrient starvation or pheromone exposure. If the cycle resumes due to the availability of nutrients, the persistence of SBF and Mediator complexes bound to the promoter causes the HO gene to be expressed again in the G1 phase of the next cell cycle. However, if such resumption is due to the disappearance of pheromone signals, HO remains repressed in the next cycle due to the action of a lncRNA, called pHO-lncRNA [139], which is transcribed 2700 nt upstream of HO and displaces the SBF complex. In that case HO is not expressed again until the G1 phase of the second cycle. In congruence with this mechanism, pHO-lncRNA only has an effect in cis, and its reintroduction in trans in a strain in which pHO-lncRNA transcription is blocked in cis does not restore wild-type regulation.
Genes whose expression shows a high variability or responds to the presence of a certain signal frequently overlap with antisense RNAs, suggesting their participation in its regulation. This was demonstrated for the SUR7 gene, which is largely expressed in media containing galactose and is repressed following stimulation with the α-factor pheromone, while its antisense, SUT719, is expressed equally in both conditions [135]. The elimination of SUT719 expression leads to derepression of the SUR7 gene in the presence of the α-factor. The result indicated that SUT719 expression leads to threshold-dependent regulation of SUR7 expression, specifically inhibiting it when induced at low levels.
4.1.4. Genome Integrity
The S. cerevisiae genome contains Ty transposable elements [156]. One of them is the Ty1 retrotransposon, whose activity is controlled by an antisense RNA, produced by polymerase II and covering from the middle of one of the transposon genes to the 5′ LTR [107]. Its mode of action consists of silencing gene expression by promoting histone deacetylation and methylation in the transposon region. The silencing activity of this antisense RNA is attenuated by its destabilization by the 5′ RNA degradation pathway through its exoribonuclease Xrn1. This is therefore an epigenetic regulatory mechanism in which the possible hybridization between sense and antisense RNA plays no role.
Sets of lncRNAs transcribed from centromeric regions play an important role in kinetochore formation [157,158], which is necessary for proper chromosome segregation in cell divisions. The centromeres of S. cerevisae are very short, about 125 bp and lncRNAs transcribed from the centromeres, called cenRNAs, are polyadenylated and have a longer size [140]. Such transcription is induced in the S phase of the cell cycle, is dependent on centromere-specific binding of CENP-A, a histone H3 variant, and replication, and is inhibited by the kinetechore protein Cbf1 and a variant of histone H2A. The amount of cenRNAs is finely regulated through the balance between their transcription and their degradation by the nuclear RNA decay pathway and is important for proper chromosomal stability and segregation [159].
Two lncRNAs play important cellular functions through their roles in telomerase activity and maintenance [160], the telomerase RNA TLC1, and the telomeric repeat-containing RNA TERRA. TLC1, known as TER1 in S. pombe [161,162], is used as a template for reverse transcription to synthesize telomeric DNA and also acts as a flexible scaffold for the telomerase and as a tether for holoenzyme protein subunits [141]. On the other hand, TERRA is transcribed from subtelomeric regions towards the telomeres and includes subtelomeric sequences at the 5′ segments [109]. TERRA length varies due to the different TSSs for RNA polymerase II and the processing of the 3′ end, which at least in some cases are polyadenylated. TERRA is transcribed from the C-rich strand, so the RNA itself is G-rich, allowing it to hybridize to the telomerase RNA template and inhibit telomerase activity in vitro. Changes in TERRA expression can cause functional alterations in telomeres, altering their length, and interfering with replicative machinery. TERRA co-localizes in vivo with telomerase and can form RNA:DNA hybrid structures at telomeric ends called R-loops, which have been linked to the onset of the initiation of senescence and to an alternative form of telomere elongation [163]. TERRA is conserved from yeast to humans, where its role as a telomere-associated lncRNA implicates it in telomere length regulation and in diseases such as cancer [164].
A synthetic genetic array procedure, which allows to assign RNAs to specific cellular processes, led to the identification of SUT457 as a lncRNA involved in telomere organization [142]. This was confirmed by studying the effect of its deletion, which accelerated senescence in telomerase deficient cells and led to the accumulation of single stranded telomeric DNA, an effect that requires Exo1 function. Ectopic expression of SUT457 in the deletion strain suppressed telomeric overhang accumulation, indicating a trans-acting role for this lncRNA in telomere overhang homeostasis.
4.2. Schizosaccaromyces pombe
Like S. cerevisiae, S. pombe is a very simple and amenable eukaryotic model microorganism with a compact genome, widely used in research, and in recent years also in lncRNA studies. LncRNAs investigated to date in this yeast (Table 3) are described in this section.

Table 3.
Specific lncRNAs investigated in S. pombe.
4.2.1. Metabolism, Cellular Functions and Stress Responses
The expression of the fbp1 gene, encoding fructose-1,6-bisphosphatase, is a well-known model for the relationship between chromatin structure and transcription factor accessibility, uncovering a multilevel regulatory process in which lncRNAs play an important role [176]. Transcription of fbp1 is strongly increased under glucose starvation, and such activation is facilitated by chromatin remodeling. This is produced by histone acetylation [177] triggered by the successive transcription of three lncRNAs, mlonRNA-a, mlonRNA-b, and mlonRNA-c [165]. Consequently, the insertion of a transcription terminator upstream of the regulatory region prevents the transcriptional cascade of lncRNAs and thus the opening of chromatin. On the other hand, transcription of these lncRNAs, which overlap upstream of the promoter and coding region of fbp1, promote in the absence of glucose the formation of double strand breaks in the fbp1 region by facilitating access of the topoisomerase Rec12 [178], which make it a hotspot of recombination in meiosis. A similar regulatory situation of opening of epigenetic activation mediated by the transcription of lncRNAs was observed at the ade6-M26 locus, also a recombination hotspot [166].
A lncRNA, prt (from phosphate repressing transcript), is involved in phosphate homeostasis in S. pombe through control of the expression of the pho1 gene, orthologous to PHO5 of S. cerevisiae, which encodes a secreted acid phosphatase involved in phosphate uptake. This is accomplished by activation of the pho1 gene under phosphate starvation, a regulation carried out by the Pho7 transcription factor [105,179]. The lncRNA prt, which is transcribed from an upstream promoter on the same strand as pho1, participates actively in this regulation by repressing pho1 transcription when phosphate is at sufficient levels. Prt transcription leads to RNAi-dependent histone methylation across the pho1 locus, resulting in transient heterochromatinization. As observed for the meiotic gene mei2, under conditions of phosphate excess, Mmi1 interacts with prt through its DSR (Selective Removal Determinant) sequences to trigger its exosomal degradation, which is coupled to the termination of prt transcription [167]. Thus, disruption of prt DSR sequences results in pho1 over-repression. Interestingly, mutations of the CTD domain of PolII do not affect the activity of the prt promoter, but affect the regulatory effect on pho1 expression, indicating an involvement of the CTD in the control of prt [180]. The regulation of Pho1 by prt requires its complete transcription. In mutants of the erh1 gene, which encodes a small nuclear protein that interacts with Mmi1, pho1 expression is derepressed [181]. This is because in this mutant, prt transcription is prematurely terminated, assigning Erh1 an antagonistic role in prt transcription termination, while Mmi1 promotes it. These data provide insight into the regulatory complexity of prt, in which other proteins involved in the progress and termination of its transcription participate.
Two other pho1 adjacent genes, which also respond to the presence of phosphate, are also regulated by lncRNAs and form a regulon with pho1: the phosphate transporter gene pho84 and the glycerophosphate transporter gene tgp1 [105]. Mutations in the promoter of lncRNA prt2 cause derepression of pho84, indicating the participation of prt2 in the expression of this gene [168]. Interestingly, this regulatory alteration also affects expression of the adjacent downstream prt1 and pho1 genes, and therefore partially derepresses pho1 as well, indicating a coordinated action of both lncRNAs in the same regulatory system. In addition, the gene tgp1 is negatively regulated by its upstream lncRNA nc-tgp1 [182]. As a mechanism of action, the transcription of nc-tgp1 on the tgp1 promoter increases the density of nucleosomes, blocking the access of activating transcription factor and silencing tgp1. However, rather than involving transient heterochromatin formation triggered by targeted lncRNA degradation, the regulation of tgp1 by nc-tgp1 RNA seems to be mediated by transcriptional interference. Reduced transcription of nc-tgp1 allows tgp1 expression during phosphate starvation, and loss of nc-tgp1 induces tgp1 even under repressive conditions. The polyadenylation site of nc-tgp1 coincides with the binding element of the Pho7 regulator, so its transcription can negatively affect tgp1 activation [169]. This lncRNA can be polyadenylated at an upstream site, resulting in a shorter transcript, and removal of this polyadenylation site also alters tgp expression. The choice of polyadenylation site is governed by the CTD of PolII, adding an additional novel control in the mechanism of action of prt2.
A combination of transcriptome analysis by RNA-seq and proteome analysis by mass spectrometry in response to stress allowed to associate changes in non-coding transcripts to specific protein changes [170]. To confirm this connection in one of them, deletion of the locus for the putative lncRNA SPNCRNA.1164 resulted in reduced Atf1 protein levels and altered sensitivity to oxidative stress, indicating that SPNCRNA.1164 is a lncRNA involved in the regulation of stress responses in this yeast. Interestingly, SPNCRNA.1164 has two small antisense transcripts within the locus: SPNCRNA.1165 and SPNCRNA.1166/prl6, which are longer than 200 nt and could participate in or even control its own function.
The elimination of Exo2/Xrn1 activity allowed the identification of numerous antisense lncRNAs whose increase in the mutant leads to the inactivation of their associated genes. This is the case of the catalase gene ctt1 and its antisense lncRNA XUT0794 [66]. The ctt1 gene strongly increases its expression in the presence of H2O2, but the increase is significantly lower in the Exo2/Xrn1-deficient mutant, which shows higher levels of XUT0794. Expression of XUT0794 is also stimulated by H2O2, suggesting a fine modulation of ctt1 expression by its antisense RNA. The attenuation of ctt1 by XUT0794 is accompanied by low RNAPII-ser5 phosphorylation and involves the recruitment of histone acetylation HDAC machinery. This requires Set2-dependent H3K36me3 marks, as indicated by the similar effect produced by the absence of both HDAC and Set2 function.
4.2.2. Sexual Cycle and Development
An interesting role recently identified for lncRNAs is the organization of nuclear structures [183]. The regulation of spatial positioning within the nucleus of eukaryotic cells is necessary for its proper functioning. The interaction between their components is facilitated by a correct compartmentalization in subnuclear domains or nuclear bodies [184]. This is the case of the lncRNA meiRNA in S. pombe, involved in the formation of a nuclear dot structure at the sme2 locus, with its protein-binding partner Mei2 [185,186]. The sme2 gene encodes two lncRNAs, meiRNA-S and meiRNA-L, that are required for Mei2 dot formation and play an important role in recognition of homologous chromosomes in meiosis [110,111]. The Mei2 structure promotes meiosis progression by sequestering free Mmi1, a factor involved in inducing selective degradation of meiosis-specific transcripts, thereby inhibiting their function [187]. Although the molecular details are not fully understood, it is assumed that the meiRNA functions as a lure to attract Mmi1. In fact, the localization of meiRNA at sme2 depends on its association with Mmi1. Moreover, one of the multiple foci of Mmi1 in mitotic cells localizes to the sme2 locus, and meiRNA overexpression promotes Mmi1 accumulation at the sme2 locus even in the absence of Mei2 and reduces Mmi1 activity.
In a search of potential lncRNAs, those that passed several computational filters and were conserved in closely related species were identified, suggesting that they may have relevant functions [171]. The effects of deleting 12 intergenic lncRNAs on growth and development under different culture and stress conditions were analyzed. The mutant phenotype of one of them, nc1995, was noteworthy, as it initiates sexual development under conditions in which it should not, although it does not enter meiosis because it is not diploid. This phenotype is due to a repressing effect of nc1995 on the expression of the ste11 gene in the presence of nitrogen which causes deregulation of ste11 in the mutant. The transition of S. pombe vegetative cells to meiosis and haploid ascospore formation is triggered by nutrient starvation. This process, which involves conjugation between h+ and h- cells and subsequent entry into meiosis, is regulated by the product of the ste11 gene [188].
Expression of ste11 is also negatively regulated by the lncRNA rse1, which is divergently transcribed from the ste11 promoter [172]. Its mechanism of action is in cis, functioning as a scaffold to recruit a repression complex that promotes SET3C-dependent histone deacetylation. Upstream to rse1 there is a smaller ncRNA, rce1, probably also a lncRNA, that could interfere with rse1 transcription. Another lncRNA that acts in the control of the transition from mitosis to meiosis is mamRNA, which acts as a scaffold in a specific nuclear body to facilitate the interaction between the proteins Mmi1 and Mei2 and their mutual inhibition [173]. In this interaction, Mmi1 downregulates Mei2, inducing its ubiquitinylation by another complex, while Mei2 removes part of Mmi1, with the remainder being available to facilitate the degradation of the lncRNA meiRNA—already mentioned above—and meiosis mRNAs. Under conditions of meiosis stimulation, there is a greater amount of meiRNA, which sequesters Mei2 and Mmi1, so that it stimulates the translation of the mRNAs of the genes necessary to enter meiosis.
The detection of RNAs by the Mmi1 protein determines their degradation by nuclear exosomes. In the search for Mmi1 RNA targets, lncRNA nam1 (from non-coding RNA associated with Mmi1) was identified, which also participates in the regulation of sexual differentiation [174]. The binding of Mmi1 to nam1 not only promotes its degradation but also stops its transcription when it binds to the nascent RNA, thus activating the downstream gene byr2, in whose expression the complete transcription of nam1 interferes negatively. Byr2 is a mitogen-activated MAPKKKK that plays a key role in sexual differentiation. In addition, Mmi1 arrests the transcription of other lncRNA genes in pericentromeric regions, contributing to their silencing as heterochromatin. In the mechanism of action of the inactivation of byr2 by the upstream transcription of nam1, an important role is played by a cryptic intron found in the sequence of nam1 overlapping with byr2, which forms a scaffold for recruiting splicing machinery and other proteins, such as Pir2, which plays various roles in RNA metabolism [189]. As a result, chromatin-modifying activities involved in gene silencing are formed, leading to gene repression. The same mechanism seems to operate for other genes in S. pombe, such as the regulation of Pho1 by the lncRNA prt, mentioned above.
In S. pombe, about 70 genes affecting its chronological lifespan have been identified, some of them related to ribosome function [190]. One lncRNA, aal1 (from aging-associated lncRNA), is expressed in quiescent cells and lengthens their lifespan by attenuating their translation activity [175]. This action is exerted in trans, since its ectopic overexpression further extends the lifespan, reducing the number of ribosomes, while its deletion shortens it by increasing its translational activity. Its mechanism of action consists of binding to the mRNA rirpl1901, which encodes a ribosomal protein, reducing its availability for ribosome formation. In congruence, aal1 is mainly detected in the cytoplasm, with a tendency to be associated with ribosomes. Therefore, this lncRNA plays an important evolutionary role, as it reduces the translational activity of cells when they are at rest, waiting for suitable conditions to resume their life cycle.
4.3. Other Yeasts
Research on specific lncRNAs is rare in other yeasts. In the study conducted in P. pastoris, mentioned in Section 2.1, three lncRNAs that stood out for their changes in expression due to overexpression of phospholipase A2 and the presence of methanol, TCONS_00004115, TCONS_00000958, and TCONS_00003668, were deleted for analysis of their possible function [50]. When analyzing the effects on possible target genes, in cis based on physical proximity or in trans based on coexpression analysis under the tested conditions, only changes in some of the possible target genes were found in the TCONS_00004115 mutant, related to small GAPase signal transduction, presumably involved in the adaptation to the physiological conditions tested.
4.4. Dimorphic Fungi
LncRNAs play also important roles in the regulatory mechanisms of dimorphic fungi. Many of them are pathogenic in animals, and lncRNAs can modulate key processes in their pathogenic activity, such as dimorphic transition, virulence, stress response and host adaptation. Their study may open new perspectives for understanding the phenotypic plasticity of these fungi and could reveal new therapeutic targets against fungal infections. Table 4 lists the lncRNAs that have received most attention in this group of fungi and whose mechanisms of action are summarized in this section.

Table 4.
Specific lncRNAs investigated in dimorphic fungi.
4.4.1. Ustilago maydis
Some of the noncoding RNAs identified in the U. maydis cDNA libraries show differential expression at different stages of development and may be associated with the pathogenesis process. This prediction was confirmed for one of them, called ncRNA1 [70]. Deletion of the gene for this putative lncRNA does not produce detectable phenotypic changes, except for a reduction in virulence on maize. The mechanism by which ncRNA1 is involved in pathogenesis remains to be elucidated, but its deletion results in increased expression of the um12316 gene, which encodes a secreted protein [194]. Moreover, ncRNA1 partially overlaps in antisense orientation with gene um02150, which itself possesses an antisense RNA, as-um12316, and whose function could also be associated with pathogenesis [195]. Engineered-induced as-um12316 levels in haploid cells, in which it is normally barely expressed, results in double levels of its complementary um12316 mRNA, presumably by stabilization in two-stranded forms, which could be experimentally detected. However, doubling of um12316 content does not lead to increased amounts of the encoded protein, probably because the additional mRNA is not available for translation.
Another antisense RNA investigated in detail in U. maydis is as-ssm1 [108]. The complementary gene, ssm1, encodes a mitochondrial seryl-tRNA synthetase, that catalyzes the transfer of L-serine to tRNA and its deletion causes cell lysis, making it a necessary gene for the integrity and viability of the fungus. The ssm1 gene is induced in the dikaryotic stage, and its ectopic expression in haploid cells slows growth, reduces virulence and alters mitochondrial activity, as evidenced by reduced oxygen consumption. Similarly to what was observed with the um12316 gene, ssm1 mRNA levels increase and two-stranded as-ssm1/ssm1 forms are detected, without changing the amount of Ssm1 protein. The data suggest a regulatory role of this antisense RNA reducing mitochondrial function in the teliospores, which may be involved in its dormancy state. Thirty-three other antisense mitochondrial gene transcripts, with an average length of 720 nt, have been detected, making it likely that as-ssm1 functions in coordination with other antisense transcripts to reduce mitochondrial activity in a more general mechanism associated with the dormancy state in this group of fungi.
Telomerase RNA production varies in the different eukaryotic kingdoms, with differences in processing or in the transcription machinery involved, polymerases II in animals or some fungi (see Section 4.1.4) or polymerase III in plants. In U. maydis the telomeric RNA TER is not derived from a separate transcript, but from a long UTR region of the emi1 gene transcript, which encodes the “early meiotic induction protein” [191]. The emi1 mRNA is subject to typical capping, polyadenylation and intron splicing, and may be subject to alternative maturation, including processing to produce the 1291 nt TER.
4.4.2. Candida auris
Dimorphism in Candida plays an important role in its pathogenicity. Thus, the filamentous form is more effective as an invasive agent than the yeast form during infection, although the unicellular form is more effective for dispersal. Therefore, in addition to other factors, as biofilm formation or enzymes or toxins secretion, the control of the yeast/hyphae switch plays an important role in its biological cycle [196]. In a screening of C. auris mutants obtained by insertion with a transposon, a mutant was identified that develops only in the filamentous form [192]. The mutant is affected in a lncRNA, called DINOR (from DNA damage-inducible non-coding RNA), and has higher levels of DNA damage due to the DNA checkpoint kinase RAD53 being hyperphosphorylated. This phenotype is related to the filamentous growth-inducing effect of DNA alkylating agents, which induce DINOR expression. This lncRNA modulates the response to different stress agents, including the presence of certain antifungals, H2O2, SDS, or macrophages. These stressing conditions upregulate DINOR, indicating that this lncRNA integrates different stress signals, as well as having a role in the maintenance of genome integrity.
4.4.3. Cryptococcus neoformans
C. neoformans is another pathogenic yeast in which the morphological change from yeast to hyphae plays a relevant role in pathogenesis, in addition to other processes, such as the sexual cycle. A major role in this morphological transition is played by the transcription factor Znf2 [197]. An Agrobacterium-mediated insertional mutation screening in this species allowed the identification of a mutant with a yeast-like phenotype, similar to that shown by the mutant lacking Znf2. The Ti insertion in the mutant affects a gene for a lncRNA located 2.5 kb upstream of ZNF2, which was named RZE1 [193]. This lncRNA is functionally restricted to its own nucleus, as revealed in a heterokaryon assay, and exerts its function mainly in cis. Transcriptomic and RNA localization analysis showed that the mutant lacking RZE1 has lower levels of znf2 mRNA and it accumulates in greater proportion in the nucleus, so that this lncRNA facilitates both its transcription and its passage to the cytoplasm, allowing a sufficient amount of functional Znf2 protein. RZE1 transcript is found in all subspecies of the Cryptococcus neoformans species complex, indicating a conserved role in this pathogenic fungal group.
4.4.4. Ustilaginoidea virens
In the study of lncRNAs by RNA-seq carried out in U. virens (see Section 2.3), one of the identified lncRNAs, Uvlnc001515, was studied in more detail [53]. This was renamed UvlncNAT-MFS because its 3′ end is antisense to the 3′ end of transcript of gene UvMFS (Uv-07373), which encodes a putative transporter of the Major Facility Superfamily, probably involved in zinc transport. The two transcripts showed similar expression patterns throughout different stages of rice infection, and parallel transcript levels were also found in strains silenced or overexpressing UvlncNAT-MFS or UcMFS, evidencing an interdependent expression. Either repression or overexpression of both transcripts produced abnormal fungal growth, indicating the importance of fine-tuned regulation of their transcripts. However, none of these changes led to alterations in pathogenesis. The detail of the mechanism of action of the regulation of UvlncNAT-MFS on UcMFS is unknown, except that it was demonstrated that they form a duplex RNA structure in vivo and they co-localize on in situ hybridizations.
4.5. Filamentous Fungi
Although the morphological complexity and metabolic versatility of filamentous fungi make them models of great interest for research, and some of them are very well-stablished research objects, studies on specific lncRNAs in filamentous species are not abundant, although their number continues to grow. Major findings on specific lncRNAs in these fungi, listed in Table 5, are summarized below.

Table 5.
Specific lncRNAs investigated in filamentous fungi.
4.5.1. Neurospora crassa
One of the aspects in which Neurospora crassa has stood out as a research model is the study of circadian rhythms [212]. Several lncRNAs have been identified that modulate the expression of genes involved in the regulation of the oscillation of the circadian machinery. One of the best known is the antisense transcription of the frq (frequency) gene, a central component of the circadian clock. A lncRNA antisense to the frq gene, termed qrf, regulates frq sense mRNA expression through transcriptional interference [199]. Subsequent work that has deepened on the mechanism of action observed that qrf transcription initially promotes frq expression by facilitating a suitable chromatin environment but subsequently leads to its inactivation by heterochromatinization [200]. The transcription of qrf contributes to modulating the amplitude and duration of frq expression, whereas frq transcription inhibits qrf expression and drives the antiphase rhythm of both transcripts. Thus, mutual inhibition of frq and qrf transcription forms a double negative feedback loop that is part of the core of circadian rhythm functioning [198]. In turn, qrf is not essential for circadian rhythmicity, and its transcriptional rhythm is mediated by the morning-specific repressor CSP-1 [213]. This scenario provides an example of how lncRNAs can act as regulators of biological oscillators through interference mechanisms.
4.5.2. Trichoderma reesei
In different strains of T. reesei, known for their capacity to produce cellulases, a lncRNA called HAX1 has been found whose length varies in different strains, being longer in hypercellulolytic strains [201]. A correlation was observed between the expression of this lncRNA and cellulolytic activity, the latter being increased in strains in which HAX1 is overexpressed, which confers biotechnological interest to this lncRNA. The mechanism of action is by interacting with a set of specific regulators of these enzymes, as seems to be the case for the xylanase activator Xyr1 [112]. It was observed that Xyr1 can inhibit its own transcription by a feed-back mechanism; a regulatory model has been proposed in which HAX1 physically interacts with Xyr1, reducing this retroinhibition and increasing its availability to increase the expression of its target genes, including the cellulase genes cbh1 and cbh2.
4.5.3. Fusarium fujikuroi and Fusarium oxysporum
Carotenoid synthesis in Fusarium fujikuroi and Fusarium oxysporum is activated by light and certain stress conditions, and is negatively regulated by a protein of the RING finger family, called CarS [214]. Mutants with a loss of function of the CarS protein show strong pigmentation due to upregulated expression of the carotenoid pathway genes. Transcriptomic analyses of the effect of light and the carS mutation, which affects hundreds of genes [215], detected upstream of the carS gene a lncRNA in both species, called Ff-carP and Fo-carP [202]. Deletion of either of both lncRNAs resulted in a sharp decrease in the mRNA levels of carotenoid biosynthetic genes and an albino phenotype (Figure 3). In addition to lacking assignable coding function, the non-coding nature was evidenced by the absence of significant overlapping ORFs when comparing Ff-carP and Fo-carP, due to the presence of small deletions/insertions, despite high sequence conservation and coincidence in the phenotypic effects of their mutations.
Figure 3.
Identification and effect of deletion lncRNA carP in Fusarium fujikuroi. (a) Transcript readings according to RNA-Seq data in the genomic region between the Ffuj_08715 and carS (Ffuj_08714) genes in the wild-type (WT) and carS mutant SG39 of F. fujikuroi. The readings were represented with the IGV program. The carP transcript is marked with a dashed box. (b) Colonies of the wild type and a representative ΔcarP mutant grown on minimal medium for 1 week under light. (c) Carotenoid content in the wild-type and the same ΔcarP mutant grown for 1 week in the dark (blue bars) or under light (orange bars). (d) Transcript levels for the carotenoid gene carB, and neighbor gene carS in the same strains grown in the dark or exposed for 1 h to light. RT-qPCR data show the mean and standard error of three independent experiments. Relative mRNA levels refer to the mRNA content of the wild-type strain in darkness. Adapted from Parra-Rivero et al. [202].
Ff-carP and Fo-carP are transcribed from the same strand than carS in their respective genomes, and the deletion of any of them upregulates carS mRNA levels, so it is proposed that the action of these lncRNAs is at the level of interfering with carS transcription. Consistently, overexpression of carS in F. fujikuroi results in an albino phenotype similar to that produced by Ff-carP deletion [216]. In support, when Ff-carP gene was reintroduced into the deletion mutants the carotenoid synthesis phenotype was only recovered when inserted upstream of carS [203]. In this work, the effect of its deletion on the transcriptome was also investigated and found to affect the transcript levels of several hundred genes. Most changes could be explained by its action on carS, but strong alterations were found in the expression of some genes that were not affected by the carS mutation. So, it is not excluded that Ff-carP may play other regulatory functions in trans in addition to its regulatory effect on carS in cis.
4.5.4. Fusarium graminearum
Several lncRNAs related to development and pathogenesis have been investigated in Fusarium graminearum (Gibberella zeae), a pathogen of maize, also infecting wheat and barley. In this fungus, the antisense lncRNA GzmetE-AS, which is transcribed from the complementary strand of the GzmetE gene, encoding a homoserine O-acetyltransferase involved in sexual development and pathogenesis, has been described [204]. The expression of GzmetE, low during conidiation and high at the end of the sexual cycle, is opposite to that of GzmetE-AS. In confirmation, an inverse correlation between both transcripts was observed in strains overexpressing this lncRNA or with premature termination in its 5′ region. In fact, strains lacking GzmetE-AS by the presence of the 5′ terminator show a phenotype similar to that of GzmetE-overexpressing strains, including an increased production of sexual structures. The regulation of this antisense lncRNA is carried out through the destruction of GzmetE mRNA by the RNA interference (RNAi) system, as evidenced by the fact that the reduction in GzmetE mRNA in GzmetE-AS-overexpressing strains is not observed in mutants lacking RNAi Dicer proteins.
The synthesis of deoxynivalenol (DON), a mycotoxin contaminating food and cereals infected by this fungus, is regulated by different proteins, including transcription factors such as Tri6 and Tri10. The study of their function by RNA-seq analysis of tri6 and tri10 mutants allowed the identification of two non-coding RNAs involved in the regulation of tri5, which encodes a key enzyme in the synthesis of these metabolites [205]. On the one hand, a tri5 antisense RNA is transcribed, which negatively regulates tri5 levels, as evidenced by its reduction when this antisense RNA is overexpressed. On the other hand, tri5 is regulated by the lncRNA RNA5P, which is transcribed in the same direction in its promoter region. Loss of RNA5P transcription results in increased expression of tri5, pointing to RNA5P as a negative regulator interfering with tri5 transcription. Consistent with this hypothesis, RNA5P exerts its action in cis, since its ectopic overexpression has no consequences on tri5 expression.
In a previous study, another lncRNA involved in the regulation of DON synthesis was found through its effect on the Fgsp1 gene, which encodes a protein of the sugar transporter family. Mutation of this gene has pleiotropic effects, including alterations in the sexual cycle (in ascospore release), virulence, and DON synthesis. Interestingly, a similar phenotype is shown by mutants lacking a lncRNA called lncRsp1, detected in the promoter region of rsp1 and transcribed from the same strand [206]. Both Fgsp1 and lncRsp1 negatively regulate the expression of several DON biosynthesis genes, TRI4, TRI5, TRI6, and TRI13. Loss of lncRsp1 transcription reduces rsp1 mRNA levels, so by an unknown mechanism its expression favors transcription from the same promoter.
A study of alternative splicing and polyadenylation in F. graminearum led to identify the lncRNA LncRsn, located between coding genes FgSna and FgPta [207]. FgSna contains a Longin domain, usually associated with vesicle trafficking, and FgPta is a phospholipid-transporting ATP-ase. LncRsn is antisense to FgSna and is transcribed from the same strand as FgPta, and its deletion reduced the number of perithecia and the number of ascospores [208]. However, conidia formation and germination were not affected by the deletion, supporting that is not necessary for asexual reproduction. LncRsn has an important role in the infection of wheat because the deletion mutant showed a lower disease index and reduced length of the lesions in the coleoptiles and in the corn silks. This lncRNA is involved in the regulation of the biosynthesis of DON, as the mutants produced double the amount of DON than the wild type. When the expression of the flanking genes were analyzed by RT-qPCR, FgSna and FgPta were found to be strongly and partially downregulated in the mutant, respectively, reinforcing the regulatory function of this lncRNA.
4.5.5. Cochliobolus heterostrophus
Dark pigmentation in the maize pathogen Cochliobolus heterostrophus, due to melanin accumulation, increases under illumination or hyperosmotic stress, and is controlled by the transcription factor Cmr1, whose gene expression is regulated by the mitogen-activated protein kinases (MAPKs) Chk1 and Mps1 [209]. Interestingly, the cmr1 gene is transcribed from both strands, giving rise to sense and antisense cmr1 transcripts, both of which are upregulated by Chk1 and Mps1. An epigenetic regulation at the level of the chromatin organization of cmr1 expression was proposed, but the involvement of the cmr1 antisense transcript in this mechanism, or its possible neutralizing role on the sense cmr1 mRNA, has not been elucidated.
4.5.6. Metharhizium robertsii
Strand-specific ARN-seq analysis in the insect pathogen Metarhizium robertsii led to the identification of 1655 lncRNAs in germinated conidia after a heat treatment at 40 °C, of which 1081 were differentially expressed [87]. Expression level in the conidia of one of them, mrLncRNA1, increased ca. 200-fold after the heat treatment, and was chosen for a more detailed study. Deletion of this lncRNA resulted in a reduction in germination of 31% compared to the wild-type and complemented strains, suggesting that mrLncRNA1 is involved in conidial thermotolerance through an unknown mechanism.
4.5.7. Phytophthora infestans
Phytophthora infestans, the causal agent of potato late blight disease, produces important economic losses [217]. For this reason, much effort has been devoted to investigating the mechanism of pathogenesis by this fungus. In one of the works, the analysis of a P. infestans cDNA library enriched with sequences regulated during potato infection led to the identification of a new gene family of predicted non-coding sequences named Pinci1 (from P. infestans non-coding infection-specific family 1). This family consists of more than 400 Pinci1-like copies, with high similarity to each other, organized in clusters at more than 90 locations in the P. infestans genome [210]. The coding features were investigated in detail in two of the Pinci1 sequences, Pinci1-1 and Pinci1-5, about 700 nt in length. Lack of coding function was checked by bioinformatic means (Testcode algorithm) and confirmed by the lack of detection of FLAG-tagged proteins after expression of three putative FLAG-tagged Pinci1-ORFs. They are thus a family of related non-coding polyadenylated transcripts that are upregulated during potato infection. However, their possible role in the mechanism of pathogenesis remains unknown.
4.5.8. Pyricularia oryzae
RNA-seq data in hyphae and conidia in P. oryzae showed differential expression for the MGG_15773 gene, with much higher mRNA levels in conidia than in hyphae. The lncRNA G003973, which is transcribed from the same strand upstream of MGG_15773, in its promoter region, shows an opposite pattern of expression, consistent with this lncRNA being a negative regulator of its transcription [59]. Deletion of G003973 resulted in increased expression of the MGG_15773 gene in three independent mutants, which showed slower development of their colonies than the control strain, indicating that lncRNA G003973 plays a regulatory role on hyphal development through attenuation of transcription of its neighboring gene MGG_15773.
4.5.9. Rhizopus delemar
In Rhizopus delemar, an opportunistic pathogen that causes mucormycosis, weighted gene co-expression network analysis (WGCNA) was done to identify lncRNAs involved in the germination of conidia [211]. Most of the cell wall modifying enzymes were predicted to interact with lncRNAs. For example, lncRNAs MSTRG.10396.1 and MSTRG.13812.1 were foreseen to interact with RO3G_03340 mRNA coding for a chitin β-1–6-glucanosyl transferase, and lncRNAs MSTRG.13812.1 and MSTRG.13812.1 were predicted to interact with RO3G_01689 and RO3G_04985 mRNAs, coding for α-mannosyltransferases. Although such interactions remain to be experimentally demonstrated, the one with the highest probability (lowest free energy) is included in Table 5.
4.5.10. Verticillium dahliae
In the analysis on the occurrence of lncRNAs involved in pathogenesis in Verticillium dahliae (see Section 2.4), identified by changes in their expression patterns in mutants of genes for cell wall lytic enzymes, the involvement of two intergenic lncRNAs was investigated through the effect of their overexpression [58]. Strains overexpressing LNC_01522 (Gene id. XLOC_006536) showed a 10-fold increase in the expression of the VDAG_09366 and VDAG_01782 genes, encoding respectively for an endo-polygalacturonase and a pectin esterase, and a 4-fold increase in the VDAG_01205 gene, encoding an endoglucanase. These strains produce more severe leaf damage, demonstrating the participation of LNC_01522 in cotton plant pathogenesis. On the other hand, overexpression of LNC_000201 (Gene id. XLOC_000836) produced a 6-fold increase in the expression of the VDAG_09366 gene, encoding for an endo-polygalacturonase, but this alteration was not sufficient to show an apparent change in virulence.
In a parallel work, other lncRNAs were identified showing expression changes during the infection process and overexpressed to test the consequences on pathogenesis [82]. Virulence assays with the engineered strains showed that overexpression of lncRNA012077 resulted in more severe disease symptoms compared to the wild type strain, whereas overexpression of lncRNA009491 or lncRNA007722 resulted in reduced virulence, the effect being more pronounced in the case of lncRNA009491. However, no apparent effects were found in the case of other lncRNAs. These results indicate a collective action between numerous lncRNAs in the infection process, some acting as positive and others as negative regulators, with a more relevant participation for some of them.
5. Conclusions and Future Prospects
Global transcriptomic analysis techniques have enabled new approaches to the study of lncRNAs by contrasting transcriptomes with annotated genomes. Over the last decade, the number of lncRNAs investigated in fungi has exploded, most of which have regulatory functions. From their location respective to protein coding genes, they are basically distributed into two types, intergenic and antisense lncRNAs. The cases investigated in more detail reveal a wide diversity of mechanisms of action, greatly increasing the complexity of gene regulatory systems in fungi. Interference with transcription or promotion of changes in chromatin structure are frequent in intergenic lncRNAs, and interference with the mRNA with which they overlap is a predictable consequence in antisense lncRNAs, but other studies reveal numerous variations on these themes, including facilitation of recruitment of other effector proteins or trans-acting effects.
The development of new techniques and the improvement of CRISPR-directed mutagenesis methodologies is allowing more ambitious strategies in the study of lncRNA functions. An example is the massive study of the function of 141 intergenic lncRNAs carried out in S. pombe by systematic deletion, allowing phenotypes to be found either visually or by microscopy or flow cytometry for almost 60% of the lncRNAs analyzed [218]. This work also showed the diversity of biological processes to which they are associated, demonstrating that fungi use this regulatory tool in a massive and generalized way, considerably increasing the sophistication of their gene regulation systems. In addition, the efficiency of regulation by lncRNAs opens new perspectives for their biotechnological applications. In S. pombe, a pho1 reporter gene regulated by the lncRNA nc-tgp1, whose transcription is regulated by the thiamine-activatable nmt1 promoter, has been tested [219]. This hybrid regulatory system allows for strong and rapid induction of any gene of interest, enabling the adjustment of its transcription through the concentration of thiamine in the medium. Another case, already mentioned in Section 4.1.1, is the increase in the yield of heterologous protein production by S. cerevisiae due to the deletion of certain intergenic lncRNAs [123]. These are just two examples of the vast field of possibilities opened by the study of the mechanisms of action of lncRNAs, with implications in such diverse areas as biotechnological production or biocontrol and treatment of fungal diseases.
Despite advances in their identification, the study of lncRNAs in fungi is an area of research with significant constraints. First, knowledge about the precise function of many of the lncRNAs that have been investigated remains limited. This is largely due to their diversity and the difficulty in identifying their molecular targets and mechanisms of action, especially in the case of intergenic lncRNAs. Their study faces several challenges, including their low evolutionary conservation, as they tend to be very species-specific, making extrapolation of results difficult, and their often low or transient expression (partially resolved by the use of mutants of the degradation machinery), making it difficult to distinguish functional lncRNAs from transcriptional noise. Progress in this field is expected to be enhanced using new technologies, e.g., in structural biology, to visualize how lncRNAs fold and interact with target proteins, or in bioinformatics, with the growing support of artificial intelligence, to predict the formation of secondary structures or interaction with other molecules with increasing reliability. Methodologies for identifying lncRNA partners in which advances are expected include high-resolution in vivo RNA imaging, identification of RNA-binding proteins (ChIRP), capture hybridization analysis of RNA targets (CHART), or different purification or immunoprecipitation techniques), fostered by the increasing importance of lncRNAs in biomedicine [220]. Advances are also expected in single-cell omics techniques, which should allow the analysis of expression variability in heterogeneous cell populations, of special interest in yeasts.
An important aspect in lncRNA research is the assumed absence of coding functions. Demonstrating this absence requires the use of unusual techniques, such as ribosome profiling, which, due to their technical complexity, are not often addressed in lncRNA studies. However, just as there is pervasive transcription, there is also pervasive translation. This is not unexpected, since XUTs are subject to NMD, implying that they are recognized by ribosomes [221]. Pervasive translation has been confirmed in recent studies in S. cerevisiae, which show that many lncRNAs can be recognized by ribosomes and give rise to small proteins [222,223], expanding the proteome of this yeast and blurring the boundary between coding and non-coding RNAs. The study of the possible functions of these proteins is an experimental challenge which is beginning to be addressed. An important aspect of these novel peptides is that they may co-evolve with other proteins in the genome, and they may play an evolutionary role in the generation of new coding genes [224,225,226].
The available data on lncRNAs in fungi is heavily biased towards yeasts, mostly due to their experimental advantages, especially outstanding in S. cerevisiae. It is expected that in the future there will be increasing attention to the study of lncRNAs in filamentous fungi for different reasons. Many of their species are particularly relevant as pathogens, especially in agriculture, or have biotechnological interest due to their metabolic versatility or their use as biocontrol agents. They also stand out for their ability to adapt to changing environments and respond to adverse conditions. In addition, they have more complex morphologies and development programs than yeasts, not only due to the formation and branching of hyphae, but also to the processes of sporulation or fruiting body formation [227]. All of this is reflected in the larger size of their genomes, often in the range of 40–50 Mb, while yeast genomes are typically around 10–15 Mb in size. Therefore, greater regulatory complexity mediated by lncRNAs can be expected in filamentous fungi, making research in this area particularly promising.
Author Contributions
Conceptualization, J.A.; writing—original draft preparation, J.A.; writing—review and editing, M.C.L.; Mechanisms interpretation, A.P.-B.; funding acquisition, M.C.L. and J.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by I + D + I project PID2022-140414NB-I00 funded by MICIU/AEI/10.13039/501100011033 and by ERDF/EU.
Informed Consent Statement
Not applicable.
Acknowledgments
J.A., A.P.B., and M.C.L. are members of the CaRed Excellence Network from the Spanish Ministry of Science and Innovation (RED2022-134577-T).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CTD | Carboxy terminal domain |
| CUT | Cryptic unstable transcript |
| DON | Deoxynivalenol |
| DSR | Determinant of selective removal |
| DUT | Dicer-sensitive unstable transcript |
| EST | Expressed sequence tag |
| lncRNA | Long non-coding RNA |
| lincRNA | Long intergenic non-coding RNA |
| MUT | Meiotic unannotated transcripts |
| ncRNA | Non-coding RNA |
| NMD | Nonsense-mediated decay |
| NUT | Nrd1-unterminated transcript |
| snoRNA | small nucleolar RNA |
| SUT | Stable unannotated transcript |
| TSS | Transcription start site |
| XUT | Xrn1-sensitive unstable transcripts XUT |
| cenRNA | Centromeric RNA |
References
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Rutenberg-Schoenberg, M.; Sexton, A.N.; Simon, M.D. The Properties of Long Noncoding RNAs That Regulate Chromatin. Annu. Rev. Genomics Hum. Genet. 2016, 17, 69–94. [Google Scholar] [CrossRef]
- Bester, A.C. Deciphering the Hidden Language of Long Non-Coding RNAs: Recent Findings and Challenges. In Genetics; Dabas, P., Ed.; IntechOpen: London, UK, 2024; Volume 2, ISBN 978-1-80356-710-5. [Google Scholar]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the Classification of Long Non-Coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Tani, H. Biomolecules Interacting with Long Noncoding RNAs. Biology 2025, 14, 442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, Y.; Liao, Q. Long Noncoding RNA: A Crosslink in Biological Regulatory Network. Brief. Bioinform. 2018, 19, 930–945. [Google Scholar] [CrossRef]
- Munroe, S.H.; Zhu, J. Overlapping Transcripts, Double-Stranded RNA and Antisense Regulation: A Genomic Perspective. Cell. Mol. Life Sci. CMLS 2006, 63, 2102–2118. [Google Scholar] [CrossRef]
- Szachnowski, U.; Becker, E.; Stuparević, I.; Wery, M.; Sallou, O.; Boudet, M.; Bretaudeau, A.; Morillon, A.; Primig, M. Pervasive Formation of Double-Stranded RNAs by Overlapping Sense/Antisense Transcripts in Budding Yeast Mitosis and Meiosis. RNA 2025, 31, 497–513. [Google Scholar] [CrossRef]
- Lamping, J.-P.; Krebber, H. The Hidden Power of Antisense Long Non-Coding RNAs: A Dive into a Novel Regulatory Layer Mediated by Double-Stranded RNA Formation. RNA Biol. 2025, 22, 1–16. [Google Scholar] [CrossRef]
- Wery, M.; Descrimes, M.; Vogt, N.; Dallongeville, A.-S.; Gautheret, D.; Morillon, A. Nonsense-Mediated Decay Restricts lncRNA Levels in Yeast Unless Blocked by Double-Stranded RNA Structure. Mol. Cell 2016, 61, 379–392. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- de Poloni, J.F.; de Oliveira, F.H.S.; Feltes, B.C. Localization Is the Key to Action: Regulatory Peculiarities of lncRNAs. Front. Genet. 2024, 15, 1478352. [Google Scholar] [CrossRef]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and Functional Analysis of lncRNAs: Methodologies to Investigate an Uncharacterized Transcriptome. Biochim. Biophys. Acta 2016, 1859, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.-C.; Carninci, P. Expanded ENCODE Delivers Invaluable Genomic Encyclopedia. Nature 2020, 583, 685–686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An Updated Database Dedicated to Long Non-Coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef]
- Meijueiro, M.L.; Santoyo, F.; Ramírez, L.; Pisabarro, A.G. Transcriptome Characteristics of Filamentous Fungi Deduced Using High-Throughput Analytical Technologies. Brief. Funct. Genomics 2014, 13, 440–450. [Google Scholar] [CrossRef]
- Werner, A.; Kanhere, A.; Wahlestedt, C.; Mattick, J.S. Natural Antisense Transcripts as Versatile Regulators of Gene Expression. Nat. Rev. Genet. 2024, 25, 730–744. [Google Scholar] [CrossRef]
- Mockler, T.C.; Chan, S.; Sundaresan, A.; Chen, H.; Jacobsen, S.E.; Ecker, J.R. Applications of DNA Tiling Arrays for Whole-Genome Analysis. Genomics 2005, 85, 1–15. [Google Scholar] [CrossRef]
- David, L.; Huber, W.; Granovskaia, M.; Toedling, J.; Palm, C.J.; Bofkin, L.; Jones, T.; Davis, R.W.; Steinmetz, L.M. A High-Resolution Map of Transcription in the Yeast Genome. Proc. Natl. Acad. Sci. USA 2006, 103, 5320–5325. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Miura, F.; Onda, M. Unexpected Complexity of the Budding Yeast Transcriptome. IUBMB Life 2008, 60, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Miura, F.; Kawaguchi, N.; Sese, J.; Toyoda, A.; Hattori, M.; Morishita, S.; Ito, T. A Large-Scale Full-Length cDNA Analysis to Explore the Budding Yeast Transcriptome. Proc. Natl. Acad. Sci. USA 2006, 103, 17846–17851. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef]
- Yassour, M.; Pfiffner, J.; Levin, J.Z.; Adiconis, X.; Gnirke, A.; Nusbaum, C.; Thompson, D.-A.; Friedman, N.; Regev, A. Strand-Specific RNA Sequencing Reveals Extensive Regulated Long Antisense Transcripts That Are Conserved across Yeast Species. Genome Biol. 2010, 11, R87. [Google Scholar] [CrossRef]
- Li, D.; Dong, Y.; Jiang, Y.; Jiang, H.; Cai, J.; Wang, W. A de Novo Originated Gene Depresses Budding Yeast Mating Pathway and Is Repressed by the Protein Encoded by Its Antisense Strand. Cell Res. 2010, 20, 408–420. [Google Scholar] [CrossRef]
- Huber, F.; Bunina, D.; Gupta, I.; Khmelinskii, A.; Meurer, M.; Theer, P.; Steinmetz, L.M.; Knop, M. Protein Abundance Control by Non-Coding Antisense Transcription. Cell Rep. 2016, 15, 2625–2636. [Google Scholar] [CrossRef]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.-C.; Dufour, M.-E.; Boulay, J.; Régnault, B.; Devaux, F.; Namane, A.; Séraphin, B.; et al. Cryptic Pol II Transcripts Are Degraded by a Nuclear Quality Control Pathway Involving a New Poly(A) Polymerase. Cell 2005, 121, 725–737. [Google Scholar] [CrossRef]
- Gudipati, R.K.; Xu, Z.; Lebreton, A.; Séraphin, B.; Steinmetz, L.M.; Jacquier, A.; Libri, D. Extensive Degradation of RNA Precursors by the Exosome in Wild-Type Cells. Mol. Cell 2012, 48, 409–421. [Google Scholar] [CrossRef]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Münster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L.M. Bidirectional Promoters Generate Pervasive Transcription in Yeast. Nature 2009, 457, 1033–1037. [Google Scholar] [CrossRef]
- Neil, H.; Malabat, C.; d’Aubenton-Carafa, Y.; Xu, Z.; Steinmetz, L.M.; Jacquier, A. Widespread Bidirectional Promoters Are the Major Source of Cryptic Transcripts in Yeast. Nature 2009, 457, 1038–1042. [Google Scholar] [CrossRef]
- Thiebaut, M.; Kisseleva-Romanova, E.; Rougemaille, M.; Boulay, J.; Libri, D. Transcription Termination and Nuclear Degradation of Cryptic Unstable Transcripts: A Role for the Nrd1-Nab3 Pathway in Genome Surveillance. Mol. Cell 2006, 23, 853–864. [Google Scholar] [CrossRef]
- Arigo, J.T.; Eyler, D.E.; Carroll, K.L.; Corden, J.L. Termination of Cryptic Unstable Transcripts Is Directed by Yeast RNA-Binding Proteins Nrd1 and Nab3. Mol. Cell 2006, 23, 841–851. [Google Scholar] [CrossRef]
- Vasiljeva, L.; Kim, M.; Terzi, N.; Soares, L.M.; Buratowski, S. Transcription Termination and RNA Degradation Contribute to Silencing of RNA Polymerase II Transcription within Heterochromatin. Mol. Cell 2008, 29, 313–323. [Google Scholar] [CrossRef]
- Schulz, D.; Schwalb, B.; Kiesel, A.; Baejen, C.; Torkler, P.; Gagneur, J.; Soeding, J.; Cramer, P. Transcriptome Surveillance by Selective Termination of Noncoding RNA Synthesis. Cell 2013, 155, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.P.; Tongprasit, W.; Sethi, H.; Chin, C.-S.; Stolc, V. Global Identification of Noncoding RNAs in Saccharomyces cerevisiae by Modulating an Essential RNA Processing Pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 4192–4197. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.J.; Mosley, A.L. Rrp6: Integrated Roles in Nuclear RNA Metabolism and Transcription Termination. Wiley Interdiscip. Rev. RNA 2016, 7, 91–104. [Google Scholar] [CrossRef]
- Davis, C.A.; Ares, M. Accumulation of Unstable Promoter-Associated Transcripts upon Loss of the Nuclear Exosome Subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2006, 103, 3262–3267. [Google Scholar] [CrossRef]
- Lardenois, A.; Liu, Y.; Walther, T.; Chalmel, F.; Evrard, B.; Granovskaia, M.; Chu, A.; Davis, R.W.; Steinmetz, L.M.; Primig, M. Execution of the Meiotic Noncoding RNA Expression Program and the Onset of Gametogenesis in Yeast Require the Conserved Exosome Subunit Rrp6. Proc. Natl. Acad. Sci. USA 2011, 108, 1058–1063. [Google Scholar] [CrossRef]
- Castelnuovo, M.; Zaugg, J.B.; Guffanti, E.; Maffioletti, A.; Camblong, J.; Xu, Z.; Clauder-Münster, S.; Steinmetz, L.M.; Luscombe, N.M.; Stutz, F. Role of Histone Modifications and Early Termination in Pervasive Transcription and Antisense-Mediated Gene Silencing in Yeast. Nucleic Acids Res. 2014, 42, 4348–4362. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-Mediated mRNA Decay: An Intricate Machinery That Shapes Transcriptomes. Nat. Rev. Mol. Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Chen, C.L.; d’Aubenton-Carafa, Y.; Gourvennec, S.; Kwapisz, M.; Roche, V.; Bertrand, C.; Silvain, M.; Legoix-Né, P.; Loeillet, S.; et al. XUTs Are a Class of Xrn1-Sensitive Antisense Regulatory Non-Coding RNA in Yeast. Nature 2011, 475, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Andjus, S.; Morillon, A.; Wery, M. From Yeast to Mammals, the Nonsense-Mediated mRNA Decay as a Master Regulator of Long Non-Coding RNAs Functional Trajectory. Non-Coding RNA 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Malabat, C.; Feuerbach, F.; Ma, L.; Saveanu, C.; Jacquier, A. Quality Control of Transcription Start Site Selection by Nonsense-Mediated-mRNA Decay. eLife 2015, 4, e06722. [Google Scholar] [CrossRef] [PubMed]
- Wery, M.; Szachnowski, U.; Andjus, S.; de Andres-Pablo, A.; Morillon, A. The RNA Helicases Dbp2 and Mtr4 Regulate the Expression of Xrn1-Sensitive Long Non-Coding RNAs in Yeast. Front. RNA Res. 2023, 1, 1244554. [Google Scholar] [CrossRef]
- Wilhelm, B.T.; Marguerat, S.; Watt, S.; Schubert, F.; Wood, V.; Goodhead, I.; Penkett, C.J.; Rogers, J.; Bähler, J. Dynamic Repertoire of a Eukaryotic Transcriptome Surveyed at Single-Nucleotide Resolution. Nature 2008, 453, 1239–1243. [Google Scholar] [CrossRef]
- Rhind, N.; Chen, Z.; Yassour, M.; Thompson, D.A.; Haas, B.J.; Habib, N.; Wapinski, I.; Roy, S.; Lin, M.F.; Heiman, D.I.; et al. Comparative Functional Genomics of the Fission Yeasts. Science 2011, 332, 930–936. [Google Scholar] [CrossRef]
- Sun, W.-H.; Wang, Y.-Z.; Xu, Y.; Yu, X.-W. Genome-Wide Analysis of Long Non-Coding RNAs in Pichia pastoris during Stress by RNA Sequencing. Genomics 2019, 111, 398–406. [Google Scholar] [CrossRef]
- Ho, E.C.H.; Cahill, M.J.; Saville, B.J. Gene Discovery and Transcript Analyses in the Corn Smut Pathogen Ustilago maydis: Expressed Sequence Tag and Genome Sequence Comparison. BMC Genomics 2007, 8, 334. [Google Scholar] [CrossRef]
- Donaldson, M.E.; Ostrowski, L.A.; Goulet, K.M.; Saville, B.J. Transcriptome Analysis of Smut Fungi Reveals Widespread Intergenic Transcription and Conserved Antisense Transcript Expression. BMC Genom. 2017, 18, 340. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, X.; Yan, Y.; Huang, J.; Luo, C.; Tom, H.; Zheng, L. Comprehensive Transcriptome Profiling Reveals Abundant Long Non-Coding RNAs Associated with Development of the Rice False Smut Fungus, Ustilaginoidea virens. Environ. Microbiol. 2021, 23, 4998–5013. [Google Scholar] [CrossRef] [PubMed]
- Arthanari, Y.; Heintzen, C.; Griffiths-Jones, S.; Crosthwaite, S.K. Natural Antisense Transcripts and Long Non-Coding RNA in Neurospora crassa. PLoS ONE 2014, 9, e91353. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, B.; Yin, C.; Guo, Y.; Lin, Y.; Pan, L.; Wang, B. Characterization of Natural Antisense Transcript, Sclerotia Development and Secondary Metabolism by Strand-Specific RNA Sequencing of Aspergillus flavus. PLoS ONE 2014, 9, e97814. [Google Scholar] [CrossRef]
- Choi, G.; Jeon, J.; Lee, H.; Zhou, S.; Lee, Y.-H. Genome-Wide Profiling of Long Non-Coding RNA of the Rice Blast Fungus Magnaporthe oryzae during Infection. BMC Genom. 2022, 23, 132. [Google Scholar] [CrossRef]
- Shi, H.; Ding, G.; Wang, Y.; Wang, J.; Wang, X.; Wang, D.; Lu, P. Genome-Wide Identification of Long Non-Coding RNA for Botrytis cinerea during Infection to Tomato (Solanum lycopersicum) Leaves. BMC Genom. 2025, 26, 7. [Google Scholar] [CrossRef]
- Liu, N.; Wang, P.; Li, X.; Pei, Y.; Sun, Y.; Ma, X.; Ge, X.; Zhu, Y.; Li, F.; Hou, Y. Long Non-Coding RNAs Profiling in Pathogenesis of Verticillium dahliae: New Insights in the Host-Pathogen Interaction. Plant Sci. Int. J. Exp. Plant Biol. 2022, 314, 111098. [Google Scholar] [CrossRef]
- Li, Z.; Yang, J.; Peng, J.; Cheng, Z.; Liu, X.; Zhang, Z.; Bhadauria, V.; Zhao, W.; Peng, Y.-L. Transcriptional Landscapes of Long Non-Coding RNAs and Alternative Splicing in Pyricularia oryzae Revealed by RNA-Seq. Front. Plant Sci. 2021, 12, 723636. [Google Scholar] [CrossRef]
- Kim, W.; Miguel-Rojas, C.; Wang, J.; Townsend, J.P.; Trail, F. Developmental Dynamics of Long Noncoding RNA Expression during Sexual Fruiting Body Formation in Fusarium graminearum. mBio 2018, 9, e01292-18. [Google Scholar] [CrossRef]
- Alcid, E.A.; Tsukiyama, T. Expansion of Antisense lncRNA Transcriptomes in Budding Yeast Species since the Loss of RNAi. Nat. Struct. Mol. Biol. 2016, 23, 450–455. [Google Scholar] [CrossRef]
- Szachnowski, U.; Andjus, S.; Foretek, D.; Morillon, A.; Wery, M. Endogenous RNAi Pathway Evolutionarily Shapes the Destiny of the Antisense lncRNAs Transcriptome. Life Sci. Alliance 2019, 2, e201900407. [Google Scholar] [CrossRef] [PubMed]
- Quintales, L.; Sánchez, M.; Antequera, F. Analysis of DNA Strand-Specific Differential Expression with High Density Tiling Microarrays. BMC Bioinform. 2010, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.R.; Marguerat, S.; Bitton, D.A.; Rodríguez-López, M.; Rallis, C.; Lemay, J.-F.; Cotobal, C.; Malecki, M.; Smialowski, P.; Mata, J.; et al. Long Noncoding RNA Repertoire and Targeting by Nuclear Exosome, Cytoplasmic Exonuclease, and RNAi in Fission Yeast. RNA 2018, 24, 1195–1213. [Google Scholar] [CrossRef] [PubMed]
- DeGennaro, C.M.; Alver, B.H.; Marguerat, S.; Stepanova, E.; Davis, C.P.; Bähler, J.; Park, P.J.; Winston, F. Spt6 Regulates Intragenic and Antisense Transcription, Nucleosome Positioning, and Histone Modifications Genome-Wide in Fission Yeast. Mol. Cell. Biol. 2013, 33, 4779–4792. [Google Scholar] [CrossRef]
- Wery, M.; Gautier, C.; Descrimes, M.; Yoda, M.; Migeot, V.; Hermand, D.; Morillon, A. Bases of Antisense lncRNA-Associated Regulation of Gene Expression in Fission Yeast. PLoS Genet. 2018, 14, e1007465. [Google Scholar] [CrossRef]
- Watts, B.R.; Wittmann, S.; Wery, M.; Gautier, C.; Kus, K.; Birot, A.; Heo, D.-H.; Kilchert, C.; Morillon, A.; Vasiljeva, L. Histone Deacetylation Promotes Transcriptional Silencing at Facultative Heterochromatin. Nucleic Acids Res. 2018, 46, 5426–5440. [Google Scholar] [CrossRef]
- Eser, P.; Wachutka, L.; Maier, K.C.; Demel, C.; Boroni, M.; Iyer, S.; Cramer, P.; Gagneur, J. Determinants of RNA Metabolism in the Schizosaccharomyces pombe Genome. Mol. Syst. Biol. 2016, 12, 857. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Perlin, M.H.; Garcia-Pedrajas, M.; Covert, S.F.; Gold, S.E. Genetics of Morphogenesis and Pathogenic Development of Ustilago maydis. Adv. Genet. 2007, 57, 1–47. [Google Scholar] [CrossRef]
- Morrison, E.N.; Donaldson, M.E.; Saville, B.J. Identification and Analysis of Genes Expressed in the Ustilago maydis Dikaryon: Uncovering a Novel Class of Pathogenesis Genes. Can. J. Plant Pathol. 2012, 34, 417–435. [Google Scholar] [CrossRef]
- Hovhannisyan, H.; Gabaldón, T. The Long Non-Coding RNA Landscape of Candida Yeast Pathogens. Nat. Commun. 2021, 12, 7317. [Google Scholar] [CrossRef]
- Roche, C.M.; Loros, J.J.; McCluskey, K.; Glass, N.L. Neurospora crassa: Looking Back and Looking Forward at a Model Microbe. Am. J. Bot. 2014, 101, 2022–2035. [Google Scholar] [CrossRef]
- Dunlap, J.C.; Borkovich, K.A.; Henn, M.R.; Turner, G.E.; Sachs, M.S.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W.; et al. Enabling a Community to Dissect an Organism: Overview of the Neurospora Functional Genomics Project. Adv. Genet. 2007, 57, 49–96. [Google Scholar] [CrossRef]
- Cemel, I.A.; Ha, N.; Schermann, G.; Yonekawa, S.; Brunner, M. The Coding and Noncoding Transcriptome of Neurospora crassa. BMC Genom. 2017, 18, 978. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Robertson, D.; Yates, B.; Nielsen, D.M.; Brown, D.; Dean, R.A.; Payne, G.A. The Effect of Temperature on Natural Antisense Transcript (NAT) Expression in Aspergillus flavus. Curr. Genet. 2008, 54, 241–269. [Google Scholar] [CrossRef] [PubMed]
- Davati, N.; Ghorbani, A. Discovery of Long Non-Coding RNAs in Aspergillus flavus Response to Water Activity, CO2 Concentration, and Temperature Changes. Sci. Rep. 2023, 13, 10330. [Google Scholar] [CrossRef]
- Wang, Y.; Shao, Y.; Zhu, Y.; Wang, K.; Ma, B.; Zhou, Q.; Chen, A.; Chen, H. XRN1-Associated Long Non-Coding RNAs May Contribute to Fungal Virulence and Sexual Development in Entomopathogenic Fungus Cordyceps militaris. Pest Manag. Sci. 2019, 75, 3302–3311. [Google Scholar] [CrossRef]
- Smith, J.E.; Baker, K.E. Nonsense-Mediated RNA Decay—A Switch and Dial for Regulating Gene Expression. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015, 37, 612–623. [Google Scholar] [CrossRef]
- Veloso, J.; van Kan, J.A.L. Many Shades of Grey in Botrytis-Host Plant Interactions. Trends Plant Sci. 2018, 23, 613–622. [Google Scholar] [CrossRef]
- Van Kan, J.A.L.; Stassen, J.H.M.; Mosbach, A.; Van Der Lee, T.A.J.; Faino, L.; Farmer, A.D.; Papasotiriou, D.G.; Zhou, S.; Seidl, M.F.; Cottam, E.; et al. A Gapless Genome Sequence of the Fungus Botrytis cinerea. Mol. Plant Pathol. 2017, 18, 75–89. [Google Scholar] [CrossRef]
- Li, R.; Xue, H.-S.; Zhang, D.-D.; Wang, D.; Song, J.; Subbarao, K.V.; Klosterman, S.J.; Chen, J.-Y.; Dai, X.-F. Identification of Long Non-Coding RNAs in Verticillium dahliae Following Inoculation of Cotton. Microbiol. Res. 2022, 257, 126962. [Google Scholar] [CrossRef]
- Glad, H.M.; Tralamazza, S.M.; Croll, D. The Expression Landscape and Pangenome of Long Non-Coding RNA in the Fungal Wheat Pathogen Zymoseptoria tritici. Microb. Genomics 2023, 9, 001136. [Google Scholar] [CrossRef]
- Menegidio, F.B.; Aciole Barbosa, D.; Alencar, V.C.; Vilas Boas, R.O.; Costa de Oliveira, R.; Jabes, D.L.; Nunes, L.R. Transcriptomic Profiling Identifies Novel Transcripts, Isomorphs, and Noncoding RNAs in Paracoccidioides brasiliensis. Med. Mycol. 2021, 59, 197–200. [Google Scholar] [CrossRef]
- Kalem, M.C.; Panepinto, J.C. Long Non-Noding RNAs in Cryptococcus neoformans: Insights into Fungal Pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 858317. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, W.; Wang, Y. Genome-Wide Identification of Long Non-Coding RNAs Suggests a Potential Association with Effector Gene Transcription in Phytophthora sojae. Mol. Plant Pathol. 2018, 19, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, Y.; Wu, H.; Xie, X.; Huang, B. Genome-Wide Identification and Functional Prediction of Long Non-Coding RNAs Involved in the Heat Stress Response in Metarhizium robertsii. Front. Microbiol. 2019, 10, 2336. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Zhang, L.; Zhou, X. Long Noncoding RNAs Are Potentially Involved in the Degeneration of Virulence in an Aphid-Obligate Pathogen, Conidiobolus obscurus (Entomophthoromycotina). Virulence 2021, 12, 1705–1716. [Google Scholar] [CrossRef]
- Lai, T.; Yu, Q.; Pan, J.; Wang, J.; Tang, Z.; Bai, X.; Shi, L.; Zhou, T. The Identification and Comparative Analysis of Non-Coding RNAs in Spores and Mycelia of Penicillium expansum. J. Fungi Basel Switz. 2023, 9, 999. [Google Scholar] [CrossRef]
- Silva, R.G.; Amaral, P.P.; Franco, G.R.; Góes-Neto, A. Exploring the Hidden Hot World of Long Non-Coding RNAs in Thermophilic Fungus Using a Robust Computational Pipeline. Sci. Rep. 2024, 14, 19797. [Google Scholar] [CrossRef]
- Song, X.; Zhang, M.; Chen, M.; Shang, X.; Zhou, F.; Yu, H.; Song, C.; Tan, Q. Transcriptomic Communication between Nucleus and Mitochondria during the Browning Process of Lentinula edodes. J. Agric. Food Chem. 2024, 72, 23592–23605. [Google Scholar] [CrossRef]
- Coltman, B.L.; Motheramgari, K.; Tatto, N.; Gasser, B. Identification and Functional Analysis of Growth Rate Associated Long Non-Coding RNAs in Komagataella phaffii. Comput. Struct. Biotechnol. J. 2025, 27, 1693–1705. [Google Scholar] [CrossRef]
- Cao, W.; Pan, X.; Yu, R.; Sheng, Y.; Zhang, H. Genome-Wide Identification of Long Non-Coding RNAs Reveals Potential Association with Phytophthora infestans Asexual and Sexual Development. Microbiol. Spectr. 2025, 13, e0199824. [Google Scholar] [CrossRef]
- Till, P.; Mach, R.L.; Mach-Aigner, A.R. A Current View on Long Noncoding RNAs in Yeast and Filamentous Fungi. Appl. Microbiol. Biotechnol. 2018, 102, 7319–7331. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Yin, Z.; Hu, Z.; Zhang, K.-Q. An Overview on Identification and Regulatory Mechanisms of Long Non-Coding RNAs in Fungi. Front. Microbiol. 2021, 12, 638617. [Google Scholar] [CrossRef]
- Yamashita, A.; Shichino, Y.; Yamamoto, M. The Long Non-Coding RNA World in Yeasts. Biochim. Biophys. Acta 2016, 1859, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Niederer, R.O.; Hass, E.P.; Zappulla, D.C. Long Noncoding RNAs in the Yeast S. cerevisiae. Adv. Exp. Med. Biol. 2017, 1008, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.; Váchová, L.; Hlaváček, O.; Maršíková, J.; Gilfillan, G.D.; Palková, Z. Long Noncoding RNAs in Yeast Cells and Differentiated Subpopulations of Yeast Colonies and Biofilms. Oxid. Med. Cell. Longev. 2018, 2018, 4950591. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, M.E.; Saville, B.J. Natural Antisense Transcripts in Fungi. Mol. Microbiol. 2012, 85, 405–417. [Google Scholar] [CrossRef]
- Hongay, C.F.; Grisafi, P.L.; Galitski, T.; Fink, G.R. Antisense Transcription Controls Cell Fate in Saccharomyces cerevisiae. Cell 2006, 127, 735–745. [Google Scholar] [CrossRef]
- Uhler, J.P.; Hertel, C.; Svejstrup, J.Q. A Role for Noncoding Transcription in Activation of the Yeast PHO5 Gene. Proc. Natl. Acad. Sci. USA 2007, 104, 8011–8016. [Google Scholar] [CrossRef]
- Soudet, J.; Beyrouthy, N.; Pastucha, A.M.; Maffioletti, A.; Menéndez, D.; Bakir, Z.; Stutz, F. Antisense-Mediated Repression of SAGA-Dependent Genes Involves the HIR Histone Chaperone. Nucleic Acids Res. 2022, 50, 4515–4528. [Google Scholar] [CrossRef]
- Camblong, J.; Iglesias, N.; Fickentscher, C.; Dieppois, G.; Stutz, F. Antisense RNA Stabilization Induces Transcriptional Gene Silencing via Histone Deacetylation in S. cerevisiae. Cell 2007, 131, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Hainer, S.J.; Pruneski, J.A.; Mitchell, R.D.; Monteverde, R.M.; Martens, J.A. Intergenic Transcription Causes Repression by Directing Nucleosome Assembly. Genes Dev. 2011, 25, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Shuman, S. Transcriptional Interference at Tandem lncRNA and Protein-Coding Genes: An Emerging Theme in Regulation of Cellular Nutrient Homeostasis. Nucleic Acids Res. 2020, 48, 8243–8254. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.A.; Wu, P.-Y.J.; Winston, F. Regulation of an Intergenic Transcript Controls Adjacent Gene Transcription in Saccharomyces cerevisiae. Genes Dev. 2005, 19, 2695–2704. [Google Scholar] [CrossRef]
- Berretta, J.; Pinskaya, M.; Morillon, A. A Cryptic Unstable Transcript Mediates Transcriptional Trans-Silencing of the Ty1 Retrotransposon in S. cerevisiae. Genes Dev. 2008, 22, 615–626. [Google Scholar] [CrossRef]
- Ostrowski, L.A.; Saville, B.J. Natural Antisense Transcripts Are Linked to the Modulation of Mitochondrial Function and Teliospore Dormancy in Ustilago maydis. Mol. Microbiol. 2017, 103, 745–763. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, L.; Lu, S. Role of TERRA in the Regulation of Telomere Length. Int. J. Biol. Sci. 2015, 11, 316–323. [Google Scholar] [CrossRef]
- Ding, D.-Q.; Okamasa, K.; Yamane, M.; Tsutsumi, C.; Haraguchi, T.; Yamamoto, M.; Hiraoka, Y. Meiosis-Specific Noncoding RNA Mediates Robust Pairing of Homologous Chromosomes in Meiosis. Science 2012, 336, 732–736. [Google Scholar] [CrossRef]
- Ding, D.-Q.; Okamasa, K.; Katou, Y.; Oya, E.; Nakayama, J.-I.; Chikashige, Y.; Shirahige, K.; Haraguchi, T.; Hiraoka, Y. Chromosome-Associated RNA-Protein Complexes Promote Pairing of Homologous Chromosomes during Meiosis in Schizosaccharomyces pombe. Nat. Commun. 2019, 10, 5598. [Google Scholar] [CrossRef]
- Till, P.; Derntl, C.; Kiesenhofer, D.P.; Mach, R.L.; Yaver, D.; Mach-Aigner, A.R. Regulation of Gene Expression by the Action of a Fungal lncRNA on a Transactivator. RNA Biol. 2020, 17, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Tisseur, M.; Kwapisz, M.; Morillon, A. Pervasive Transcription—Lessons from Yeast. Biochimie 2011, 93, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.J.; Daugharthy, E.R.; Kim, J. Pervasive Antisense Transcription Is Evolutionarily Conserved in Budding Yeast. Mol. Biol. Evol. 2013, 30, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V. From Transcriptional Complexity to Cellular Phenotypes: Lessons from Yeast. Yeast Chichester Engl. 2017, 34, 475–482. [Google Scholar] [CrossRef]
- Thiebaut, M.; Colin, J.; Neil, H.; Jacquier, A.; Séraphin, B.; Lacroute, F.; Libri, D. Futile Cycle of Transcription Initiation and Termination Modulates the Response to Nucleotide Shortage in S. cerevisiae. Mol. Cell 2008, 31, 671–682. [Google Scholar] [CrossRef]
- Kuehner, J.N.; Brow, D.A. Regulation of a Eukaryotic Gene by GTP-Dependent Start Site Selection and Transcription Attenuation. Mol. Cell 2008, 31, 201–211. [Google Scholar] [CrossRef]
- Houseley, J.; Rubbi, L.; Grunstein, M.; Tollervey, D.; Vogelauer, M. A ncRNA Modulates Histone Modification and mRNA Induction in the Yeast GAL Gene Cluster. Mol. Cell 2008, 32, 685–695. [Google Scholar] [CrossRef]
- Pinskaya, M.; Gourvennec, S.; Morillon, A. H3 Lysine 4 Di- and Tri-Methylation Deposited by Cryptic Transcription Attenuates Promoter Activation. EMBO J. 2009, 28, 1697–1707. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Chen, H.-T.; Teng, S.-C. Intragenic Transcription of a Noncoding RNA Modulates Expression of ASP3 in Budding Yeast. RNA 2010, 16, 2085–2093. [Google Scholar] [CrossRef]
- Awasthi, A.; Nain, V.; Srikanth, C.V.; Puria, R. A Regulatory Circuit between lncRNA and TOR Directs Amino Acid Uptake in Yeast. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118680. [Google Scholar] [CrossRef]
- du Mee, D.J.M.; Ivanov, M.; Parker, J.P.; Buratowski, S.; Marquardt, S. Efficient Termination of Nuclear lncRNA Transcription Promotes Mitochondrial Genome Maintenance. eLife 2018, 7, e31989. [Google Scholar] [CrossRef]
- Qin, L.; Pan, Y.; Xue, S.; Yan, Z.; Xiao, C.; Liu, X.; Yuan, D.; Hou, J.; Huang, M. Multi-Omics Analysis Reveals Impacts of lincRNA Deletion on Yeast Protein Synthesis. Adv. Sci. Weinh. Baden-Wurtt. Ger. 2025, 12, e2406873. [Google Scholar] [CrossRef]
- Balarezo-Cisneros, L.N.; Parker, S.; Fraczek, M.G.; Timouma, S.; Wang, P.; O’Keefe, R.T.; Millar, C.B.; Delneri, D. Functional and Transcriptional Profiling of Non-Coding RNAs in Yeast Reveal Context-Dependent Phenotypes and in Trans Effects on the Protein Regulatory Network. PLoS Genet. 2021, 17, e1008761. [Google Scholar] [CrossRef]
- Granovskaia, M.V.; Jensen, L.J.; Ritchie, M.E.; Toedling, J.; Ning, Y.; Bork, P.; Huber, W.; Steinmetz, L.M. High-Resolution Transcription Atlas of the Mitotic Cell Cycle in Budding Yeast. Genome Biol. 2010, 11, R24. [Google Scholar] [CrossRef]
- Nevers, A.; Doyen, A.; Malabat, C.; Néron, B.; Kergrohen, T.; Jacquier, A.; Badis, G. Antisense Transcriptional Interference Mediates Condition-Specific Gene Repression in Budding Yeast. Nucleic Acids Res. 2018, 46, 6009–6025. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Ribelles, M.; Solé, C.; Xu, Z.; Steinmetz, L.M.; de Nadal, E.; Posas, F. Control of Cdc28 CDK1 by a Stress-Induced lncRNA. Mol. Cell 2014, 53, 549–561. [Google Scholar] [CrossRef]
- Lázari, L.C.; Wolf, I.R.; Schnepper, A.P.; Valente, G.T. LncRNAs of Saccharomyces cerevisiae Bypass the Cell Cycle Arrest Imposed by Ethanol Stress. PLoS Comput. Biol. 2022, 18, e1010081. [Google Scholar] [CrossRef] [PubMed]
- van Werven, F.J.; Neuert, G.; Hendrick, N.; Lardenois, A.; Buratowski, S.; van Oudenaarden, A.; Primig, M.; Amon, A. Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast. Cell 2012, 150, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Moretto, F.; van Werven, F.J. Transcription of the Mating-Type-Regulated lncRNA IRT1 Is Governed by TORC1 and PKA. Curr. Genet. 2017, 63, 325–329. [Google Scholar] [CrossRef]
- Moretto, F.; Wood, N.E.; Kelly, G.; Doncic, A.; van Werven, F.J. A Regulatory Circuit of Two lncRNAs and a Master Regulator Directs Cell Fate in Yeast. Nat. Commun. 2018, 9, 780. [Google Scholar] [CrossRef]
- Moretto, F.; Wood, N.E.; Chia, M.; Li, C.; Luscombe, N.M.; van Werven, F.J. Transcription Levels of a Noncoding RNA Orchestrate Opposing Regulatory and Cell Fate Outcomes in Yeast. Cell Rep. 2021, 34, 108643. [Google Scholar] [CrossRef]
- Gelfand, B.; Mead, J.; Bruning, A.; Apostolopoulos, N.; Tadigotla, V.; Nagaraj, V.; Sengupta, A.M.; Vershon, A.K. Regulated Antisense Transcription Controls Expression of Cell-Type-Specific Genes in Yeast. Mol. Cell. Biol. 2011, 31, 1701–1709. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bumgarner, S.L.; Dowell, R.D.; Grisafi, P.; Gifford, D.K.; Fink, G.R. Toggle Involving Cis-Interfering Noncoding RNAs Controls Variegated Gene Expression in Yeast. Proc. Natl. Acad. Sci. USA 2009, 106, 18321–18326. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wei, W.; Gagneur, J.; Clauder-Münster, S.; Smolik, M.; Huber, W.; Steinmetz, L.M. Antisense Expression Increases Gene Expression Variability and Locus Interdependency. Mol. Syst. Biol. 2011, 7, 468. [Google Scholar] [CrossRef] [PubMed]
- Raupach, E.A.; Martens, J.A.; Arndt, K.M. Evidence for Regulation of ECM3 Expression by Methylation of Histone H3 Lysine 4 and Intergenic Transcription in Saccharomyces cerevisiae. G3 Bethesda Md 2016, 6, 2971–2981. [Google Scholar] [CrossRef]
- Novačić, A.; Vučenović, I.; Primig, M.; Stuparević, I. Non-Coding RNAs as Cell Wall Regulators in Saccharomyces cerevisiae. Crit. Rev. Microbiol. 2020, 46, 15–25. [Google Scholar] [CrossRef]
- Bunina, D.; Štefl, M.; Huber, F.; Khmelinskii, A.; Meurer, M.; Barry, J.D.; Kats, I.; Kirrmaier, D.; Huber, W.; Knop, M. Upregulation of SPS100 Gene Expression by an Antisense RNA via a Switch of mRNA Isoforms with Different Stabilities. Nucleic Acids Res. 2017, 45, 11144–11158. [Google Scholar] [CrossRef][Green Version]
- Yu, Y.; Yarrington, R.M.; Chuong, E.B.; Elde, N.C.; Stillman, D.J. Disruption of Promoter Memory by Synthesis of a Long Noncoding RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 9575–9580. [Google Scholar] [CrossRef]
- Ling, Y.H.; Yuen, K.W.Y. Point Centromere Activity Requires an Optimal Level of Centromeric Noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 6270–6279. [Google Scholar] [CrossRef]
- Zappulla, D.C. Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Mol. Basel Switz. 2020, 25, 2750. [Google Scholar] [CrossRef]
- Kyriakou, D.; Stavrou, E.; Demosthenous, P.; Angelidou, G.; San Luis, B.-J.; Boone, C.; Promponas, V.J.; Kirmizis, A. Functional Characterisation of Long Intergenic Non-Coding RNAs through Genetic Interaction Profiling in Saccharomyces cerevisiae. BMC Biol. 2016, 14, 106. [Google Scholar] [CrossRef]
- Thebault, P.; Boutin, G.; Bhat, W.; Rufiange, A.; Martens, J.; Nourani, A. Transcription Regulation by the Noncoding RNA SRG1 Requires Spt2-Dependent Chromatin Deposition in the Wake of RNA Polymerase II. Mol. Cell. Biol. 2011, 31, 1288–1300. [Google Scholar] [CrossRef]
- Geisler, S.; Lojek, L.; Khalil, A.M.; Baker, K.E.; Coller, J. Decapping of Long Noncoding RNAs Regulates Inducible Genes. Mol. Cell 2012, 45, 279–291. [Google Scholar] [CrossRef]
- Kim, H.J.; Szurgot, M.R.; van Eeuwen, T.; Ricketts, M.D.; Basnet, P.; Zhang, A.L.; Vogt, A.; Sharmin, S.; Kaplan, C.D.; Garcia, B.A.; et al. Structure of the Hir Histone Chaperone Complex. Mol. Cell 2024, 84, 2601–2617.e12. [Google Scholar] [CrossRef] [PubMed]
- Korber, P.; Barbaric, S. The Yeast PHO5 Promoter: From Single Locus to Systems Biology of a Paradigm for Gene Regulation through Chromatin. Nucleic Acids Res. 2014, 42, 10888–10902. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.; Hall, M.N. TOR Signalling and Control of Cell Growth. Curr. Opin. Cell Biol. 1997, 9, 782–787. [Google Scholar] [CrossRef] [PubMed]
- de Nadal, E.; Posas, F. The HOG Pathway and the Regulation of Osmoadaptive Responses in Yeast. FEMS Yeast Res. 2022, 22, foac013. [Google Scholar] [CrossRef]
- Wolf, I.R.; Marques, L.F.; de Almeida, L.F.; Lázari, L.C.; de Moraes, L.N.; Cardoso, L.H.; de Alves, C.C.O.; Nakajima, R.T.; Schnepper, A.P.; de Golim, M.A.; et al. Integrative Analysis of the Ethanol Tolerance of Saccharomyces cerevisiae. Int. J. Mol. Sci. 2023, 24, 5646. [Google Scholar] [CrossRef]
- Schnepper, A.P.; Marques, L.F.; Wolf, I.R.; Kubo, A.M.S.; Valente, G.T. Potential Global Cis and Trans Regulation of lncRNAs in Saccharomyces cerevisiae Subjected to Ethanol Stress. Gene 2024, 920, 148521. [Google Scholar] [CrossRef]
- Schnepper, A.P.; Kubo, A.M.S.; Pinto, C.M.; Gomes, R.H.M.; Fioretto, M.N.; Justulin, L.A.; Braz, A.M.M.; de Golim, M.A.; Grotto, R.M.T.; Valente, G.T. Long Noncoding RNAs Responding to Ethanol Stress in Yeast Seem Associated with Protein Synthesis and Membrane Integrity. Genes 2025, 16, 170. [Google Scholar] [CrossRef]
- Horsey, E.W.; Jakovljevic, J.; Miles, T.D.; Harnpicharnchai, P.; Woolford, J.L. Role of the Yeast Rrp1 Protein in the Dynamics of Pre-Ribosome Maturation. RNA 2004, 10, 813–827. [Google Scholar] [CrossRef]
- Halme, A.; Bumgarner, S.; Styles, C.; Fink, G.R. Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast. Cell 2004, 116, 405–415. [Google Scholar] [CrossRef]
- Lee, C.-S.; Haber, J.E. Mating-Type Gene Switching in Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, MDNA3-0013-2014. [Google Scholar] [CrossRef]
- Yarrington, R.M.; Rudd, J.S.; Stillman, D.J. Spatiotemporal Cascade of Transcription Factor Binding Required for Promoter Activation. Mol. Cell. Biol. 2015, 35, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Bleykasten-Grosshans, C.; Friedrich, A.; Schacherer, J. Genome-Wide Analysis of Intraspecific Transposon Diversity in Yeast. BMC Genom. 2013, 14, 399. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Transcribing Centromeres: Noncoding RNAs and Kinetochore Assembly. Trends Genet. TIG 2018, 34, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Corless, S.; Höcker, S.; Erhardt, S. Centromeric RNA and Its Function at and beyond Centromeric Chromatin. J. Mol. Biol. 2020, 432, 4257–4269. [Google Scholar] [CrossRef]
- Ling, Y.H.; Yuen, K.W.Y. Centromeric Non-Coding RNA as a Hidden Epigenetic Factor of the Point Centromere. Curr. Genet. 2019, 65, 1165–1171. [Google Scholar] [CrossRef]
- Wu, R.A.; Upton, H.E.; Vogan, J.M.; Collins, K. Telomerase Mechanism of Telomere Synthesis. Annu. Rev. Biochem. 2017, 86, 439–460. [Google Scholar] [CrossRef]
- Leonardi, J.; Box, J.A.; Bunch, J.T.; Baumann, P. TER1, the RNA Subunit of Fission Yeast Telomerase. Nat. Struct. Mol. Biol. 2008, 15, 26–33. [Google Scholar] [CrossRef]
- McMurdie, K.; Peeney, A.N.; Mefford, M.A.; Baumann, P.; Zappulla, D.C. S. Pombe Telomerase RNA: Secondary Structure and Flexible-Scaffold Function. BioRxiv Prepr. Serv. Biol. 2025; preprint. [Google Scholar] [CrossRef]
- Zeinoun, B.; Teixeira, M.T.; Barascu, A. TERRA and Telomere Maintenance in the Yeast Saccharomyces cerevisiae. Genes 2023, 14, 618. [Google Scholar] [CrossRef] [PubMed]
- Cusanelli, E.; Chartrand, P. Telomeric Repeat-Containing RNA TERRA: A Noncoding RNA Connecting Telomere Biology to Genome Integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Miyoshi, T.; Kugou, K.; Hoffman, C.S.; Shibata, T.; Ohta, K. Stepwise Chromatin Remodelling by a Cascade of Transcription Initiation of Non-Coding RNAs. Nature 2008, 456, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Ohta, K. Cascade Transcription of mRNA-Type Long Non-Coding RNAs (mlonRNAs) and Local Chromatin Remodeling. Epigenetics 2009, 4, 5–7. [Google Scholar] [CrossRef][Green Version]
- Shah, S.; Wittmann, S.; Kilchert, C.; Vasiljeva, L. lncRNA Recruits RNAi and the Exosome to Dynamically Regulate Pho1 Expression in Response to Phosphate Levels in Fission Yeast. Genes Dev. 2014, 28, 231–244. [Google Scholar] [CrossRef]
- Garg, A.; Sanchez, A.M.; Shuman, S.; Schwer, B. A Long Noncoding (Lnc)RNA Governs Expression of the Phosphate Transporter Pho84 in Fission Yeast and Has Cascading Effects on the Flanking Prt lncRNA and Pho1 Genes. J. Biol. Chem. 2018, 293, 4456–4467. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Shuman, S.; Schwer, B. Poly(A) Site Choice and Pol2 CTD Serine-5 Status Govern lncRNA Control of Phosphate-Responsive Tgp1 Gene Expression in Fission Yeast. RNA 2018, 24, 237–250. [Google Scholar] [CrossRef]
- Leong, H.S.; Dawson, K.; Wirth, C.; Li, Y.; Connolly, Y.; Smith, D.L.; Wilkinson, C.R.M.; Miller, C.J. A Global Non-Coding RNA System Modulates Fission Yeast Protein Levels in Response to Stress. Nat. Commun. 2014, 5, 3947. [Google Scholar] [CrossRef]
- Ono, Y.; Katayama, K.; Onuma, T.; Kubo, K.; Tsuyuzaki, H.; Hamada, M.; Sato, M. Structure-Based Screening for Functional Non-Coding RNAs in Fission Yeast Identifies a Factor Repressing Untimely Initiation of Sexual Differentiation. Nucleic Acids Res. 2022, 50, 11229–11242. [Google Scholar] [CrossRef]
- Fauquenoy, S.; Migeot, V.; Finet, O.; Yague-Sanz, C.; Khorosjutina, O.; Ekwall, K.; Hermand, D. Repression of Cell Differentiation by a Cis-Acting lincRNA in Fission Yeast. Curr. Biol. CB 2018, 28, 383–391.e3. [Google Scholar] [CrossRef]
- Andric, V.; Nevers, A.; Hazra, D.; Auxilien, S.; Menant, A.; Graille, M.; Palancade, B.; Rougemaille, M. A Scaffold lncRNA Shapes the Mitosis to Meiosis Switch. Nat. Commun. 2021, 12, 770. [Google Scholar] [CrossRef]
- Touat-Todeschini, L.; Shichino, Y.; Dangin, M.; Thierry-Mieg, N.; Gilquin, B.; Hiriart, E.; Sachidanandam, R.; Lambert, E.; Brettschneider, J.; Reuter, M.; et al. Selective Termination of lncRNA Transcription Promotes Heterochromatin Silencing and Cell Differentiation. EMBO J. 2017, 36, 2626–2641. [Google Scholar] [CrossRef]
- Anver, S.; Sumit, A.F.; Sun, X.-M.; Hatimy, A.; Thalassinos, K.; Marguerat, S.; Alic, N.; Bähler, J. Ageing-Associated Long Non-Coding RNA Extends Lifespan and Reduces Translation in Non-Dividing Cells. EMBO Rep. 2024, 25, 4921–4949. [Google Scholar] [CrossRef]
- Asada, R.; Hirota, K. Multi-Layered Regulations on the Chromatin Architectures: Establishing the Tight and Specific Responses of Fission Yeast Fbp1 Gene Transcription. Biomolecules 2022, 12, 1642. [Google Scholar] [CrossRef]
- Adachi, A.; Senmatsu, S.; Asada, R.; Abe, T.; Hoffman, C.S.; Ohta, K.; Hirota, K. Interplay between Chromatin Modulators and Histone Acetylation Regulates the Formation of Accessible Chromatin in the Upstream Regulatory Region of Fission Yeast Fbp1. Genes Genet. Syst. 2018, 92, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, Y.; Senmatsu, S.; Oe, H.; Hoffman, C.S.; Hirota, K. Metabolic Stress-Induced Long ncRNA Transcription Governs the Formation of Meiotic DNA Breaks in the Fission Yeast Fbp1 Gene. PLoS ONE 2024, 19, e0294191. [Google Scholar] [CrossRef] [PubMed]
- Henry, T.C.; Power, J.E.; Kerwin, C.L.; Mohammed, A.; Weissman, J.S.; Cameron, D.M.; Wykoff, D.D. Systematic Screen of Schizosaccharomyces pombe Deletion Collection Uncovers Parallel Evolution of the Phosphate Signal Transduction Pathway in Yeasts. Eukaryot. Cell 2011, 10, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Sanchez, A.M.; Goldgur, Y.; Shuman, S.; Schwer, B. Transcription of lncRNA Prt, Clustered Prt RNA Sites for Mmi1 Binding, and RNA Polymerase II CTD Phospho-Sites Govern the Repression of Pho1 Gene Expression under Phosphate-Replete Conditions in Fission Yeast. RNA 2016, 22, 1011–1025. [Google Scholar] [CrossRef]
- Schwer, B.; Sanchez, A.M.; Shuman, S. Inactivation of Fission Yeast Erh1 De-Represses Pho1 Expression: Evidence That Erh1 Is a Negative Regulator of Prt lncRNA Termination. RNA 2020, 26, 1334–1344. [Google Scholar] [CrossRef]
- Ard, R.; Tong, P.; Allshire, R.C. Long Non-Coding RNA-Mediated Transcriptional Interference of a Permease Gene Confers Drug Tolerance in Fission Yeast. Nat. Commun. 2014, 5, 5576. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Spector, D.L. Long Non-Coding RNAs: Modulators of Nuclear Structure and Function. Curr. Opin. Cell Biol. 2014, 26, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Li, P.; Yu, H.; Chen, L.-L. Emerging Roles of Nuclear Bodies in Genome Spatial Organization. Trends Cell Biol. 2024, 34, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Yamamoto, M.S. Pombe Mei2+ Encodes an RNA-Binding Protein Essential for Premeiotic DNA Synthesis and Meiosis I, Which Cooperates with a Novel RNA Species meiRNA. Cell 1994, 78, 487–498. [Google Scholar] [CrossRef]
- Watanabe, Y.; Shinozaki-Yabana, S.; Chikashige, Y.; Hiraoka, Y.; Yamamoto, M. Phosphorylation of RNA-Binding Protein Controls Cell Cycle Switch from Mitotic to Meiotic in Fission Yeast. Nature 1997, 386, 187–190. [Google Scholar] [CrossRef]
- Shichino, Y.; Yamashita, A.; Yamamoto, M. Meiotic Long Non-Coding meiRNA Accumulates as a Dot at Its Genetic Locus Facilitated by Mmi1 and Plays as a Decoy to Lure Mmi1. Open Biol. 2014, 4, 140022. [Google Scholar] [CrossRef]
- Kawamukai, M. Regulation of Sexual Differentiation Initiation in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 2024, 88, 475–492. [Google Scholar] [CrossRef]
- Thillainadesan, G.; Xiao, H.; Holla, S.; Dhakshnamoorthy, J.; Jenkins, L.M.M.; Wheeler, D.; Grewal, S.I.S. Conserved Protein Pir2ARS2 Mediates Gene Repression through Cryptic Introns in lncRNAs. Nat. Commun. 2020, 11, 2412. [Google Scholar] [CrossRef]
- Ohtsuka, H.; Shimasaki, T.; Aiba, H. Genes Affecting the Extension of Chronological Lifespan in Schizosaccharomyces pombe (Fission Yeast). Mol. Microbiol. 2021, 115, 623–642. [Google Scholar] [CrossRef]
- Logeswaran, D.; Li, Y.; Akhter, K.; Podlevsky, J.D.; Olson, T.L.; Forsberg, K.; Chen, J.J.-L. Biogenesis of Telomerase RNA from a Protein-Coding mRNA Precursor. Proc. Natl. Acad. Sci. USA 2022, 119, e2204636119. [Google Scholar] [CrossRef]
- Gao, J.; Chow, E.W.L.; Wang, H.; Xu, X.; Cai, C.; Song, Y.; Wang, J.; Wang, Y. LncRNA DINOR Is a Virulence Factor and Global Regulator of Stress Responses in Candida auris. Nat. Microbiol. 2021, 6, 842–851. [Google Scholar] [CrossRef]
- Chacko, N.; Zhao, Y.; Yang, E.; Wang, L.; Cai, J.J.; Lin, X. The lncRNA RZE1 Controls Cryptococcal Morphological Transition. PLoS Genet. 2015, 11, e1005692. [Google Scholar] [CrossRef] [PubMed]
- Mueller, O.; Kahmann, R.; Aguilar, G.; Trejo-Aguilar, B.; Wu, A.; de Vries, R.P. The Secretome of the Maize Pathogen Ustilago maydis. Fungal Genet. Biol. FG B 2008, 45 (Suppl. S1), S63–S70. [Google Scholar] [CrossRef]
- Donaldson, M.E.; Saville, B.J. Ustilago maydis Natural Antisense Transcript Expression Alters mRNA Stability and Pathogenesis. Mol. Microbiol. 2013, 89, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M. Virulence Factors in Candida Species. Curr. Protein Pept. Sci. 2020, 21, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Jackson, J.C.; Feretzaki, M.; Xue, C.; Heitman, J. Transcription Factors Mat2 and Znf2 Operate Cellular Circuits Orchestrating Opposite- and Same-Sex Mating in Cryptococcus neoformans. PLoS Genet. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Xue, Z.; Ye, Q.; Anson, S.R.; Yang, J.; Xiao, G.; Kowbel, D.; Glass, N.L.; Crosthwaite, S.K.; Liu, Y. Transcriptional Interference by Antisense RNA Is Required for Circadian Clock Function. Nature 2014, 514, 650–653. [Google Scholar] [CrossRef]
- Kramer, C.; Loros, J.J.; Dunlap, J.C.; Crosthwaite, S.K. Role for Antisense RNA in Regulating Circadian Clock Function in Neurospora crassa. Nature 2003, 421, 948–952. [Google Scholar] [CrossRef]
- Li, N.; Joska, T.M.; Ruesch, C.E.; Coster, S.J.; Belden, W.J. The Frequency Natural Antisense Transcript First Promotes, Then Represses, Frequency Gene Expression via Facultative Heterochromatin. Proc. Natl. Acad. Sci. USA 2015, 112, 4357–4362. [Google Scholar] [CrossRef]
- Till, P.; Pucher, M.E.; Mach, R.L.; Mach-Aigner, A.R. A Long Noncoding RNA Promotes Cellulase Expression in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 78. [Google Scholar] [CrossRef]
- Parra-Rivero, O.; Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. A Novel lncRNA as a Positive Regulator of Carotenoid Biosynthesis in Fusarium. Sci. Rep. 2020, 10, 678. [Google Scholar] [CrossRef]
- Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. The carP lncRNA Is a carS-Related Regulatory Element with Broad Effects on the Fusarium fujikuroi Transcriptome. Non-Coding RNA 2021, 7, 46. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, W.; Xie, J.; Fu, Y.; Jiang, D.; Lin, Y.; Chen, W.; Cheng, J. A Novel Antisense Long Non-coding RNA Participates in Asexual and Sexual Reproduction by Regulating the Expression of GzmetE in Fusarium graminearum. Environ. Microbiol. 2021, 23, 4939–4955. [Google Scholar] [CrossRef]
- Huang, P.; Yu, X.; Liu, H.; Ding, M.; Wang, Z.; Xu, J.-R.; Jiang, C. Regulation of TRI5 Expression and Deoxynivalenol Biosynthesis by a Long Non-Coding RNA in Fusarium graminearum. Nat. Commun. 2024, 15, 1216. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, W.; Cheng, J.; Xie, J.; Fu, Y.; Jiang, D.; Lin, Y. lncRsp1, a Long Noncoding RNA, Influences Fgsp1 Expression and Sexual Reproduction in Fusarium graminearum. Mol. Plant Pathol. 2022, 23, 265–277. [Google Scholar] [CrossRef]
- Lu, P.; Chen, D.; Qi, Z.; Wang, H.; Chen, Y.; Wang, Q.; Jiang, C.; Xu, J.-R.; Liu, H. Landscape and Regulation of Alternative Splicing and Alternative Polyadenylation in a Plant Pathogenic Fungus. New Phytol. 2022, 235, 674–689. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Chen, Y.; Cai, G.; Peng, H.; Wang, X.; Li, P.; Gu, A.; Li, Y.; Ma, D. An Antisense Long Lon-Coding RNA, lncRsn, Is Involved in Sexual Reproduction and Full Virulence in Fusarium graminearum. J. Fungi Basel Switz. 2024, 10, 692. [Google Scholar] [CrossRef]
- Eliahu, N.; Igbaria, A.; Rose, M.S.; Horwitz, B.A.; Lev, S. Melanin Biosynthesis in the Maize Pathogen Cochliobolus heterostrophus Depends on Two Mitogen-Activated Protein Kinases, Chk1 and Mps1, and the Transcription Factor Cmr1. Eukaryot. Cell 2007, 6, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Avrova, A.O.; Whisson, S.C.; Pritchard, L.; Venter, E.; De Luca, S.; Hein, I.; Birch, P.R.J. A Novel Non-Protein-Coding Infection-Specific Gene Family Is Clustered throughout the Genome of Phytophthora infestans. Microbiol. Read. Engl. 2007, 153, 747–759. [Google Scholar] [CrossRef]
- Kalita, B.; Roy, A.; Jayaprakash, A.; Arunachalam, A.; Lakshmi, P.T.V. Identification of lncRNA and Weighted Gene Coexpression Network Analysis of Germinating Rhizopus delemar Causing Mucormycosis. Mycology 2023, 14, 344–357. [Google Scholar] [CrossRef]
- Baker, C.L.; Loros, J.J.; Dunlap, J.C. The Circadian Clock of Neurospora crassa. FEMS Microbiol. Rev. 2012, 36, 95–110. [Google Scholar] [CrossRef]
- Cemel, I.A.; Diernfellner, A.C.R.; Brunner, M. Antisense Transcription of the Neurospora Frequency Gene Is Rhythmically Regulated by CSP-1 Repressor but Dispensable for Clock Function. J. Biol. Rhythms 2023, 38, 259–268. [Google Scholar] [CrossRef]
- Avalos, J.; Pardo-Medina, J.; Parra-Rivero, O.; Ruger-Herreros, M.; Rodríguez-Ortiz, R.; Hornero-Méndez, D.; Limón, M.C. Carotenoid Biosynthesis in Fusarium. J. Fungi 2017, 3, 39. [Google Scholar] [CrossRef]
- Ruger-Herreros, M.; Parra-Rivero, O.; Pardo-Medina, J.; Romero-Campero, F.J.; Limón, M.C.; Avalos, J. Comparative Transcriptomic Analysis Unveils Interactions between the Regulatory CarS Protein and Light Response in Fusarium. BMC Genomics 2019, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Marente, J.; Avalos, J.; Limón, M.C. Controlled Transcription of Regulator Gene carS by Tet-on or by a Strong Promoter Confirms Its Role as a Repressor of Carotenoid Biosynthesis in Fusarium fujikuroi. Microorganisms 2020, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.A.; Ukladov, E.O.; Golubeva, T.S. Phytophthora infestans: An Overview of Methods and Attempts to Combat Late Blight. J. Fungi Basel Switz. 2021, 7, 1071. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lopez, M.; Anver, S.; Cotobal, C.; Kamrad, S.; Malecki, M.; Correia-Melo, C.; Hoti, M.; Townsend, S.; Marguerat, S.; Pong, S.K.; et al. Functional Profiling of Long Intergenic Non-Coding RNAs in Fission Yeast. eLife 2022, 11, e76000. [Google Scholar] [CrossRef]
- Garg, A. A lncRNA-Regulated Gene Expression System with Rapid Induction Kinetics in the Fission Yeast Schizosaccharomyces pombe. RNA 2020, 26, 1743–1752. [Google Scholar] [CrossRef]
- Cao, M.; Zhao, J.; Hu, G. Genome-Wide Methods for Investigating Long Noncoding RNAs. Biomed. Pharmacother. 2019, 111, 395–401. [Google Scholar] [CrossRef]
- Andjus, S.; Szachnowski, U.; Vogt, N.; Gioftsidi, S.; Hatin, I.; Cornu, D.; Papadopoulos, C.; Lopes, A.; Namy, O.; Wery, M.; et al. Pervasive Translation of Xrn1-Sensitive Unstable Long Noncoding RNAs in Yeast. RNA 2024, 30, 662–679. [Google Scholar] [CrossRef]
- Smith, J.E.; Alvarez-Dominguez, J.R.; Kline, N.; Huynh, N.J.; Geisler, S.; Hu, W.; Coller, J.; Baker, K.E. Translation of Small Open Reading Frames within Unannotated RNA Transcripts in Saccharomyces cerevisiae. Cell Rep. 2014, 7, 1858–1866. [Google Scholar] [CrossRef]
- Papadopoulos, C.; Arbes, H.; Cornu, D.; Chevrollier, N.; Blanchet, S.; Roginski, P.; Rabier, C.; Atia, S.; Lespinet, O.; Namy, O.; et al. The Ribosome Profiling Landscape of Yeast Reveals a High Diversity in Pervasive Translation. Genome Biol. 2024, 25, 268. [Google Scholar] [CrossRef]
- Blevins, W.R.; Ruiz-Orera, J.; Messeguer, X.; Blasco-Moreno, B.; Villanueva-Cañas, J.L.; Espinar, L.; Díez, J.; Carey, L.B.; Albà, M.M. Uncovering de Novo Gene Birth in Yeast Using Deep Transcriptomics. Nat. Commun. 2021, 12, 604. [Google Scholar] [CrossRef]
- Papadopoulos, C.; Callebaut, I.; Gelly, J.-C.; Hatin, I.; Namy, O.; Renard, M.; Lespinet, O.; Lopes, A. Intergenic ORFs as Elementary Structural Modules of de Novo Gene Birth and Protein Evolution. Genome Res. 2021, 31, 2303–2315. [Google Scholar] [CrossRef]
- Wacholder, A.; Parikh, S.B.; Coelho, N.C.; Acar, O.; Houghton, C.; Chou, L.; Carvunis, A.-R. A Vast Evolutionarily Transient Translatome Contributes to Phenotype and Fitness. Cell Syst. 2023, 14, 363–381.e8. [Google Scholar] [CrossRef]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal Evolution: Cellular, Genomic and Metabolic Complexity. Biol. Rev. Camb. Philos. Soc. 2020, 95, 1198–1232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).