Partners in Silencing: Decoding the Mammalian Argonaute Interactome
Abstract
1. Introduction
2. The Argonaute Family of Proteins
2.1. Sumoylation of AGO
2.2. Phosphorylation of AGO
2.3. Ubiquitination of AGO
2.4. Acetylation of AGO
2.5. Poly-ADP-Ribosylation of AGO
2.6. Hydroxylation of AGO
| miRISC Interactors | Interacting AGO | Functionality | Remarks | References |
|---|---|---|---|---|
| Sumoylation of AGO | ||||
| RANBP2 | AGO 1-2 | Associates with E2 enzyme UBC9 and induces the sumoylation of AGO1/2 | - | [62] |
| PIAS3 | AGO2 | E3 SUMO protein ligase that catalyzes the final steps of the sumoylation of K402-AGO2 | - | [64] |
| Phosphorylation of AGO | ||||
| CSNK1A1 | AGO 1-3 | Phosphorylation of the EI region of AGO proteins | - | [66,69] |
| ANKRD52 | AGO 2 | Dephosphorylation of EI region of AGO2 | - | [66] |
| TBK1 | AGO2 | Phosphorylates S417-AGO2 | Promotes the formation of pS417-AGO2-miR-21-RISC in NSCLC | [70] |
| EGFR | AGO2 | Phosphorylates Y103-AGO2 | During hypoxia, decreased pY103-AGO2 and DICER interaction | [76] |
| C-Src | AGO2 | Phosphorylates Y393/529/749-AGO2 | pY393 sustains interaction with DICER | [77] |
| PTP1B | AGO2 | De-phosphorylates Y393-AGO2 | - | [78] |
| E-cadherins | AGO2 | Mediates pAGO2 by ER-kinase signaling | - | [79] |
| Ubiquitination of AGO | ||||
| STUB1 | AGO 1-4 | Adds polyubiquitin chains to K48-AGO | - | [81] |
| CRL-ZSWIM8 | AGO2 | Presumably ubiquitinates K493-AGO2 | ZSWIM8 mediates TDMD in a tailing and trimming independent manner | [85] |
| LUBAC system | AGO2 | Couples Met1 linear poly-ubiquitin chain to K820-AGO2 | This mechanism occurs mainly during hypoxia | [87] |
| OTULIN | AGO2 | De-linear Deubiquitinating enzyme of AGO2 | - | [87] |
| Acetylation of AGO | ||||
| CBP | AGO2 | Acetylates K720/K493/K355-AGO2 | The acetylation of K493/K720-AGO2 promotes the biogenesis of pre-miR-19b | [88] |
| HDAC7 | AGO2 | De-acetylate AGO2 | - | [88] |
| Poly ADP-Ribosylation | ||||
| PARP | AGO 1-4 | Catalyzes Poly (ADP-ribose) polymerization of AGO proteins | Mainly present in stress granules | [89] |
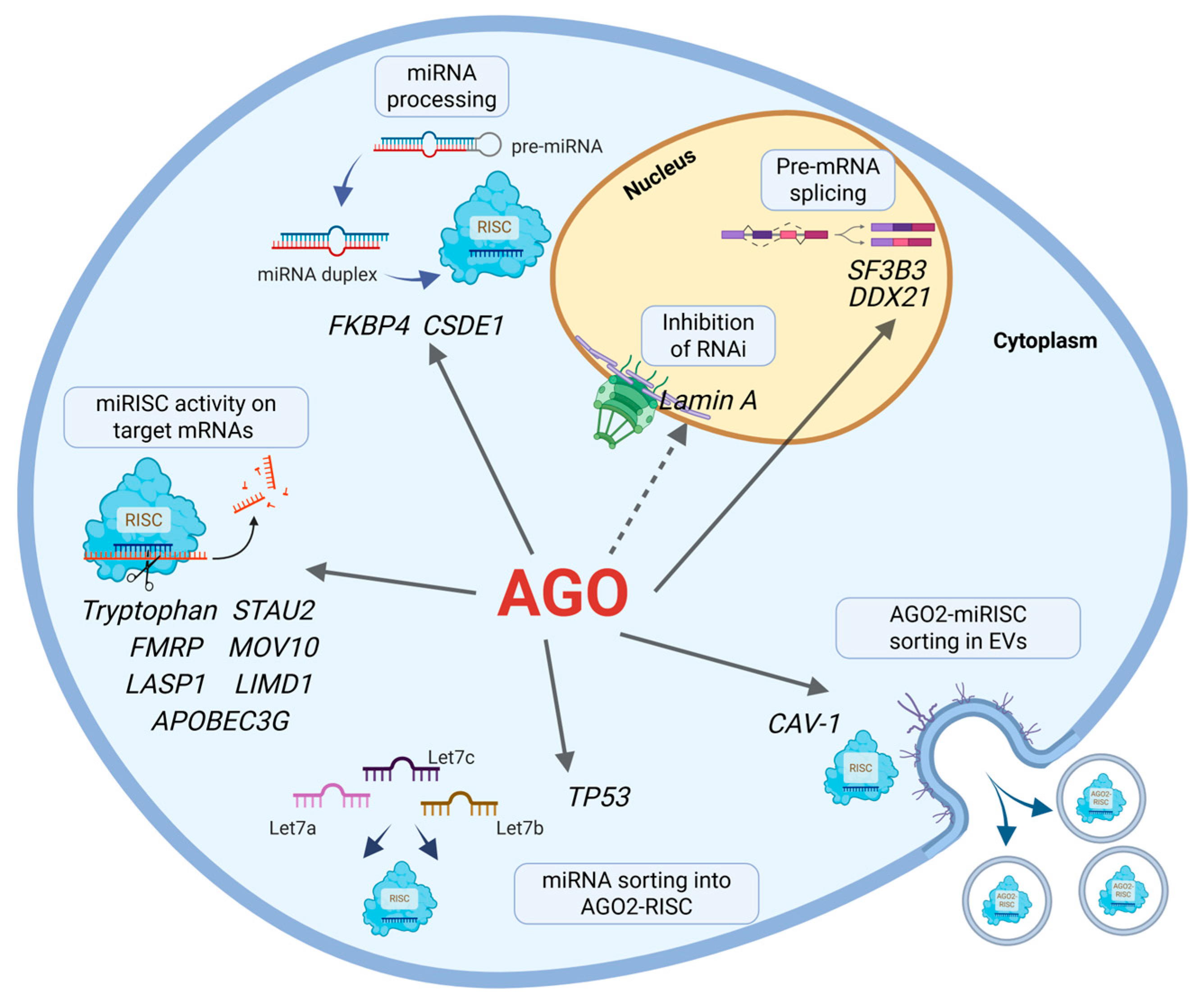
3. Unexpected Functions of miRISC
3.1. Interactors of AGO Proteins That Modulate miRISC Activities
3.2. Splicing-Associated Factors
3.3. mRNA Translation Associated Factors
3.4. RISC Associated Proteins That Control Localization and miRNA Sorting
3.5. Miscellaneous Factors Regulating RISC Activities
4. Conclusions
Funding
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–845. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Robert Horvitz, H.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs—genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, S.; Deng, H.; Zhang, S.; Wang, J.; Song, X.; Yu, D.; Zhang, Y.; Deng, W. STAT3 promotes RNA polymerase III-directed transcription by controlling the miR-106a-5p/TP73 axis. Elife 2023, 12, e82826. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Li, S.C.; Tang, P.; Lin, W.C. Intronic microRNA: Discovery and biological implications. DNA Cell Biol. 2007, 26, 195–207. [Google Scholar] [CrossRef]
- Marsico, A.; Huska, M.R.; Lasserre, J.; Hu, H.; Vucicevic, D.; Musahl, A.; Orom, U.A.; Vingron, M. PROmiRNA a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 2013, 14, R84. [Google Scholar] [CrossRef]
- Monteys, A.M.; Spengler, R.M.; Wan, J.; Tecedor, L.; Lennox, K.A.; Xing, Y.; Davidson, B.L. Structure and activity of putative intronic miRNA promoters. RNA 2010, 16, 495–505. [Google Scholar] [CrossRef]
- Stavast, C.J.; van Zuijen, I.; Karkoulia, E.; Ozcelik, A.; van Hoven-Beijen, A.; Leon, L.G.; Voerman, J.S.A.; Janssen, G.M.C.; van Veelen, P.A.; Burocziova, M.; et al. The tumor suppressor MIR139 is silenced by POLR2M to promote AML oncogenesis. Leukemia 2022, 36, 687–700. [Google Scholar] [CrossRef]
- Michlewski, G.; Caceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef]
- Yoshida, T.; Asano, Y.; Ui-Tei, K. Modulation of MicroRNA Processing by Dicer via Its Associated dsRNA Binding Proteins. Noncoding RNA 2021, 7, 57. [Google Scholar] [CrossRef]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Tsuboyama, K.; Tadakuma, H.; Tomari, Y. Conformational Activation of Argonaute by Distinct yet Coordinated Actions of the Hsp70 and Hsp90 Chaperone Systems. Mol. Cell 2018, 70, 722–729.e4. [Google Scholar] [CrossRef]
- Pare, J.M.; LaPointe, P.; Hobman, T.C. Hsp90 cochaperones p23 and FKBP4 physically interact with hAgo2 and activate RNA interference-mediated silencing in mammalian cells. Mol. Biol. Cell 2013, 24, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Huang, N.; Liu, Y.; Paroo, Z.; Huerta, C.; Li, P.; Chen, S.; Liu, Q.; Zhang, H. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 2011, 18, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Zamore, P.D. Why do miRNAs live in the miRNP? Genes. Dev. 2002, 16, 1025–1031. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Cheng, Z.; Zhu, Q. AGO2 and its partners: A silencing complex, a chromatin modulator, and new features. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 33–53. [Google Scholar] [CrossRef]
- Fabian, M.R.; Mathonnet, G.; Sundermeier, T.; Mathys, H.; Zipprich, J.T.; Svitkin, Y.V.; Rivas, F.; Jinek, M.; Wohlschlegel, J.; Doudna, J.A.; et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 2009, 35, 868–880. [Google Scholar] [CrossRef]
- Wahle, E.; Winkler, G.S. RNA decay machines: Deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 2013, 1829, 561–570. [Google Scholar] [CrossRef]
- Chen, C.A.; Zhang, Y.; Xiang, Y.; Han, L.; Chang, J.T.; Shyu, A.B. Antagonistic actions of two human Pan3 isoforms on global mRNA turnover. RNA 2017, 23, 1404–1418. [Google Scholar] [CrossRef]
- Zhang, X.; Devany, E.; Murphy, M.R.; Glazman, G.; Persaud, M.; Kleiman, F.E. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic Acids Res. 2015, 43, 10925–10938. [Google Scholar] [CrossRef]
- Rouya, C.; Siddiqui, N.; Morita, M.; Duchaine, T.F.; Fabian, M.R.; Sonenberg, N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 2014, 20, 1398–1409. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.T.; Muthukumar, S.; Weber, R.; Levdansky, Y.; Chen, Y.; Bhandari, D.; Igreja, C.; Wohlbold, L.; Valkov, E.; Izaurralde, E. A low-complexity region in human XRN1 directly recruits deadenylation and decapping factors in 5′-3′ messenger RNA decay. Nucleic Acids Res. 2019, 47, 9282–9295. [Google Scholar] [CrossRef]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biochim. Biophys. Acta 2013, 1829, 552–560. [Google Scholar] [CrossRef]
- Leitao, A.L.; Enguita, F.J. A Structural View of miRNA Biogenesis and Function. Noncoding RNA 2022, 8, 10. [Google Scholar] [CrossRef]
- Paturi, S.; Deshmukh, M.V. A Glimpse of “Dicer Biology” Through the Structural and Functional Perspective. Front. Mol. Biosci. 2021, 8, 643657. [Google Scholar] [CrossRef]
- Kuzuoglu-Ozturk, D.; Bhandari, D.; Huntzinger, E.; Fauser, M.; Helms, S.; Izaurralde, E. miRISC and the CCR4-NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J. 2016, 35, 1186–1203. [Google Scholar] [CrossRef]
- Wu, P.H.; Isaji, M.; Carthew, R.W. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell 2013, 52, 113–123. [Google Scholar] [CrossRef]
- Inada, T.; Makino, S. Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front. Genet. 2014, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Jungers, C.F.; Djuranovic, S. Modulation of miRISC-Mediated Gene Silencing in Eukaryotes. Front. Mol. Biosci. 2022, 9, 832916. [Google Scholar] [CrossRef]
- Akgul, B.; Erdogan, I. Intracytoplasmic Re-localization of miRISC Complexes. Front. Genet. 2018, 9, 403. [Google Scholar] [CrossRef]
- Yoda, M.; Cifuentes, D.; Izumi, N.; Sakaguchi, Y.; Suzuki, T.; Giraldez, A.J.; Tomari, Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013, 5, 715–726. [Google Scholar] [CrossRef]
- Robb, G.B.; Rana, T.M. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell 2007, 26, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Brothers, W.R.; Ali, F.; Kajjo, S.; Fabian, M.R. The EDC4-XRN1 interaction controls P-body dynamics to link mRNA decapping with decay. EMBO J. 2023, 42, e113933. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Winter, J.; Diederichs, S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011, 8, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Kingston, E.R.; Bartel, D.P. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 2019, 29, 1777–1790. [Google Scholar] [CrossRef]
- Smibert, P.; Yang, J.S.; Azzam, G.; Liu, J.L.; Lai, E.C. Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 2013, 20, 789–795. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef]
- Kakumani, P.K.; Ko, Y.; Ramakrishna, S.; Christopher, G.; Dodgson, M.; Shrinet, J.; Harvey, L.M.; Shin, C.; Simard, M.J. CSDE1 promotes miR-451 biogenesis. Nucleic Acids Res. 2023, 51, 9385–9396. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Simmini, S.; Abreu-Goodger, C.; Bartonicek, N.; Di Giacomo, M.; Bilbao-Cortes, D.; Horos, R.; Von Lindern, M.; Enright, A.J.; O’Carroll, D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010, 207, 1351–1358. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef]
- Turchinovich, A.; Burwinkel, B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012, 9, 1066–1075. [Google Scholar] [CrossRef]
- Morita, S.; Horii, T.; Kimura, M.; Goto, Y.; Ochiya, T.; Hatada, I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 2007, 89, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Hajarnis, S.; Yheskel, M.; Williams, D.; Brefort, T.; Glaudemans, B.; Debaix, H.; Baum, M.; Devuyst, O.; Patel, V. Suppression of microRNA Activity in Kidney Collecting Ducts Induces Partial Loss of Epithelial Phenotype and Renal Fibrosis. J. Am. Soc. Nephrol. 2018, 29, 518–531. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Park, M.S.; Sim, G.; Kehling, A.C.; Nakanishi, K. Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 28576–28578. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K. Are Argonaute-Associated Tiny RNAs Junk, Inferior miRNAs, or a New Type of Functional RNAs? Front. Mol. Biosci. 2021, 8, 795356. [Google Scholar] [CrossRef]
- Nakanishi, K. Anatomy of four human Argonaute proteins. Nucleic Acids Res. 2022, 50, 6618–6638. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, Y.E.; Truong, M.; Mahadevan, K.; Wu, J.J.; Zhang, H.; Li, J.; Smith, H.W.; Smibert, C.A.; Palazzo, A.F. RanBP2/Nup358 enhances miRNA activity by sumoylating Argonautes. PLoS Genet. 2021, 17, e1009378. [Google Scholar] [CrossRef]
- Sahin, U.; Lapaquette, P.; Andrieux, A.; Faure, G.; Dejean, A. Sumoylation of human argonaute 2 at lysine-402 regulates its stability. PLoS ONE 2014, 9, e102957. [Google Scholar] [CrossRef]
- Josa-Prado, F.; Henley, J.M.; Wilkinson, K.A. SUMOylation of Argonaute-2 regulates RNA interference activity. Biochem. Biophys. Res. Commun. 2015, 464, 1066–1071. [Google Scholar] [CrossRef]
- Sahoo, M.R.; Gaikwad, S.; Khuperkar, D.; Ashok, M.; Helen, M.; Yadav, S.K.; Singh, A.; Magre, I.; Deshmukh, P.; Dhanvijay, S.; et al. Nup358 binds to AGO proteins through its SUMO-interacting motifs and promotes the association of target mRNA with miRISC. EMBO Rep. 2017, 18, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Golden, R.J.; Chen, B.; Li, T.; Braun, J.; Manjunath, H.; Chen, X.; Wu, J.; Schmid, V.; Chang, T.C.; Kopp, F.; et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 2017, 542, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Quevillon Huberdeau, M.; Zeitler, D.M.; Hauptmann, J.; Bruckmann, A.; Fressigne, L.; Danner, J.; Piquet, S.; Strieder, N.; Engelmann, J.C.; Jannot, G.; et al. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017, 36, 2088–2106. [Google Scholar] [CrossRef]
- Bibel, B.; Elkayam, E.; Silletti, S.; Komives, E.A.; Joshua-Tor, L. Target binding triggers hierarchical phosphorylation of human Argonaute-2 to promote target release. Elife 2022, 11, e76908. [Google Scholar] [CrossRef]
- Shah, V.N.; Neumeier, J.; Huberdeau, M.Q.; Zeitler, D.M.; Bruckmann, A.; Meister, G.; Simard, M.J. Casein kinase 1 and 2 phosphorylate Argonaute proteins to regulate miRNA-mediated gene silencing. EMBO Rep. 2023, 24, e57250. [Google Scholar] [CrossRef]
- Zhao, X.; Cao, Y.; Lu, R.; Zhou, Z.; Huang, C.; Li, L.; Huang, J.; Chen, R.; Wang, Y.; Huang, J.; et al. Phosphorylation of AGO2 by TBK1 Promotes the Formation of Oncogenic miRISC in NSCLC. Adv. Sci. 2024, 11, e2305541. [Google Scholar] [CrossRef]
- Amirfallah, A.; Knutsdottir, H.; Arason, A.; Hilmarsdottir, B.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. Hsa-miR-21-3p associates with breast cancer patient survival and targets genes in tumor suppressive pathways. PLoS ONE 2021, 16, e0260327. [Google Scholar] [CrossRef]
- Li, B.; Ren, S.; Li, X.; Wang, Y.; Garfield, D.; Zhou, S.; Chen, X.; Su, C.; Chen, M.; Kuang, P.; et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014, 83, 146–153. [Google Scholar] [CrossRef]
- Rudel, S.; Wang, Y.; Lenobel, R.; Korner, R.; Hsiao, H.H.; Urlaub, H.; Patel, D.; Meister, G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011, 39, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, A.; Bose, M.; Chakraborty, A.; Chakrabarti, S.; Bhattacharyya, S.N. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep. 2013, 14, 1008–1016. [Google Scholar] [CrossRef]
- Muller, C.; Schafer, I.; Luhmann, H.J.; White, R. Oligodendroglial Argonaute protein Ago2 associates with molecules of the Mbp mRNA localization machinery and is a downstream target of Fyn kinase. Front. Cell Neurosci. 2015, 9, 328. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.O.; Du, Y.; Wang, Y.; Chang, W.C.; Chen, C.H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, H.; Fang, J.; Yang, Z.; Chen, R.; Wang, Y.; Zhao, X.; Ge, S.; Yu, J.; Huang, J. AGO2 phosphorylation by c-Src kinase promotes tumorigenesis. Neoplasia 2020, 22, 129–141. [Google Scholar] [CrossRef]
- Yang, M.; Haase, A.D.; Huang, F.K.; Coulis, G.; Rivera, K.D.; Dickinson, B.C.; Chang, C.J.; Pappin, D.J.; Neubert, T.A.; Hannon, G.J.; et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol. Cell 2014, 55, 782–790. [Google Scholar] [CrossRef]
- Li, J.N.; Sun, H.L.; Wang, M.Y.; Chen, P.S. E-cadherin Interacts With Posttranslationally-Modified AGO2 to Enhance miRISC Activity. Front. Cell Dev. Biol. 2021, 9, 671244. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Hadian, K.; Smirnova, L.; Wulczyn, E.A.; Michel, G.; Nitsch, R.; Krappmann, D.; Wulczyn, F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009, 11, 1411–1420. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, X.; Bie, Y.; Kong, J.; Wang, A.; Qiu, Y.; Zhou, X. STUB1 regulates antiviral RNAi through inducing ubiquitination and degradation of Dicer and AGO2 in mammals. Virol. Sin. 2022, 37, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Horwich, M.D.; Hung, J.H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef] [PubMed]
- Hiers, N.M.; Li, T.; Traugot, C.M.; Xie, M. Target-directed microRNA degradation: Mechanisms, significance, and functional implications. Wiley Interdiscip. Rev. RNA 2024, 15, e1832. [Google Scholar] [CrossRef]
- Fuchs Wightman, F.; Giono, L.E.; Fededa, J.P.; de la Mata, M. Target RNAs Strike Back on MicroRNAs. Front. Genet. 2018, 9, 435. [Google Scholar] [CrossRef]
- Han, J.; LaVigne, C.A.; Jones, B.T.; Zhang, H.; Gillett, F.; Mendell, J.T. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 2020, 370, eabc9546. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.Y.; Kingston, E.R.; Kleaveland, B.; Lin, D.H.; Stubna, M.W.; Bartel, D.P. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 2020, 370, eabc9359. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Guo, Y.; Chen, R.; He, J.; Li, L.; Qiang, Z.; Yang, Q.; Liu, X.; Huang, C.; et al. Hypoxia regulates overall mRNA homeostasis by inducing Met(1)-linked linear ubiquitination of AGO2 in cancer cells. Nat. Commun. 2021, 12, 5416. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Dou, J.; Guo, Y.; He, J.; Li, L.; Liu, X.; Chen, R.; Deng, R.; Huang, J.; et al. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019, 38, 1410–1431. [Google Scholar] [CrossRef]
- Leung, A.K.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef]
- Qi, H.H.; Ongusaha, P.P.; Myllyharju, J.; Cheng, D.; Pakkanen, O.; Shi, Y.; Lee, S.W.; Peng, J.; Shi, Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 2008, 455, 421–424. [Google Scholar] [CrossRef]
- Kakoulidis, P.; Theotoki, E.I.; Pantazopoulou, V.I.; Vlachos, I.S.; Emiris, I.Z.; Stravopodis, D.J.; Anastasiadou, E. Comparative structural insights and functional analysis for the distinct unbound states of Human AGO proteins. Sci. Rep. 2025, 15, 9432. [Google Scholar] [CrossRef]
- Singh, A.; Manjunath, L.E.; Kundu, P.; Sahoo, S.; Das, A.; Suma, H.R.; Fox, P.L.; Eswarappa, S.M. Let-7a-regulated translational readthrough of mammalian AGO1 generates a microRNA pathway inhibitor. EMBO J. 2019, 38, e100727. [Google Scholar] [CrossRef]
- Kakumani, P.K.; Harvey, L.M.; Houle, F.; Guitart, T.; Gebauer, F.; Simard, M.J. CSDE1 controls gene expression through the miRNA-mediated decay machinery. Life Sci. Alliance 2020, 3, e201900632. [Google Scholar] [CrossRef]
- Elkayam, E.; Faehnle, C.R.; Morales, M.; Sun, J.; Li, H.; Joshua-Tor, L. Multivalent Recruitment of Human Argonaute by GW182. Mol. Cell 2017, 67, 646–658.e3. [Google Scholar] [CrossRef]
- Saetrom, P.; Heale, B.S.; Snove, O., Jr.; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342. [Google Scholar] [CrossRef]
- Xu, F.; Ren, Y.; Teng, Y.; Mu, J.; Tang, J.; Sundaram, K.; Zhang, L.; Park, J.W.; Hwang, J.Y.; Yan, J.; et al. Tryptophan As a New Member of RNA-Induced Silencing Complexes Prevents Colon Cancer Liver Metastasis. Adv. Sci. 2024, 11, e2307937. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Zhang, X.; Wang, Y.; Mao, G.; Ou, Y.; Wei, C.; Hu, X.; Xiang, S. DDX21 interacts with nuclear AGO2 and regulates the alternative splicing of SMN2. Biosci. Biotechnol. Biochem. 2021, 85, 272–279. [Google Scholar] [CrossRef]
- Guidi, R.; Wedeles, C.; Xu, D.; Kolmus, K.; Headland, S.E.; Teng, G.; Guillory, J.; Zeng, Y.J.; Cheung, T.K.; Chaudhuri, S.; et al. Argonaute3-SF3B3 complex controls pre-mRNA splicing to restrain type 2 immunity. Cell Rep. 2023, 42, 113515. [Google Scholar] [CrossRef]
- Ehses, J.; Schlegel, M.; Schroger, L.; Schieweck, R.; Derdak, S.; Bilban, M.; Bauer, K.; Harner, M.; Kiebler, M.A. The dsRBP Staufen2 governs RNP assembly of neuronal Argonaute proteins. Nucleic Acids Res. 2022, 50, 7034–7047. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.J.; Zhou, H.; Kim, M.; Skariah, G.; Khetani, R.S.; Drnevich, J.; Arcila, M.L.; Kosik, K.S.; Ceman, S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014, 9, 1729–1741. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.J.; Kim, M.; Skariah, G.; Nielsen, J.; Lannom, M.C.; Ceman, S. The FMRP-MOV10 complex: A translational regulatory switch modulated by G-Quadruplexes. Nucleic Acids Res. 2020, 48, 862–878. [Google Scholar] [CrossRef]
- Nawaz, A.; Shilikbay, T.; Skariah, G.; Ceman, S. Unwinding the roles of RNA helicase MOV10. Wiley Interdiscip. Rev. RNA 2022, 13, e1682. [Google Scholar] [CrossRef]
- Lin, M.C.; Kuo, W.H.; Chen, S.Y.; Hsu, J.Y.; Lu, L.Y.; Wang, C.C.; Chen, Y.J.; Tsai, J.S.; Li, H.J. Ago2/CAV1 interaction potentiates metastasis via controlling Ago2 localization and miRNA action. EMBO Rep. 2024, 25, 2441–2478. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, K.; Liu, X.; Zhao, X.; Zhao, T.; Lu, G. Exosomal mir-625-3p derived from hypoxic lung cancer cells facilitates metastasis by targeting SCAI. Mol. Biol. Rep. 2022, 49, 9275–9281. [Google Scholar] [CrossRef] [PubMed]
- Fintha, A.; Gasparics, A.; Fang, L.; Erdei, Z.; Hamar, P.; Mozes, M.M.; Kokeny, G.; Rosivall, L.; Sebe, A. Characterization and role of SCAI during renal fibrosis and epithelial-to-mesenchymal transition. Am. J. Pathol. 2013, 182, 388–400. [Google Scholar] [CrossRef]
- Lobo, V.; Nowak, I.; Fernandez, C.; Correa Muler, A.I.; Westholm, J.O.; Huang, H.C.; Fabrik, I.; Huynh, H.T.; Shcherbinina, E.; Poyraz, M.; et al. Loss of Lamin A leads to the nuclear translocation of AGO2 and compromised RNA interference. Nucleic Acids Res. 2024, 52, 9917–9935. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, Y.; Wang, H.; Cheng, Y.; Zhao, D.; Sun, Q.; Chen, D. Lipid-mediated phase separation of AGO proteins on the ER controls nascent-peptide ubiquitination. Mol. Cell 2022, 82, 1313–1328.e8. [Google Scholar] [CrossRef] [PubMed]
- Tilley, A.M.C.; Howard, C.M.; Sridharan, S.; Subramaniyan, B.; Bearss, N.R.; Alkhalili, S.; Raman, D. The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer. Cancers 2020, 12, 2455. [Google Scholar] [CrossRef]
- Nowak, I.; Sarshad, A.A. Argonaute Proteins Take Center Stage in Cancers. Cancers 2021, 13, 788. [Google Scholar] [CrossRef]
- Butt, E.; Howard, C.M.; Raman, D. LASP1 in Cellular Signaling and Gene Expression: More than Just a Cytoskeletal Regulator. Cells 2022, 11, 3817. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Bridge, K.S.; Shah, K.M.; Li, Y.; Foxler, D.E.; Wong, S.C.K.; Miller, D.C.; Davidson, K.M.; Foster, J.G.; Rose, R.; Hodgkinson, M.R.; et al. Argonaute Utilization for miRNA Silencing Is Determined by Phosphorylation-Dependent Recruitment of LIM-Domain-Containing Proteins. Cell Rep. 2017, 20, 173–187. [Google Scholar] [CrossRef]
- Liu, Y.; Su, Z.; Tavana, O.; Gu, W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell 2024, 42, 946–967. [Google Scholar] [CrossRef]
- Krell, J.; Stebbing, J.; Carissimi, C.; Dabrowska, A.F.; de Giorgio, A.; Frampton, A.E.; Harding, V.; Fulci, V.; Macino, G.; Colombo, T.; et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA-mRNA interaction network. Genome Res. 2016, 26, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Zarnescu, D.C.; Ceman, S.; Nakamoto, M.; Mowrey, J.; Jongens, T.A.; Nelson, D.L.; Moses, K.; Warren, S.T. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004, 7, 113–117. [Google Scholar] [CrossRef]
- Li, Y.; Tang, W.; Zhang, L.R.; Zhang, C.Y. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol. Biosyst. 2014, 10, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, X.; Huang, F.; Yang, B.; Li, J.; Liu, B.; Luo, H.; Zhang, P.; Zhang, H. APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J. Biol. Chem. 2012, 287, 29373–29383. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Leslie, C.S. Learning to Predict miRNA-mRNA Interactions from AGO CLIP Sequencing and CLASH Data. PLoS Comput. Biol. 2016, 12, e1005026. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Alfaro, A.; Nicholas, S.L.N.; Sanabria, H. FRET-FCS: Advancing comprehensive insights into complex biological systems. Biophys. J. 2025, S0006-3495(25)00242-5. [Google Scholar] [CrossRef] [PubMed]

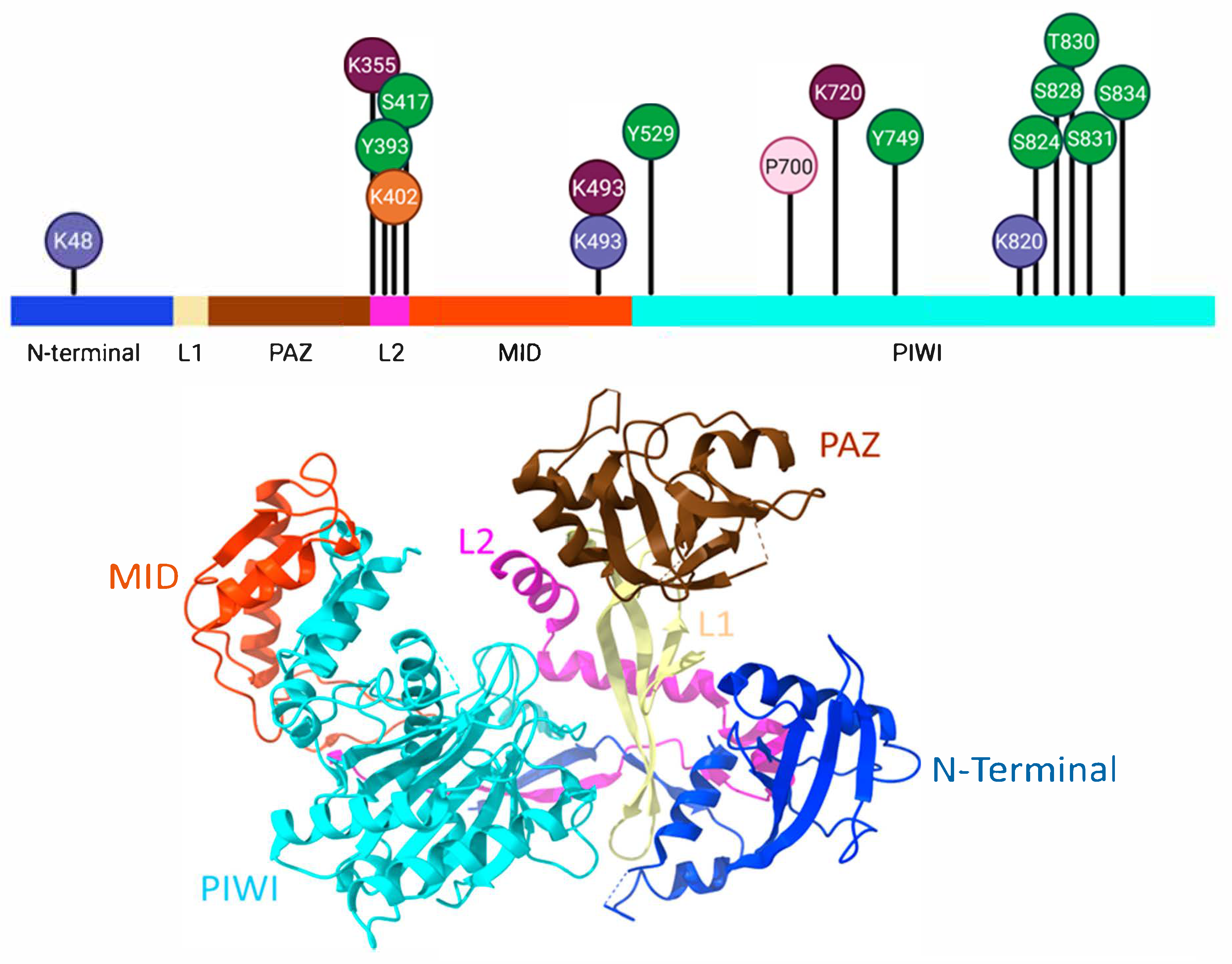
| miRISC Interactors | AGO Binding | Functionality | Remarks | Refs. |
|---|---|---|---|---|
| DICER | AGO 1-4 | Pre-miRNA processing | - | [5,13,24,34,35] |
| GW182 family (TNRC6A/B/C) | AGO 1-4 | Scaffold protein to recruit other factors of miRISC | - | [36,37] |
| CCR4-NOT complex | AGO 1-4 via GW182 proteins | Involved in de-adenylation mediated miRISC mechanisms | - | [36,38] |
| PAN2-PAN3 | AGO2 | Involved in a 2-stage de-adenylation process with CCR4-NOT complex | - | [27,39] |
| PABP | AGO 1-4 via GW182 proteins | Involved in de-adenylation mediated miRISC mechanisms | - | [26,40] |
| CAF1 | AGO 1-2 | Additional de-adenylase in context-dependent miRISC functionality | - | [26] |
| PARN | AGO2 | Involved in miR-125b-AGO2 RISC repression of TP53 | Also involved in pre-miR-451 biogenesis pathway along with AGO2 | [29,41] |
| DDX6 | AGO 2 | Functions as an activator of decapping enzymes DCP1/DCP2 | - | [30] |
| DHX9 | AGO 2-3 | RNA helicase that functions in RISC assembly and miRNA loading | - | [42] |
| DCP1/DCP2 | AGO 1-2 | Supports de-capping of target mRNAs to promote 5′ to 3′ nucleolytic activity | - | [31] |
| XRN1 | Not directly | Mediates 5′ to 3′ mRNA decay and is recruited by EDC4 | Implicated in miRISC-mediated silencing | [43] |
| TRBP | AGO2 via DICER | Promotes dsRNA binding to RLC | - | [15,44] |
| PACT | AGO2 via DICER | Promotes dsRNA binding to RLC | - | [15,17] |
| Hsc70 | AGO2 | Involved in ATP- dependent conformation change in AGO2 to enable miRNA loading | - | [18] |
| Hsp90 | AGO2 | - | [18] | |
| p23 | AGO2 | Part of the Hsc70/Hsp90 chaperone complex for miRISC loading | - | [19] |
| miRISC Interactors | AGO Binding | Core/ Auxiliary Factors | Functionality | Remarks | References |
|---|---|---|---|---|---|
| FKBP4 | AGO2 | Auxiliary | Promotes ATP-dependent loading of miRNAs in RISC | - | [19] |
| CSDE1 | AGO2 | Auxiliary | Binds pre-miR-451, AGO2 and PARN to enhance trimming of the hairpin | - | [51,93] |
| Tryptophan (Amino Acid) | AGO 1-4 | Auxiliary | Enhances nuclease activity of AGO2 | - | [96] |
| DDX21 | AGO2 | Auxiliary | Increases nuclear AGO2 levels | - | [97] |
| SF3B3 complex | AGO3 | Auxiliary | Promotes Nisch splicing to increase IL-13 expression in T cells | - | [98] |
| STAU2 | AGO2 | Auxiliary | STAU2 regulates global translation by miRISC | - | [99] |
| FMRP | AGO2 | Auxiliary | FMRP-mediates translational control by AGO2-miRISC | [117,118] | |
| MOV10 | AGO 1-3 | Auxiliary | MOV10 relieves complex secondary structures in 3′-UTRs to augment AGO-miRISC binding to MRE | - | [102,119] |
| APOBEC3G | Indirect activity | - | Binds MOV10 to inhibit AGO2-miRISC interaction to mRNAs | - | [120] |
| CAV-1 | AGO2 | Auxiliary | Sorting of AGO2 RISC into EVs | Observed in breast cancer | [103] |
| Lamin A | Indirect activity | - | Inhibits RISC activity | - | [106] |
| PI(4,5)P2 | AGO2 | - | AGO RNPs condensation on ER membranes | Controls proteasomal degradation of nascent peptides | [107] |
| LASP1 | AGO2 | Auxiliary | Enhances translational repression of Let-7a targets | This interaction is a CXCR4-dependent mechanism | [108] |
| LIMD1 | AGO2 | Auxiliary | A pS387-AGO2-dependent mechanism decreasing translation of mRNA targets | Upon LIMD1 depletion in cells, AGO3-WTIP becomes the dominant miRISC | [114] |
| TP53 | AGO2 | Auxiliary | Facilitates sorting of Let-7 miRNAs into AGO2-miRISC | TP53 may alter the conformation of AGO2 to promote loading of Let-7 miRNAs into RISC | [116] |
| FKBP4 | AGO2 | Auxiliary | Enhances ATP-dependent loading of miRNAs into miRISC | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narasimhan, S.; Erkeland, S.J. Partners in Silencing: Decoding the Mammalian Argonaute Interactome. Non-Coding RNA 2025, 11, 62. https://doi.org/10.3390/ncrna11040062
Narasimhan S, Erkeland SJ. Partners in Silencing: Decoding the Mammalian Argonaute Interactome. Non-Coding RNA. 2025; 11(4):62. https://doi.org/10.3390/ncrna11040062
Chicago/Turabian StyleNarasimhan, Srinaath, and Stefan J. Erkeland. 2025. "Partners in Silencing: Decoding the Mammalian Argonaute Interactome" Non-Coding RNA 11, no. 4: 62. https://doi.org/10.3390/ncrna11040062
APA StyleNarasimhan, S., & Erkeland, S. J. (2025). Partners in Silencing: Decoding the Mammalian Argonaute Interactome. Non-Coding RNA, 11(4), 62. https://doi.org/10.3390/ncrna11040062






