The Good, the Bad, or Both? Unveiling the Molecular Functions of LINC01133 in Tumors
Abstract
1. Introduction
2. Results
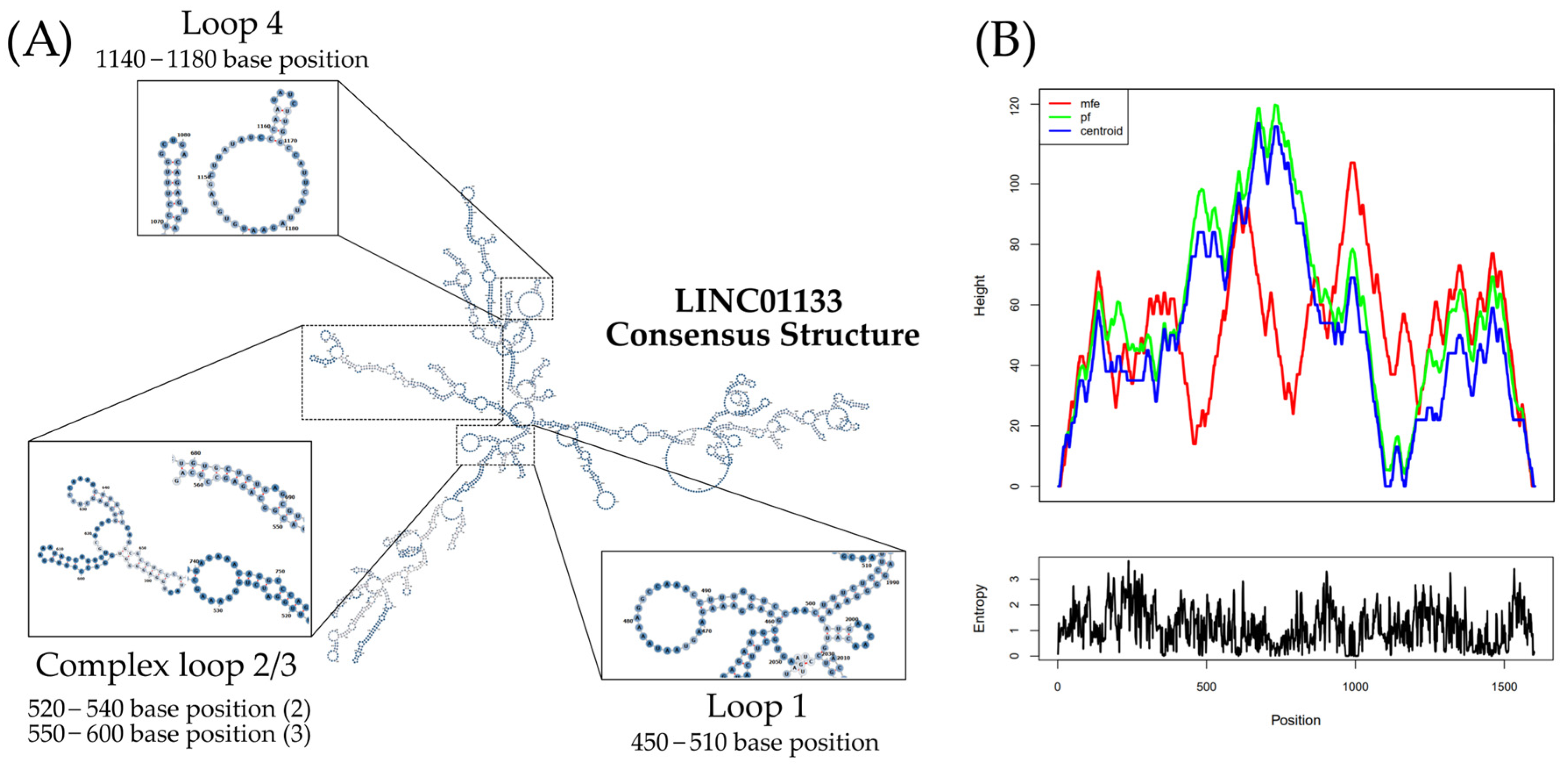
2.1. Genetic and Structural Analysis of LINC01133
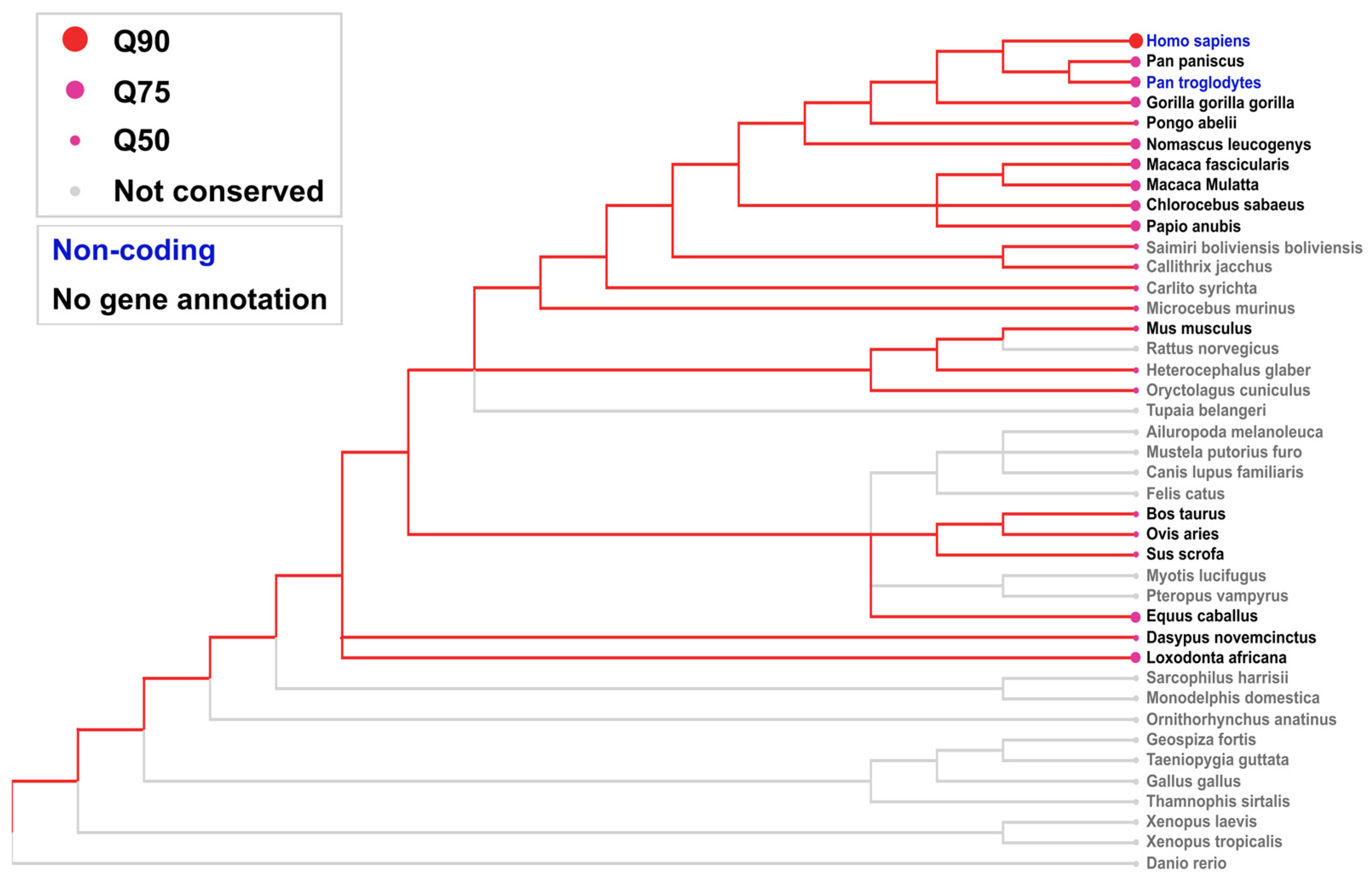
2.2. Inter-Species Conservation of LINC01133
2.3. Associations Between LINC01133 and Tumors via RNA-Seq Data
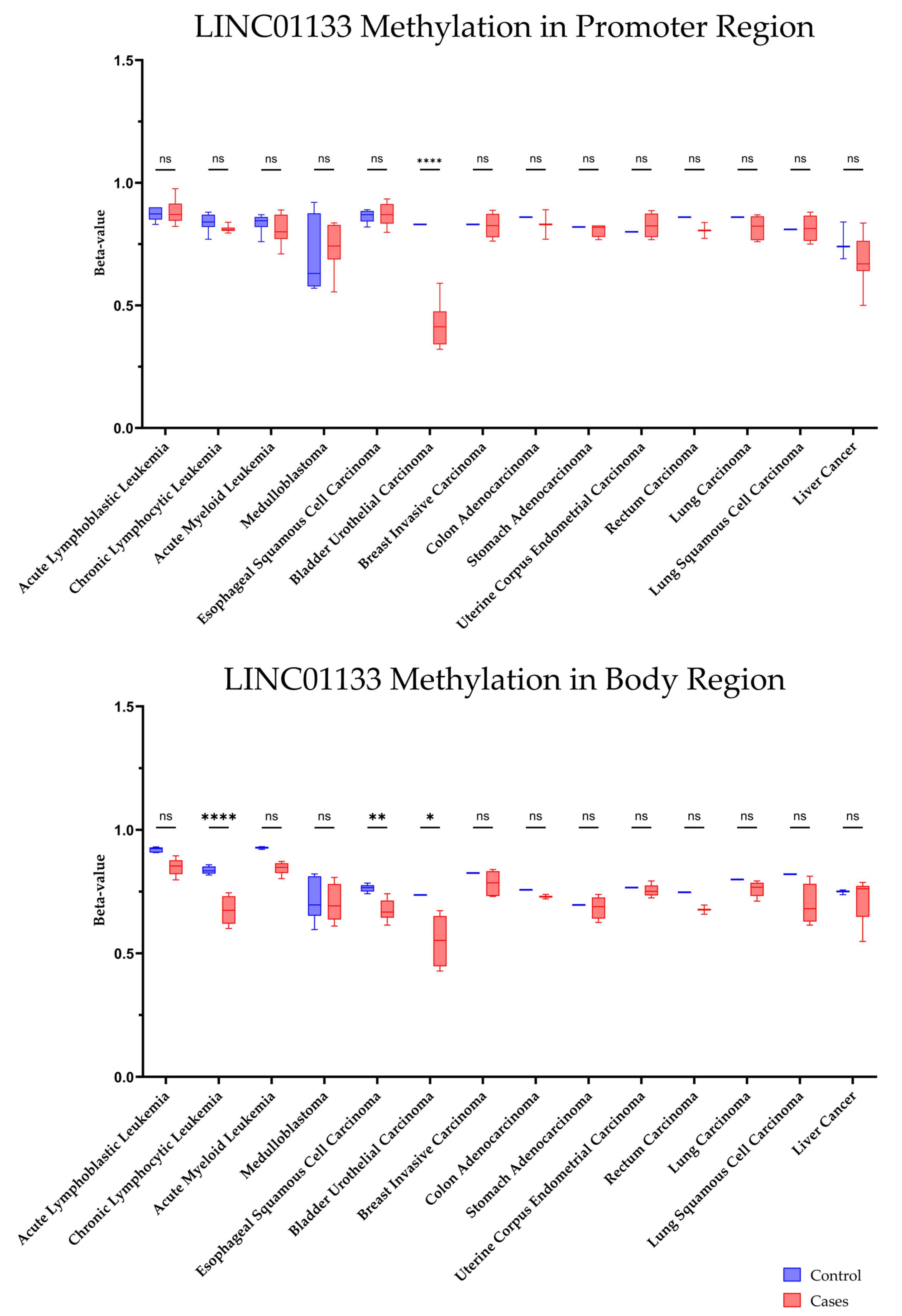
2.4. DNA Methylation Profile of the LINC01133 Gene in Different Tumor Types
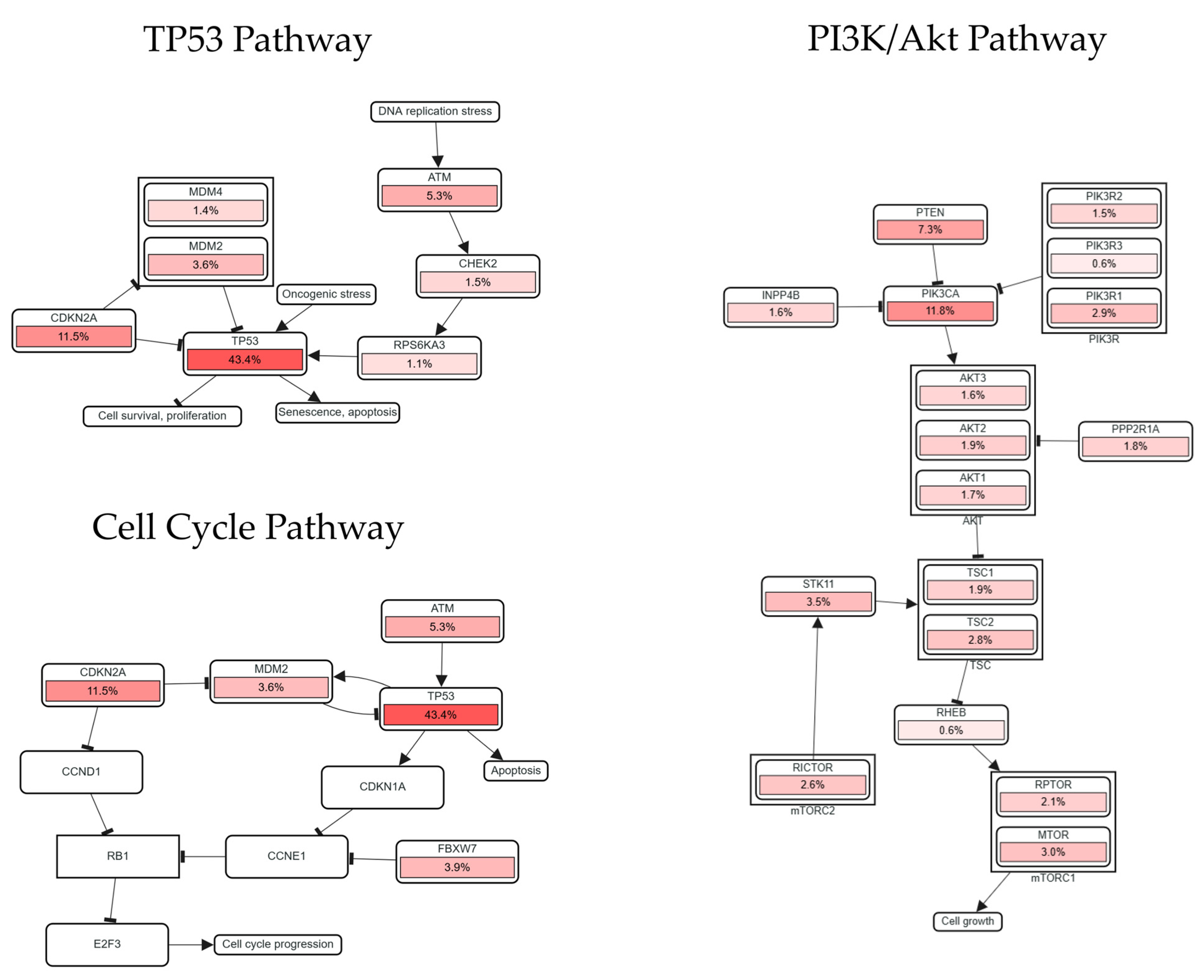
2.5. Relationships Between LINC01133 and Pathways Associated with Tumor Progression
3. Discussion
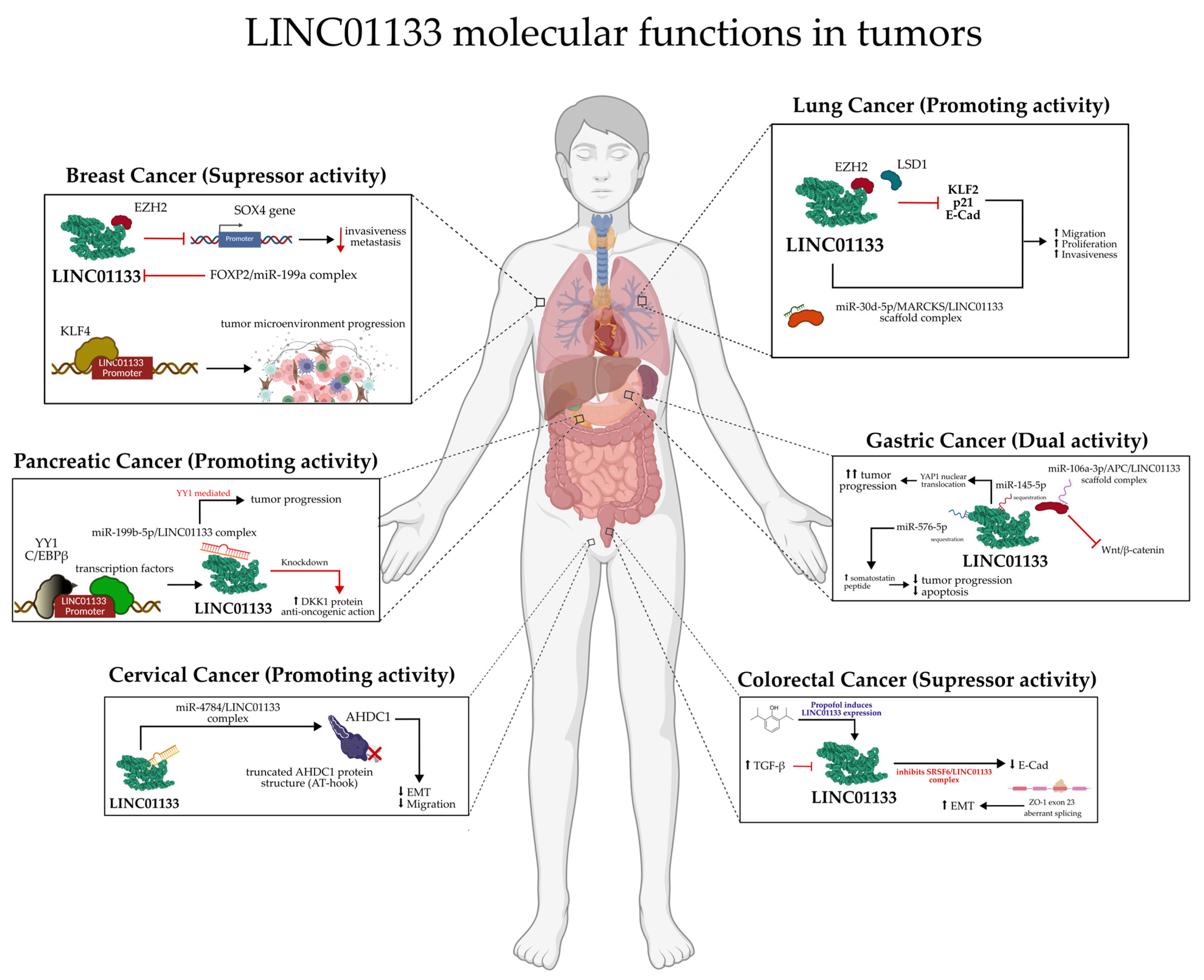
3.1. LINC01133 and Its Actions in Different Types of Tumors
3.1.1. Lung Tumors
3.1.2. Colorectal Tumors
3.1.3. Breast Tumors
3.1.4. Ovarian Tumors
3.1.5. Bladder Tumors
3.1.6. Hepatic Tumors
3.1.7. Pancreatic Tumors
3.1.8. Esophageal Tumors
3.1.9. Oral Squamous Cell Carcinomas
3.1.10. Gastric Tumors
3.1.11. Nasopharyngeal Tumors
3.1.12. Cervical Tumors
3.1.13. Renal Tumors
3.1.14. Bone Tumors
3.1.15. Endometrial Tumors
4. Methods
4.1. Sequence Acquisition
4.2. Alignment and Structural Analysis
4.3. Interspecies Conservation Analysis
4.4. Interactions Between LINC01133 and Tumorigenesis and Tumor Progression Pathways
4.5. Methylation Profile in Tumors
4.6. Narrative Literature Review
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALL | Acute Lymphoblastic Leukemia |
| AML | Acute Myeloid Leukemia |
| BC | Bone cancer/Tumors |
| BLCA | Bladder Urothelial Carcinoma |
| BLT | Bladder Tumors |
| bp | Base Pair |
| BRC | Breast Cancer/Tumor |
| BRCA | Breast Invasive Carcinoma |
| CESC | Cervical squamous cell carcinoma |
| CLL | Chronic Lymphocytic Leukemia |
| COAD | Colon Adenocarcinoma |
| CRC | Colorectal Cancer/Tumor |
| CvC | Cervical Cancer/Tumors |
| EC | Endometrium Cancer/Tumors |
| EMT | Epithelial–Mesenchymal Transition |
| EOC | Epithelial Ovarian Cancer |
| EPT | Esophageal Tumors |
| ESCC | Esophageal squamous cell carcinoma |
| GT | Gastric Cancer/Tumors |
| HCC | Hepatocellular Carcinoma |
| HpT | Hepatic Tumors |
| LINC01133 | Long intergenic non-coding RNA 01133 |
| lncRNA | long non-coding RNA |
| LSCC | Lung Squamous Cell Carcinoma |
| LT | Lung Tumors |
| LUAD | Lung Adenocarcinoma |
| LUSC | Lung Squamous Cell Carcinoma |
| MB | Medulloblastoma |
| miRNAs | Micro RNAs |
| mRNAs | Messenger RNAs |
| ncRNA | Non-Coding RNA |
| NPC | Nasopharyngeal Cancer/Tumors |
| NSCLC | Non-Small Cells Lung Cancer |
| OSCC | Oral Squamous Cell Carcinoma |
| OVT | Ovarian Tumors |
| PAAD | Pancreatic Adenocarcinoma |
| PC | Pancreatic Cancer/Tumors |
| PDAC | Pancreactic Ductal Adenocarcinoma |
| RCC | Renal Cell Carcinoma |
| READ | Rectum Adenocarcinoma |
| rRNAs | Ribosomic RNAs |
| RT | Renal Tumors |
| siRNAs | Small Interfering RNAs |
| SST | Somatostatin |
| STAD | Stomach Adenocarcinoma |
| TNBC | Triple-Negative Breast Cancer |
| tRNAs | Transporter RNAs |
| UCEC | Uterine Corpus Endometrial Carcinoma |
References
- Wishart, D.S. Is Cancer a Genetic Disease or a Metabolic Disease? EBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef]
- Yamamoto, A.; Huang, Y.; Krajina, B.A.; McBirney, M.; Doak, A.E.; Qu, S.; Wang, C.L.; Haffner, M.C.; Cheung, K.J. Metastasis from the tumor interior and necrotic core formation are regulated by breast cancer-derived angiopoietin-like 7. Proc. Natl. Acad. Sci. USA 2023, 120, e2214888120. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumor heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Yip, H.Y.K.; Papa, A. Signaling pathways in cancer: Therapeutic targets, combinatorial treatments, and new developments. Cells 2021, 10, 659. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.N.; Tian, Q.; Teng, Q.X.; Wurpel, J.N.D.; Zeng, L.; Pan, Y.; Chen, Z.S. Understanding and targeting resistance mechanisms in cancer. MedComm 2023, 4, e265. [Google Scholar] [CrossRef]
- Mendiratta, G.; Ke, E.; Aziz, M.; Liarakos, D.; Tong, M.; Stites, E.C. Cancer gene mutation frequencies for the U.S. population. Nat. Commun. 2021, 12, 5961. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef]
- Krakhmal, N.V.; Zavyalova, M.V.; Denisov, E.V.; Vtorushin, S.V.; Perelmuter, V.M. Cancer invasion: Patterns and mechanisms. Acta Naturae 2015, 7, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Han, J.D.J. LncRNAs: The missing link to senescence nuclear architecture. Trends Biochem. Sci. 2023, 48, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Agliano, F.; Rathinam, V.A.; Medvedev, A.E.; Vanaja, S.K.; Vella, A.T. Long Noncoding RNAs in Host–Pathogen Interactions. Trends Immunol. 2019, 40, 492–510. [Google Scholar] [CrossRef]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, H.; Wang, L.; Wu, W.; Zhao, J.; Li, X.; Xiong, R.; Ding, X.; Yuan, D.; Yuan, C. LncRNA MEG3: Targeting the Molecular Mechanisms and Pathogenic causes of Metabolic Diseases. Curr. Med. Chem. 2023, 31, 6140–6153. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Pourtavakoli, A.; Hussen, B.M.; Taheri, M.; Kiani, A. A review on the importance of LINC-ROR in human disorders. Pathol.-Res. Pract. 2023, 244, 154420. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Askari, A.; Moghadam, K.B.; Hussen, B.M.; Taheri, M.; Samadian, M. A review on the role of ZEB1-AS1 in human disorders. Pathol.-Res. Pract. 2023, 245, 154486. [Google Scholar] [CrossRef]
- Yang, J. Emerging roles of long non-coding RNA FOXP4-AS1 in human cancers: From molecular biology to clinical application. Heliyon 2024, 10, e39857. [Google Scholar] [CrossRef]
- Rajagopal, T.; Talluri, S.; Akshaya, R.L.; Dunna, N.R. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin. Chim. Acta 2020, 503, 1–18. [Google Scholar] [CrossRef]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef]
- Yao, F.; Wang, Q.; Wu, Q. The prognostic value and mechanisms of LNCRNA UCA1 in human cancer. Cancer Manag. Res. 2019, 11, 7685–7696. [Google Scholar] [CrossRef]
- Sekar, D.; Tusubira, D.; Ross, K. TDP-43 and NEAT long non-coding RNA: Roles in neurodegenerative disease. Front. Cell. Neurosci. 2022, 16, 954912. [Google Scholar] [CrossRef]
- Srivastava, D.; Metzler, K.R.C. Fending for a Braveheart. EMBO J. 2013, 32, 1211–1213. [Google Scholar] [CrossRef][Green Version]
- Desideri, F.; Cipriano, A.; Petrezselyova, S.; Buonaiuto, G.; Santini, T.; Kasparek, P.; Prochazka, J.; Janson, G.; Paiardini, A.; Calicchio, A.; et al. Intronic Determinants Coordinate Charme lncRNA Nuclear Activity through the Interaction with MATR3 and PTBP1. Cell Rep. 2020, 33, 108548. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Much, C.; Lasda, E.L.; Pereira, I.T.; Vallery, T.K.; Ramirez, D.; Lewandowski, J.P.; Dowell, R.D.; Smallegan, M.J.; Rinn, J.L. The temporal dynamics of lncRNA Firre-mediated epigenetic and transcriptional regulation. Nat. Commun. 2024, 15, 6821. [Google Scholar] [CrossRef]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Weiswald, L.B.; Poulain, L.; Denoyelle, C.; Meryet-Figuiere, M. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: Cytoskeleton and ECM crosstalk. J. Exp. Clin. Cancer Res. 2023, 42, 173. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Sharma, U.; Barwal, T.S.; Murmu, M.; Acharya, V.; Pant, N.; Dey, D.; Vivek; Gautam, A.; Bazala, S.; Singh, I.; et al. Clinical potential of long non-coding RNA LINC01133 as a promising biomarker and therapeutic target in cancers. Biomark. Med. 2022, 16, 349–369. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, Q.; Li, J.; Raziq, K.; Kang, X.; Liang, S.; Sun, C.; Liang, X.; Zhao, D.; Fu, S.; et al. New Sights into Long Non-Coding RNA LINC01133 in Cancer. Front. Oncol. 2022, 12, 908162. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Mokhtari, M. A review on the role of LINC01133 in cancers. Cancer Cell Int. 2022, 22, 270. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.J.; Burkard, M.E.; Turner, D.H. The energetics of small internal loops in RNA. Biopolymers 1999, 52, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Yang, K.; Xiao, Y.; Zhong, L.; Zhang, W.; Wang, P.; Ren, Y.; Shi, L. p53-regulated lncRNAs in cancers: From proliferation and metastasis to therapy. Cancer Gene. Ther. 2023, 30, 1456–1470. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.P.; Emes, R.D.; Lynch, V.J. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 2016, 5, e11994. [Google Scholar] [CrossRef]
- Chusyd, D.E.; Ackermans, N.L.; Austad, S.N.; Hof, P.R.; Mielke, M.M.; Sherwood, C.C.; Allison, D.B. Aging: What We Can Learn From Elephants. Front. Aging 2021, 2, 726714. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, F.; Wu, G.; Liu, W.; Chen, J.; Wang, B.; Chen, Y. Gene body methylation in cancer: Molecular mechanisms and clinical applications. Clin. Epigenetics 2022, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Queirós, A.C.; Beekman, R.; Martín-Subero, J.I. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 1161–1174. [Google Scholar] [CrossRef]
- Duchartre, Y.; Kim, Y.M.; Kahn, M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Rim, E.Y.; Clevers, H.; Nusse, R. The Wnt Pathway: From Signaling Mechanisms to Synthetic Modulators. Annu. Rev. Biochem. 2022, 91, 571–598. [Google Scholar] [CrossRef]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef]
- Holzem, M.; Boutros, M.; Holstein, T.W. The origin and evolution of Wnt signalling. Nat. Rev. Genet. 2024, 25, 500–512. [Google Scholar] [CrossRef]
- Ivanova, M.M.; Dao, J.; Kasaci, N.; Friedman, A.; Noll, L.; Goker-Alpan, O. Wnt signaling pathway inhibitors, sclerostin and DKK-1, correlate with pain and bone pathology in patients with Gaucher disease. Front. Endocrinol. 2022, 13, 1029130. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Strasser, A.; Kelly, G.L. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026062. [Google Scholar] [CrossRef]
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Eischen, C.M.; Lozano, G. The mdm network and its regulation of p53 activities: A rheostat of cancer risk. Hum. Mutat. 2014, 35, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef] [PubMed]
- Ghareghomi, S.; Habibi-Rezaei, M.; Arese, M.; Saso, L.; Moosavi-Movahedi, A.A. Nrf2 Modulation in Breast Cancer. Biomedicines 2022, 10, 2668. [Google Scholar] [CrossRef]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β signaling in health, disease, and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Tauriello, D.V.F.; Sancho, E.; Batlle, E. Overcoming TGFβ-mediated immune evasion in cancer. Nat. Rev. Cancer 2022, 22, 25–44. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Sundaram, M.V. Canonical RTK-Ras-ERK signaling and related alternative pathways. WormBook 2013, 11, 1–38. [Google Scholar] [CrossRef]
- Kumari, L.; Mishra, L.; Sharma, Y.; Chahar, K.; Kumar, M.; Patel, P.; Gupta, G.D.; Kurmi, B.D. NOTCH Signaling Pathway: Occurrence, Mechanism, and NOTCH-Directed Therapy for the Management of Cancer. Cancer Biother. Radiopharm. 2024, 39, 19–34. [Google Scholar] [CrossRef]
- Fischer, A.; Gessler, M. Delta-Notch-and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007, 35, 4583–4596. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Kennell, J.; Cadigan, K.M. APC and β-catenin degradation. Adv. Exp. Med. Biol. 2009, 656, 1–12. [Google Scholar] [CrossRef]
- Lü, Y.Q.; Cho, T.; Mukherjee, S.; Suarez, C.F.; Gonzalez-Foutel, N.S.; Malik, A.; Martinez, S.; Dervovic, D.; Oh, R.H.; Langille, E.; et al. Genome-wide CRISPR screens identify novel regulators of wild-type and mutant p53 stability. Mol. Syst. Biol. 2024, 20, 719–740. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, N.; Chen, X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumor Biol. 2015, 36, 7465–7471. [Google Scholar] [CrossRef] [PubMed]
- Zang, C.; Nie, F.Q.; Wang, Q.; Sun, M.; Li, W.; He, J.; Zhang, M.; Lu, K.H. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non-small cell lung cancer. Oncotarget 2016, 7, 11696–11707. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ma, X.; Zhu, C.; Guo, L.; Li, Q.; Liu, M.; Zhang, J. The prognostic value of long non coding RNAs in non-small cell lung cancer: A meta-analysis. Oncotarget 2016, 7, 81292–81304. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, W.; Chen, R.; Gu, Y.; Zhao, M.; Song, J.; Shi, Z.; Wu, J.; Chang, H.W.; Liu, M. LINC01133 regulates MARCKS expression via sponging miR-30d-5p to promote the development of lung squamous cell carcinoma. Transl. Oncol. 2024, 44, 101931. [Google Scholar] [CrossRef]
- Xin, X.; Liu, Y.; Li, Y.; Ji, X.; Yun, Y.; Yang, G.; Liu, H.; Liang, X.; Yang, S. LINC01133 contributes to the malignant phenotypes of non-small cell lung cancer by targeting miR-30b-5p/FOXA1 pathway. Cell. Mol. Biol. 2024, 70, 42–47. [Google Scholar] [CrossRef]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef]
- Zhang, J.H.; Li, A.Y.; Wei, N. Downregulation of long non-coding RNA LINC01133 is predictive of poor prognosis in colorectal cancer patients. Eur. Rev. Med. Pharm. Sci. 2017, 21, 2103–2107. Available online: https://www.europeanreview.org/wp/wp-content/uploads/2103-2107-Downregulation-of-LINC01133-in-CRC-patients.pdf (accessed on 2 January 2025).
- Yao, Y.; Zhang, F.; Liu, F.; Xia, D. Propofol-induced LINC01133 inhibits the progression of colorectal cancer via miR-186-5p/NR3C2 axis. Environ. Toxicol. 2023, 39, 2265–2284. [Google Scholar] [CrossRef]
- Tu, Z.; Schmöllerl, J.; Cuiffo, B.G.; Karnoub, A.E. Microenvironmental Regulation of Long Noncoding RNA LINC01133 Promotes Cancer Stem Cell-Like Phenotypic Traits in Triple-Negative Breast Cancers. Stem Cells 2019, 37, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhang, X.; Lin, Y.; Wei, Y.; Liang, S.; Dong, C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J. Cell Mol. Med. 2019, 23, 7554–7565. [Google Scholar] [CrossRef]
- Layeghi, S.M.; Arabpour, M.; Shakoori, A.; Naghizadeh, M.M.; Mansoori, Y.; Bazzaz, J.T.; Esmaeili, R. Expression profiles and functional prediction of long non-coding RNAs LINC01133, ZEB1-AS1 and ABHD11-AS1 in the luminal subtype of breast cancer. J. Transl. Med. 2021, 19, 364. [Google Scholar] [CrossRef]
- Liu, M.; Shen, C.; Wang, C. Long Noncoding RNA LINC01133 Confers Tumor-Suppressive Functions in Ovarian Cancer by Regulating Leucine-Rich Repeat Kinase 2 as an miR-205 Sponge. Am. J. Pathol. 2019, 189, 2323–2339. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xi, X. LINC01133 contribute to epithelial ovarian cancer metastasis by regulating miR-495-3p/TPD52 axis. Biochem. Biophys. Res. Commun. 2020, 533, 1088–1094. [Google Scholar] [CrossRef]
- Yang, H.; Qu, H.; Huang, H.; Mu, Z.; Mao, M.; Xie, Q.; Wang, K.; Hu, B. Exosomes-mediated transfer of long noncoding RNA LINC01133 represses bladder cancer progression via regulating the Wnt signaling pathway. Cell Biol. Int. 2021, 45, 1510–1522. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Hu, Z.Q.; Luo, C.B.; Wang, X.Y.; Xin, H.Y.; Sun, R.Q.; Wang, P.C.; Li, J.; Fan, J.; Zhou, Z.J.; et al. LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin. Transl. Med. 2021, 11, e409. [Google Scholar] [CrossRef]
- Zheng, Y.F.; Zhang, X.Y.; Bu, Y.Z. LINC01133 aggravates the progression of hepatocellular carcinoma by activating the PI3K/AKT pathway. J. Cell Biochem. 2019, 120, 4172–4179. [Google Scholar] [CrossRef]
- Huang, C.S.; Chu, J.; Zhu, X.X.; Li, J.H.; Huang, X.T.; Cai, J.P.; Zhao, W.; Yin, X.Y. The C/EBPβ-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett. 2018, 421, 63–72. [Google Scholar] [CrossRef]
- Giulietti, M.; Righetti, A.; Principato, G.; Piva, F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis 2018, 39, 1016–1025. [Google Scholar] [CrossRef]
- Weng, Y.C.; Ma, J.; Zhang, J.; Wang, J.C. Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol. Ther. 2019, 20, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, S.; Zhang, Y.; Yi, H.; Xu, M.; Xu, J.; Liu, H.; Ding, Z.; He, H.; Wang, H.; et al. Mir-216a-5p inhibits tumorigenesis in pancreatic cancer by targeting tpt1/mtorc1 and is mediated by linc01133. Int. J. Biol. Sci. 2020, 16, 2612–2627. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, T.; Yang, X.; Qin, P.; Wang, P.; Zhang, H.; Bai, M.; Wu, R.; Li, F. Tumor-derived exosomal long noncoding RNA LINC01133, regulated by Periostin, contributes to pancreatic ductal adenocarcinoma epithelial-mesenchymal transition through the Wnt/β-catenin pathway by silencing AXIN2. Oncogene 2021, 40, 3164–3179. [Google Scholar] [CrossRef]
- Yang, X.; Wang, L.; Zhou, F.; Ye, S.; Sun, Q. Yin Yang 1-induced activation of LINC01133 facilitates the progression of pancreatic cancer by sponging miR-199b-5p to upregulate myelin regulatory factor expression. Bioengineered 2022, 13, 13352–13365. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; He, X.; Yao, R.; Fan, L.; Zhao, L.; Lu, B.; Pang, Z. Genome instability-related LINC02577, LINC01133 and AC107464.2 are lncRNA prognostic markers correlated with immune microenvironment in pancreatic adenocarcinoma. BMC Cancer 2023, 23, 430. [Google Scholar] [CrossRef]
- Li, J.; Lin, J.; Ji, Y.; Wang, X.; Fu, D.; Wang, W.; Shen, B. A novel pyroptosis-associated lncRNA LINC01133 promotes pancreatic adenocarcinoma development via miR-30b-5p/SIRT1 axis. Cell. Oncol. 2023, 46, 1381–1398. [Google Scholar] [CrossRef]
- Li, S.; Jiang, F.; Chen, F.; Deng, Y.; Huang, H. Silencing long noncoding RNA LINC01133 suppresses pancreatic cancer through regulation of microRNA-1299-dependent IGF2BP3. J. Biochem. Mol. Toxicol. 2024, 38, e23534. [Google Scholar] [CrossRef]
- Yang, Y.; Gong, Y.; Ding, Y.; Sun, S.; Bai, R.; Zhuo, S.; Zhang, Z. LINC01133 promotes pancreatic ductal adenocarcinoma epithelial–mesenchymal transition mediated by SPP1 through binding to Arp3. Cell Death Dis. 2024, 15, 492. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; He, Q.J.; Cheng, T.T.; Chi, J.; Lei, Z.Y.; Tang, Z.; Liao, Q.X.; Zhang, H.; Zeng, L.S.; Cui, S.Z. Predictive Value of LINC01133 for Unfavorable Prognosis was Impacted by Alcohol in Esophageal Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 48, 251–262. [Google Scholar] [CrossRef]
- Kong, J.; Sun, W.; Zhu, W.; Liu, C.; Zhang, H.; Wang, H. Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J. Surg. Oncol. 2018, 118, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; Cheng, T.T.; He, Q.J.; Lei, Z.Y.; Chi, J.; Tang, Z.; Liao, Q.X.; Zhang, H.; Zeng, L.S.; Cui, S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer 2018, 17, 126. [Google Scholar] [CrossRef]
- Foroughi, K.; Amini, M.; Atashi, A.; Mahmoodzadeh, H.; Hamann, U.; Manoochehri, M. Tissue-specific down-regulation of the long non-coding RNAs PCAT18 and LINC01133 in gastric cancer development. Int. J. Mol. Sci. 2018, 19, 3881. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, K.; Zuo, Z.; Ye, F.; Cao, D.; Peng, Y.; Tang, T.; Li, X.; Zhou, S.; Duan, L. LINC01133 hampers the development of gastric cancer through increasing somatostatin via binding to microRNA-576-5p. Epigenomics 2021, 13, 1205–1219. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, Y.; He, J.; Tian, Y.; Zhang, G.; Zhao, R.; Zhu, W.J.; Gao, P. Linc01133 contributes to gastric cancer growth by enhancing YES1-dependent YAP1 nuclear translocation via sponging miR-145-5p. Cell Death Dis. 2022, 13, 51. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zhang, Q.; Hao, M.; Chen, Y. Serum LINC01133 combined with CEA and CA19-9 contributes to the diagnosis and survival prognosis of gastric cancer. Medicine 2024, 103, e40564. [Google Scholar] [CrossRef]
- Zhang, W.; Du, M.; Wang, T.; Chen, W.; Wu, J.; Li, Q.; Tian, X.; Qian, L.; Wang, Y.; Peng, F.; et al. Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am. J. Cancer Res. 2019, 9, 779–790. [Google Scholar] [PubMed]
- Feng, Y.; Qu, L.; Wang, X.; Liu, C. LINC01133 promotes the progression of cervical cancer by sponging miR-4784 to up-regulate AHDC1. Cancer Biol. Ther. 2019, 20, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Wang, D.; Zhao, M.; Sun, X.J.; Li, Y.; Lin, H.; Che, Y.Q.; Huang, C.Z. Serum lncrnas (Ccat2, linc01133, linc00511) with squamous cell carcinoma antigen panel as novel non-invasive biomarkers for detection of cervical squamous carcinoma. Cancer Manag Res 2020, 12, 9495–9502. [Google Scholar] [CrossRef]
- Ding, S.; Huang, X.; Zhu, J.; Xu, B.; Xu, L.; Gu, D.; Zhang, W. ADH7, miR-3065 and LINC01133 are associated with cervical cancer progression in different age groups. Oncol. Lett. 2020, 19, 2326–2338. [Google Scholar] [CrossRef]
- Zhai, X.; Wu, Y.; Wang, Z.; Zhao, D.; Li, H.; Chong, T.; Zhao, J. Long Noncoding RNA LINC01133 Promotes the Malignant Behaviors of Renal Cell Carcinoma by Regulating the miR-30b-5p/Rab3D Axis. Cell Transpl. 2020, 29, 0963689720964413. [Google Scholar] [CrossRef]
- Lv, W.; Li, Y.; Fu, L.; Meng, F.; Li, J. Linc01133 promotes proliferation and metastasis of human renal cell carcinoma through sponging miR-760. Cell Cycle 2022, 21, 1502–1511. [Google Scholar] [CrossRef]
- Li, Z.; Xu, D.; Chen, X.; Li, S.; Chan, M.T.V.; Wu, W.K.K. LINC01133: An emerging tumor-associated long non-coding RNA in tumor and osteosarcoma. Environ. Sci. Pollut. Res. 2020, 27, 32467–32473. [Google Scholar] [CrossRef]
- Yang, W.; Yue, Y.; Yin, F.; Qi, Z.; Guo, R.; Xu, Y. LINC01133 and LINC01243 are positively correlated with endometrial carcinoma pathogenesis. Arch. Gynecol. Obstet. 2021, 303, 207–215. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Magis, C.; Taly, J.F.; Bussotti, G.; Chang, J.M.; Di Tommaso, P.; Erb, I.; Espinosa-Carrasco, J.; Notredame, C. T-coffee: Tree-based consistency objective function for alignment evaluation. Methods Mol. Biol. 2014, 1079, 117–129. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef]
- Frankish, A.; Carbonell-Sala, S.; Diekhans, M.; Jungreis, I.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Arnan, C.; Barnes, I.; et al. GENCODE: Reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 2023, 51, D942–D949. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef]
- Lun, A.T.L.; McCarthy, D.J.; Marioni, J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 2016, 5, 2122. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
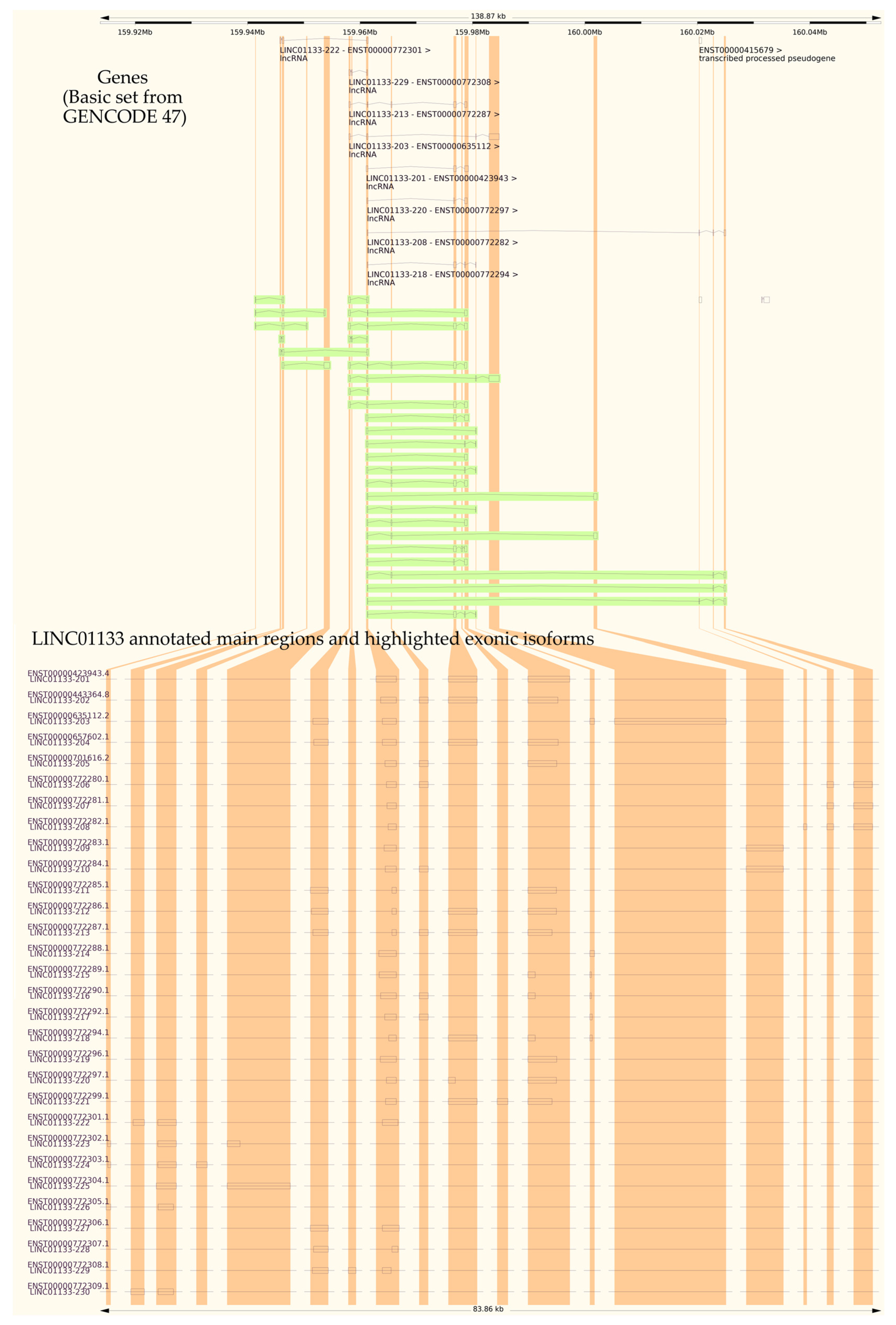



| Transcript ID | Name | Base Pairs |
|---|---|---|
| ENST00000772280.1 | LINC01133-206 | 717 |
| ENST00000635112.2 | LINC01133-203 | 2363 |
| ENST00000423943.4 | LINC01133-201 | 1478 |
| ENST00000657602.1 | LINC01133-204 | 1418 |
| ENST00000443364.8 | LINC01133-202 | 1358 |
| ENST00000772304.1 | LINC01133-225 | 1354 |
| ENST00000772287.1 | LINC01133-213 | 1325 |
| ENST00000772286.1 | LINC01133-212 | 1273 |
| ENST00000772299.1 | LINC01133-221 | 1207 |
| ENST00000772284.1 | LINC01133-210 | 932 |
| ENST00000772285.1 | LINC01133-211 | 822 |
| ENST00000772283.1 | LINC01133-209 | 804 |
| ENST00000701616.2 | LINC01133-205 | 795 |
| ENST00000772294.1 | LINC01133-218 | 741 |
| ENST00000772301.1 | LINC01133-222 | 737 |
| ENST00000772297.1 | LINC01133-220 | 734 |
| ENST00000772296.1 | LINC01133-219 | 723 |
| ENST00000772282.1 | LINC01133-208 | 601 |
| ENST00000772281.1 | LINC01133-207 | 568 |
| ENST00000772302.1 | LINC01133-223 | 564 |
| ENST00000772306.1 | LINC01133-227 | 562 |
| ENST00000772290.1 | LINC01133-216 | 539 |
| ENST00000772308.1 | LINC01133-229 | 527 |
| ENST00000772303.1 | LINC01133-224 | 522 |
| ENST00000772309.1 | LINC01133-230 | 471 |
| ENST00000772289.1 | LINC01133-215 | 413 |
| ENST00000772292.1 | LINC01133-217 | 369 |
| ENST00000772288.1 | LINC01133-214 | 357 |
| ENST00000772307.1 | LINC01133-228 | 341 |
| ENST00000772305.1 | LINC01133-226 | 325 |
| Source | Project ID | Disease Name (Short Name) | Sample Number |
|---|---|---|---|
| GEO | GSE116229 | Acute Lymphoblastic Leukemia | 38 (31 cases, 7 controls) |
| GEO | GSE135869 | Acute Myeloid Leukemia | 15 (9 cases, 6 controls) |
| GEO | GSE113336 | Chronic Lymphocytic Leukemia | 18 (11 cases, 7 controls) |
| GEO | GSE149608 | Esophageal Squamous Cell Carcinoma | 19 (10 cases, 9 controls) |
| GEO | GSE142241 | Medulloblastoma | 12 (8 cases, 4 controls) |
| GEO | GSE79799 | Liver cancer | 6 (3 cases, 3 controls) |
| TCGA | TCGA-BLCA | Bladder Urothelial Carcinoma | 7 (6 cases, 1 control) |
| TCGA | TCGA-BRCA | Breast Invasive Carcinoma | 6 (5 cases, 1 control) |
| TCGA | TCGA-COAD | Colon Adenocarcinoma | 3 (2 cases, 1 control) |
| TCGA | TCGA-LUAD | Lung Adenocarcinoma | 6 (5 cases, 1 control) |
| TCGA | TCGA-LUSC | Lung Squamous Cell Carcinoma | 5 (4 cases, 1 control) |
| TCGA | TCGA-READ | Rectal Adenocarcinoma | 3 (2 cases, 1 control) |
| TCGA | TCGA-STAD | Stomach Adenocarcinoma | 5 (4 cases, 1 control) |
| TCGA | TCGA-UCEC | Uterine Endometrial Carcinoma | 6 (5 cases, 1 control) |
| Signaling Pathway | General Function | Main Genes and Components |
|---|---|---|
| WNT/β-Catenin | Regulation of cell proliferation, polarity, differentiation, and embryonic development. Its hyperactivation is associated with uncontrolled cell proliferation, resistance to apoptosis, and cell invasion [43]. | CTNNB1 (β-catenin): main intracellular effector [44]. APC: tumor suppressor regulating β-catenin [45]. WNT ligands: activators of the pathway [46]. DKK and SFRPs: extracellular inhibitors of the pathway [47]. |
| TP53 | Regulation of genomic integrity by controlling the cell cycle and apoptosis in response to DNA damage. Its loss of function or mutation promotes genomic instability and oncogenesis [48]. | TP53: central gene of the pathway [49]. MDM2 and MDM4: negative regulators of p53 [50]. ATM: kinases that activate p53 in response to DNA damage. CDKN1A (p21): inhibits cyclin-dependent kinases (CDKs) mediated by p53 [51]. |
| PI3K/AKT | Regulation of cell survival, metabolism, angiogenesis, and proliferation. Its hyperactivation can result in resistance to apoptosis and tumor growth [52]. | PIK3CA: catalytic subunit of PI3K frequently mutated in cancer. PTEN: tumor suppressor antagonizing the pathway. AKT1/2/3: intracellular signaling mediators. MTOR: regulates protein synthesis and metabolism [52]. |
| NRF2 | Control of the antioxidant response by regulating genes that protect cells from oxidative stress. Its overexpression promotes adaptation to the stressful tumor microenvironment [53]. | NFE2L2 (NRF2): main transcription factor. KEAP1: negatively regulates NRF2, promoting its degradation. CUL3: component of the ubiquitination complex [53]. |
| TGF-β | Functional duality: in early stages of cancer, it acts as a tumor suppressor; in advanced stages, it promotes invasion, metastasis, and tumor microenvironment remodeling [54,55]. | TGFB1-3: pathway ligands. TGFBR1/2: receptors initiating signaling. SMAD2/3: intracellular mediators that translocate to the nucleus. SMAD4: coactivator of SMADs [54]. |
| MYC | Regulation of gene transcription linked to cell growth, metabolism, ribosomal biogenesis, cell cycle, and apoptosis [56]. | MYC (c-Myc): central transcription factor. MAX: forms heterodimers with Myc to activate transcription. MXD Complex: interacts with Myc to suppress proliferative genes [56]. |
| RTK-RAS | Transduction of extracellular signals for controlling cell proliferation, survival, and differentiation. Overexpression of the pathway, especially in solid tumors [57]. | EGFR, ERBB2 (HER2): frequently amplified tyrosine kinase receptors. KRAS, NRAS, and HRAS: members of the Ras family frequently mutated in cancer. RAF1/BRAF: downstream effectors activated by Ras [57]. |
| NOTCH | Control of cell fate decisions during development and homeostasis. Its deregulation leads to abnormal proliferation and resistance to apoptosis [58]. | NOTCH1-4: pathway receptors. JAG1/2: ligands that activate Notch [58]. HES/HEY: target genes regulating cell differentiation [59]. |
| Cell Cycle | Coordination of cell cycle progression and control. Alterations in checkpoint controls lead to deregulated cell division, a hallmark of tumors [48]. | FBXW7: inhibits CDK activators. RB1: regulates the cell cycle restriction point. TP53 and CDKN2A: controls the response to DNA damage and cell cycle arrest [48,49]. |
| HIPPO | Regulation of organ size, cell proliferation, and apoptosis. Its inactivation promotes tumor growth and resistance to apoptosis [60]. | LATS1/2: regulators that inhibit downstream effectors. YAP/TAZ: transcriptional effectors promoting cell growth. TEAD: transcription factor activated by YAP/TAZ. NF2 (Merlin): upstream tumor suppressor [60]. |
| Tumor | Studies | Promoting (P), Suppressing (S) or Both (B) When Expression Is Significant | References |
|---|---|---|---|
| Lung | 5 | (P) in LSCC (P) in NSCLC (P) in LUSC | [63,64,65,66,67] |
| Colorectal | 3 | (S) in CRC | [68,69,70] |
| Breast | 3 | (S) in TNBC (S) in Luminal | [71,72,73] |
| Ovarian | 2 | (B) in EOC * * (1 study in cell lines 1 study in patients’ ovarian samples) | [74,75] |
| Bladder | 1 | (S) in BC | [76] |
| Hepatocellular | 2 | (P) in HCC | [77,78] |
| Pancreas | 10 | (P) in PC (P) in PAAD (P) in PDAC | [79,80,81,82,83,84,85,86,87,88] |
| Esophagus | 1 | (S) in ESCC | [89] |
| Oral Squamous Cells | 1 | (S) in OSCC | [90] |
| Gastric | 5 | (B) in GT * * (1 study-(P)) | [91,92,93,94,95] |
| Nasopharynx | 1 | (S) in NPC | [96] |
| Cervical | 3 | (P) in CC (P) in CESC | [97,98,99] |
| Renal | 2 | (P) in RCC | [100,101] |
| Bone | 1 | (P) in BC | [102] |
| Endometrium | 1 | (P) in EC | [103] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teodoro Júnior, L.; Sogayar, M.C. The Good, the Bad, or Both? Unveiling the Molecular Functions of LINC01133 in Tumors. Non-Coding RNA 2025, 11, 58. https://doi.org/10.3390/ncrna11040058
Teodoro Júnior L, Sogayar MC. The Good, the Bad, or Both? Unveiling the Molecular Functions of LINC01133 in Tumors. Non-Coding RNA. 2025; 11(4):58. https://doi.org/10.3390/ncrna11040058
Chicago/Turabian StyleTeodoro Júnior, Leandro, and Mari Cleide Sogayar. 2025. "The Good, the Bad, or Both? Unveiling the Molecular Functions of LINC01133 in Tumors" Non-Coding RNA 11, no. 4: 58. https://doi.org/10.3390/ncrna11040058
APA StyleTeodoro Júnior, L., & Sogayar, M. C. (2025). The Good, the Bad, or Both? Unveiling the Molecular Functions of LINC01133 in Tumors. Non-Coding RNA, 11(4), 58. https://doi.org/10.3390/ncrna11040058









