Direct circRNA-mRNA Binding Controls mRNA Fate: A New Mechanism for circRNAs
Abstract
1. Introduction
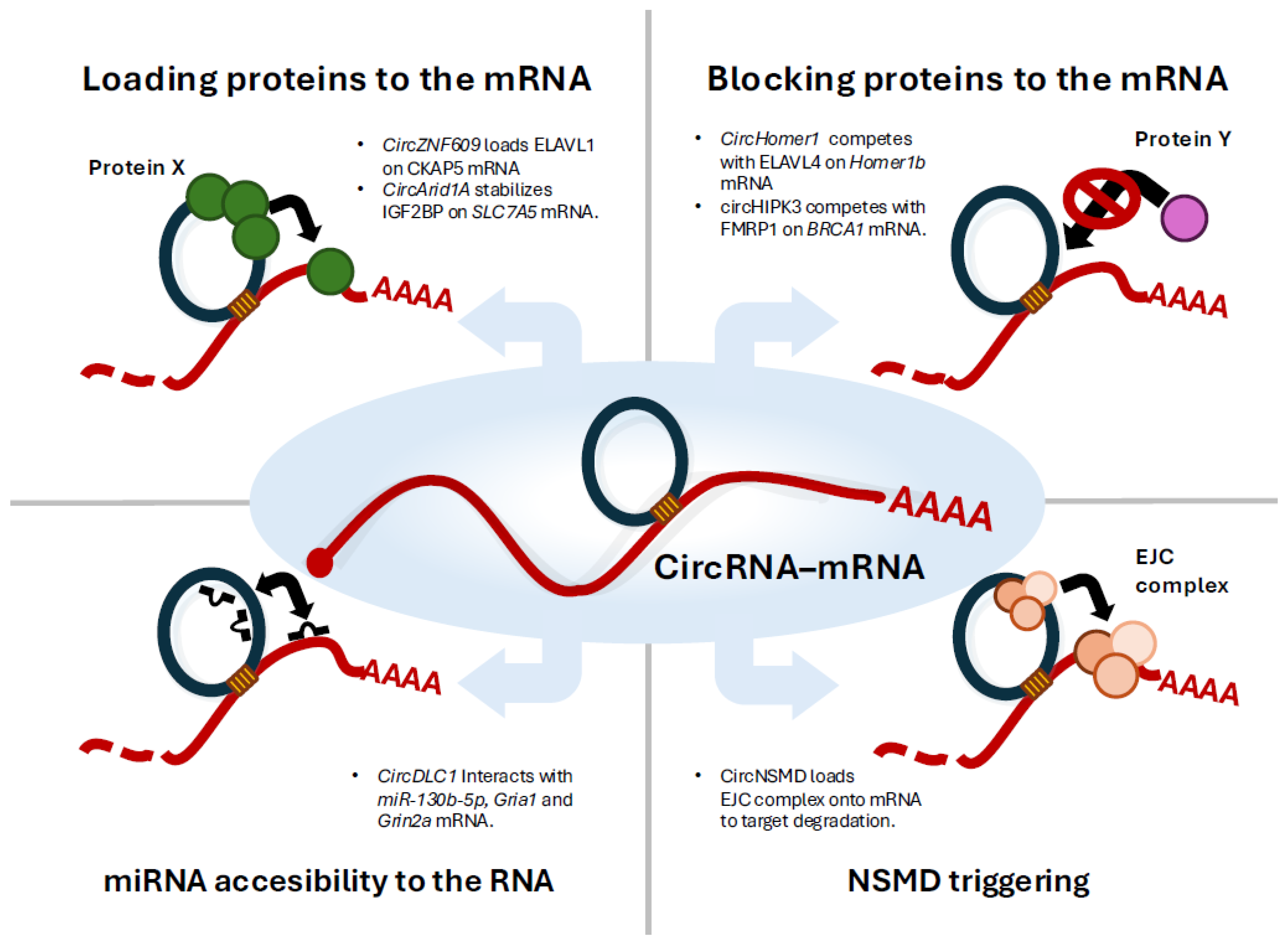
2. CircZNF609 Stabilises CKAP5 mRNA Helping the Loading of ELAVL1 Protein
3. CircHOMER1 Regulates Synaptic Localisation of HOMER1b mRNA Competing with ELAVL4 Protein
4. CircARID1A Enhances SLC7A5 mRNA Stability by Scaffolding IGF2BP3
5. CLiPPR-Seq Is a Genome-Wide Identification Technique That Detects Widespread circRNA–mRNA Interactions
6. CircDLC1 Regulates Gria1 and Grin2a mRNAs Translation Through Direct RNA–RNA Interaction and miRNA Competition
7. CircHIPK3 Regulates BRCA1 mRNA Translation Competing with FMRP
8. CircRNA-mRNA Pairings Induce Nonsense-Mediated Decay by Transferring Exon-Junction Complexes from circRNA to mRNA 3′ UTRs
9. CircFOXK2 Promotes Stabilisation and Translation of CCND1 mRNA Helping the Loading of ELAVL1 Protein
| Work | circRNA | Number of circRNA-mRNA Interactions | Methods |
|---|---|---|---|
| Rossi et al., 2022 [47] | circZNF609 | 11 | Psoralen and native circRNA pull down, LNA ASO |
| Hafez et al., 2022 [52] | circHOMER1 | 2 | Native circRNA pull down and RNA Antisense Purification |
| Ma et al., 2022 [55] | circARID1A | 1 | Native circRNA pull down |
| Singh et al., 2024 [56] | Several | +2000 | CLIPPR-seq |
| Silenzi et al., 2024 [57] | circDLC1 | 2 | Native circRNA pull down |
| Grelloni et al., 2024 [58] | circHIPK3 | 15 | Psoralen and native circRNA pull down, LNA ASO |
| Boo et al., 2024 [61] | Several | 41 | RIC-seq, LNA ASO |
| Yi et al., 2025 [67] | circFOXK2 | 1 | In vitro RNA–RNA interaction assay |
10. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLASH | Crosslinking Ligation and Sequencing of Hybrids |
| MARIO | MApping RNA Interactome in vivO |
| RIC-Seq | RNA In situ Conformation sequencing |
| AGO-CLIP | Argonaut CrossLinked ImmunoPrecipitation |
| HiCLIP | RNA Hybrid and Individual-nucleotide resolution CrossLinking Immunoprecipitation |
| PIP-seq | Protein Interaction Profile sequencing |
| PARS | Parallel Analysis of RNA Structure |
| PARTE | Parallel Analysis of RNA structures with Temperature Elevation |
| Frag-seq | FRAGmentation sequencing |
| SHAPE-seq | Selective 2′-Hydroxyl Acylation analysed by Primer Extension and sequencing |
| LIGR-seq | LIGation of interacting RNA followed by high-throughput sequencing |
| PARIS | Psoralen Analysis of RNA Interactions and Structures |
| SPLASH | sequencing of psoralen crosslinked, ligated, and selected hybrids |
| IRES | Internal Ribosome entry site |
References
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The Reference Human Genome Annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Wery, M.; Kwapisz, M.; Morillon, A. Noncoding RNAs in Gene Regulation. WIREs Syst. Biol. Med. 2011, 3, 728–738. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Guil, S.; Esteller, M. RNA–RNA Interactions in Gene Regulation: The Coding and Noncoding Players. Trends Biochem. Sci. 2015, 40, 248–256. [Google Scholar] [CrossRef]
- Singh, S.; Shyamal, S.; Panda, A.C. Detecting RNA–RNA Interactome. WIREs RNA 2022, 13, e1715. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional Interactions among MicroRNAs and Long Noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for SnoRNAs and SnoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef]
- Valadkhan, S.; Gunawardane, L.S. Role of Small Nuclear RNAs in Eukaryotic Gene Expression. Essays Biochem. 2013, 54, 79–90. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as MiRNA Sponges. In Advances in Experimental Medicine and Biology; Springer LLC: New York, NY, USA, 2018; Volume 1087, pp. 67–79. [Google Scholar]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A Census of Human RNA-Binding Proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Kudla, G.; Granneman, S.; Hahn, D.; Beggs, J.D.; Tollervey, D. Cross-Linking, Ligation, and Sequencing of Hybrids Reveals RNA–RNA Interactions in Yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 10010–10015. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP Decodes MicroRNA–MRNA Interaction Maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Chakrabarti, A.M.; Luscombe, N.M.; Ule, J. Using HiCLIP to Identify RNA Duplexes That Interact with a Specific RNA-Binding Protein. Nat. Protoc. 2017, 12, 611–637. [Google Scholar] [CrossRef]
- Foley, S.W.; Gregory, B.D. Protein Interaction Profile Sequencing (PIP-seq). Curr. Protoc. Mol. Biol. 2016, 116, 27.5.1–27.5.15. [Google Scholar] [CrossRef]
- Nguyen, T.C.; Cao, X.; Yu, P.; Xiao, S.; Lu, J.; Biase, F.H.; Sridhar, B.; Huang, N.; Zhang, K.; Zhong, S. Mapping RNA–RNA Interactome and RNA Structure in Vivo by MARIO. Nat. Commun. 2016, 7, 12023. [Google Scholar] [CrossRef]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-Seq for Global in Situ Profiling of RNA–RNA Spatial Interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Chang, H.Y. Genome-Wide Mapping of RNA Structure Using Nuclease Digestion and High-Throughput Sequencing. Nat. Protoc. 2013, 8, 849–869. [Google Scholar] [CrossRef]
- Loughrey, D.; Watters, K.E.; Settle, A.H.; Lucks, J.B. SHAPE-Seq 2.0: Systematic Optimization and Extension of High-Throughput Chemical Probing of RNA Secondary Structure with next Generation Sequencing. Nucleic Acids Res. 2014, 42, e165. [Google Scholar] [CrossRef]
- Underwood, J.G.; Uzilov, A.V.; Katzman, S.; Onodera, C.S.; Mainzer, J.E.; Mathews, D.H.; Lowe, T.M.; Salama, S.R.; Haussler, D. FragSeq: Transcriptome-Wide RNA Structure Probing Using High-Throughput Sequencing. Nat. Methods 2010, 7, 995–1001. [Google Scholar] [CrossRef]
- Lu, Z.; Gong, J.; Zhang, Q.C. PARIS: Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution. In RNA Detection: Methods and Protocols; Springer: New York, NY, USA, 2018; pp. 59–84. [Google Scholar]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.-L.; Tapsin, S.; Chan, Y.-S.; Tan, C.-P.; Sim, A.Y.L.; et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Zhao, H.; Wang, X.; Xue, Y. Technological Advancements in Deciphering RNA-RNA Interactions. Mol. Cell 2024, 84, 3722–3736. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. The Biogenesis and Emerging Roles of Circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-CircRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Patop, I.L.; Kadener, S. CircRNAs in Cancer. Curr. Opin. Genet. Dev. 2018, 48, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Conn, V.M.; Gabryelska, M.; Toubia, J.; Kirk, K.; Gantley, L.; Powell, J.A.; Cildir, G.; Marri, S.; Liu, R.; Stringer, B.W.; et al. Circular RNAs Drive Oncogenic Chromosomal Translocations within the MLL Recombinome in Leukemia. Cancer Cell 2023, 41, 1309–1326.e10. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A CircRNA from SEPALLATA3 Regulates Splicing of Its Cognate MRNA through R-Loop Formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A Length-Dependent Evolutionarily Conserved Pathway Controls Nuclear Export of Circular RNAs. Genes. Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Lin, J.; Song, Z.; Wang, Q.; Zhao, W.; Wang, Y.; Xiu, X.; Deng, Y.; Li, X.; et al. Exportin 4 Depletion Leads to Nuclear Accumulation of a Subset of Circular RNAs. Nat. Commun. 2022, 13, 5769. [Google Scholar] [CrossRef]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and MiR-7 in Cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes with P21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA That Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous MicroRNA Sponges: Evidence and Controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B. Signal and Noise in CircRNA Translation. Methods 2021, 196, 68–73. [Google Scholar] [CrossRef]
- Rossi, F.; Beltran, M.; Damizia, M.; Grelloni, C.; Colantoni, A.; Setti, A.; Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Nicoletti, C.; et al. Circular RNA ZNF609/CKAP5 MRNA Interaction Regulates Microtubule Dynamics and Tumorigenicity. Mol. Cell 2022, 82, 75–89.e9. [Google Scholar] [CrossRef]
- Rossi, F.; Legnini, I.; Megiorni, F.; Colantoni, A.; Santini, T.; Morlando, M.; Di Timoteo, G.; Dattilo, D.; Dominici, C.; Bozzoni, I. Circ-ZNF609 Regulates G1-S Progression in Rhabdomyosarcoma. Oncogene 2019, 38, 3843–3854. [Google Scholar] [CrossRef]
- Mazan-Mamczarz, K.; Galbán, S.; De Silanes, I.L.; Martindale, J.L.; Atasoy, U.; Keene, J.D.; Gorospe, M. RNA-Binding Protein HuR Enhances P53 Translation in Response to Ultraviolet Light Irradiation. Proc. Natl. Acad. Sci. USA 2003, 100, 8354–8359. [Google Scholar] [CrossRef]
- Galbán, S.; Kuwano, Y.; Pullmann, R.; Martindale, J.L.; Kim, H.H.; Lal, A.; Abdelmohsen, K.; Yang, X.; Dang, Y.; Liu, J.O.; et al. RNA-Binding Proteins HuR and PTB Promote the Translation of Hypoxia-Inducible Factor 1α. Mol. Cell Biol. 2008, 28, 93–107. [Google Scholar] [CrossRef]
- Beltran, M.; Rossi, F.; Bozzoni, I. CircZNF609 as a Prototype to Elucidate the Biological Function of CircRNA-MRNA Interactions. Mol. Cell Oncol. 2022, 9, 2055939. [Google Scholar] [CrossRef]
- Hafez, A.K.; Zimmerman, A.J.; Papageorgiou, G.; Chandrasekaran, J.; Amoah, S.K.; Lin, R.; Lozano, E.; Pierotti, C.; Dell’Orco, M.; Hartley, B.J.; et al. A Bidirectional Competitive Interaction between CircHomer1 and Homer1b within the Orbitofrontal Cortex Regulates Reversal Learning. Cell Rep. 2022, 38, 110282. [Google Scholar] [CrossRef]
- Cervera-Carles, L.; Dols-Icardo, O.; Molina-Porcel, L.; Alcolea, D.; Cervantes-Gonzalez, A.; Muñoz-Llahuna, L.; Clarimon, J. Assessing Circular RNAs in Alzheimer’s Disease and Frontotemporal Lobar Degeneration. Neurobiol. Aging 2020, 92, 7–11. [Google Scholar] [CrossRef]
- Bronicki, L.M.; Jasmin, B.J. Emerging Complexity of the HuD/ELAVl4 Gene; Implications for Neuronal Development, Function, and Dysfunction. RNA 2013, 19, 1019–1037. [Google Scholar] [CrossRef]
- Ma, Q.; Yang, F.; Huang, B.; Pan, X.; Li, W.; Yu, T.; Wang, X.; Ran, L.; Qian, K.; Li, H.; et al. CircARID1A Binds to IGF2BP3 in Gastric Cancer and Promotes Cancer Proliferation by Forming a CircARID1A-IGF2BP3-SLC7A5 RNA–Protein Ternary Complex. J. Exp. Clin. Cancer Res. 2022, 41, 251. [Google Scholar] [CrossRef]
- Singh, S.; Shyamal, S.; Das, A.; Panda, A.C. Global Identification of MRNA-Interacting Circular RNAs by CLiPPR-Seq. Nucleic Acids Res. 2024, 52, e29. [Google Scholar] [CrossRef] [PubMed]
- Silenzi, V.; D’Ambra, E.; Santini, T.; D’Uva, S.; Setti, A.; Salvi, N.; Nicoletti, C.; Scarfò, R.; Cordella, F.; Mongiardi, B.; et al. A Tripartite CircRNA/MRNA/MiRNA Interaction Regulates Glutamatergic Signaling in the Mouse Brain. Cell Rep. 2024, 43, 114766. [Google Scholar] [CrossRef]
- Grelloni, C.; Garraffo, R.; Setti, A.; Rossi, F.; Peruzzi, G.; Cinquanta, M.; Di Rosa, M.C.; Pierotti, M.A.; Beltran, M.; Bozzoni, I. BRCA1 Levels and DNA-Damage Response Are Controlled by the Competitive Binding of CircHIPK3 or FMRP to the BRCA1 MRNA. Mol. Cell 2024, 84, 4079–4094.e10. [Google Scholar] [CrossRef]
- Ascano, M.; Mukherjee, N.; Bandaru, P.; Miller, J.B.; Nusbaum, J.D.; Corcoran, D.L.; Langlois, C.; Munschauer, M.; Dewell, S.; Hafner, M.; et al. FMRP Targets Distinct MRNA Sequence Elements to Regulate Protein Expression. Nature 2012, 492, 382–386. [Google Scholar] [CrossRef]
- Kurosaki, T.; Mitsutomi, S.; Hewko, A.; Akimitsu, N.; Maquat, L.E. Integrative Omics Indicate FMRP Sequesters MRNA from Translation and Deadenylation in Human Neuronal Cells. Mol. Cell 2022, 82, 4564–4581.e11. [Google Scholar] [CrossRef]
- Boo, S.H.; Shin, M.-K.; Hwang, H.J.; Hwang, H.; Chang, S.; Kim, T.; Baek, D.; Kim, Y.K. Circular RNAs Trigger Nonsense-Mediated MRNA Decay. Mol. Cell 2024, 84, 4862–4877. [Google Scholar] [CrossRef]
- Hir, H.L.; Saulière, J.; Wang, Z. The Exon Junction Complex as a Node of Post-Transcriptional Networks. Nat. Rev. Mol. Cell Biol. 2016, 17, 41–54. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Supek, F.; Lehner, B. The Rules and Impact of Nonsense-Mediated MRNA Decay in Human Cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef]
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense Surveillance Regulates Expression of Diverse Classes of Mammalian Transcripts and Mutes Genomic Noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L. Bim: A Novel Member of the Bcl-2 Family That Promotes Apoptosis. EMBO J. 1998, 17, 384–395. [Google Scholar] [CrossRef]
- Luo, S.; Garcia-Arencibia, M.; Zhao, R.; Puri, C.; Toh, P.P.C.; Sadiq, O.; Rubinsztein, D.C. Bim Inhibits Autophagy by Recruiting Beclin 1 to Microtubules. Mol. Cell 2012, 47, 359–370. [Google Scholar] [CrossRef]
- Yi, J.; Du, J.; Chen, X.; Nie, R.; Hu, G.; Wang, L.; Zhang, Y.; Chen, S.; Wen, X.; Luo, D.; et al. A CircRNA–MRNA Pairing Mechanism Regulates Tumor Growth and Endocrine Therapy Resistance in ER-Positive Breast Cancer. Proc. Natl. Acad. Sci. USA 2025, 122, e2420383122. [Google Scholar] [CrossRef]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a Therapeutic Target in Cancer. Nat. Rev. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.-M.; Lin, X.; Kim, Y.; Wong, D.T.W.; Xiao, X. The Landscape of MicroRNA, Piwi-Interacting RNA, and Circular RNA in Human Saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef]
- Sun, M.; Yang, Y. Biological Functions and Applications of CircRNAs—Next Generation of RNA-Based Therapy. J. Mol. Cell Biol. 2023, 15, mjad031. [Google Scholar] [CrossRef]
| Work | circRNA | mRNA Interactor | Effect on mRNA | Duplex Length (nt) | Pairing Region (mRNA) |
|---|---|---|---|---|---|
| Rossi et al. [47] | circZNF609 | CKAP5 | Positive | ~60 | 3′ UTR |
| Hafez et al. [52] | circHOMER1 | HOMER1B | Negative | ~30 | 3′ UTR |
| Ma et al. [55] | circARID1A | SLC7A5 | Positive | ~90 | CDS |
| Singh et al. [56] | circACBD3 | HPCA | Negative | ~20 | 3′ UTR |
| ARFGAP1 | Negative | ~20 | 3′ UTR | ||
| circMTCL1 | Rps6kc1 | Positive | ~30 | CDS | |
| 2310039H08Rik | Negative | ~20 | CDS | ||
| Silenzi et al. [57] | circDLC1 | Gria1 | Negative | Not specified | 3′ UTR |
| Grin2a | |||||
| Grelloni et al. [58] | circHIPK3 | BRCA1 | Positive | ~40 | Last coding exon |
| Boo et al. [61] | circ_0002082 | BCL2L11 | Negative | ~30 | 3′ UTR |
| circ_0008496 | |||||
| Yi et al. [67] | circFOXK2 | CCND1 | Positive | ~80 | 3′ UTR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garraffo, R.; Beltran Nebot, M. Direct circRNA-mRNA Binding Controls mRNA Fate: A New Mechanism for circRNAs. Non-Coding RNA 2025, 11, 53. https://doi.org/10.3390/ncrna11040053
Garraffo R, Beltran Nebot M. Direct circRNA-mRNA Binding Controls mRNA Fate: A New Mechanism for circRNAs. Non-Coding RNA. 2025; 11(4):53. https://doi.org/10.3390/ncrna11040053
Chicago/Turabian StyleGarraffo, Raffaele, and Manuel Beltran Nebot. 2025. "Direct circRNA-mRNA Binding Controls mRNA Fate: A New Mechanism for circRNAs" Non-Coding RNA 11, no. 4: 53. https://doi.org/10.3390/ncrna11040053
APA StyleGarraffo, R., & Beltran Nebot, M. (2025). Direct circRNA-mRNA Binding Controls mRNA Fate: A New Mechanism for circRNAs. Non-Coding RNA, 11(4), 53. https://doi.org/10.3390/ncrna11040053






