Analysis of RNA Transcribed by RNA Polymerase III from B2 SINEs in Mouse Cells
Abstract
1. Introduction
2. Results
2.1. Detection of B2 RNA in Mouse Cell Cultures and Tissues
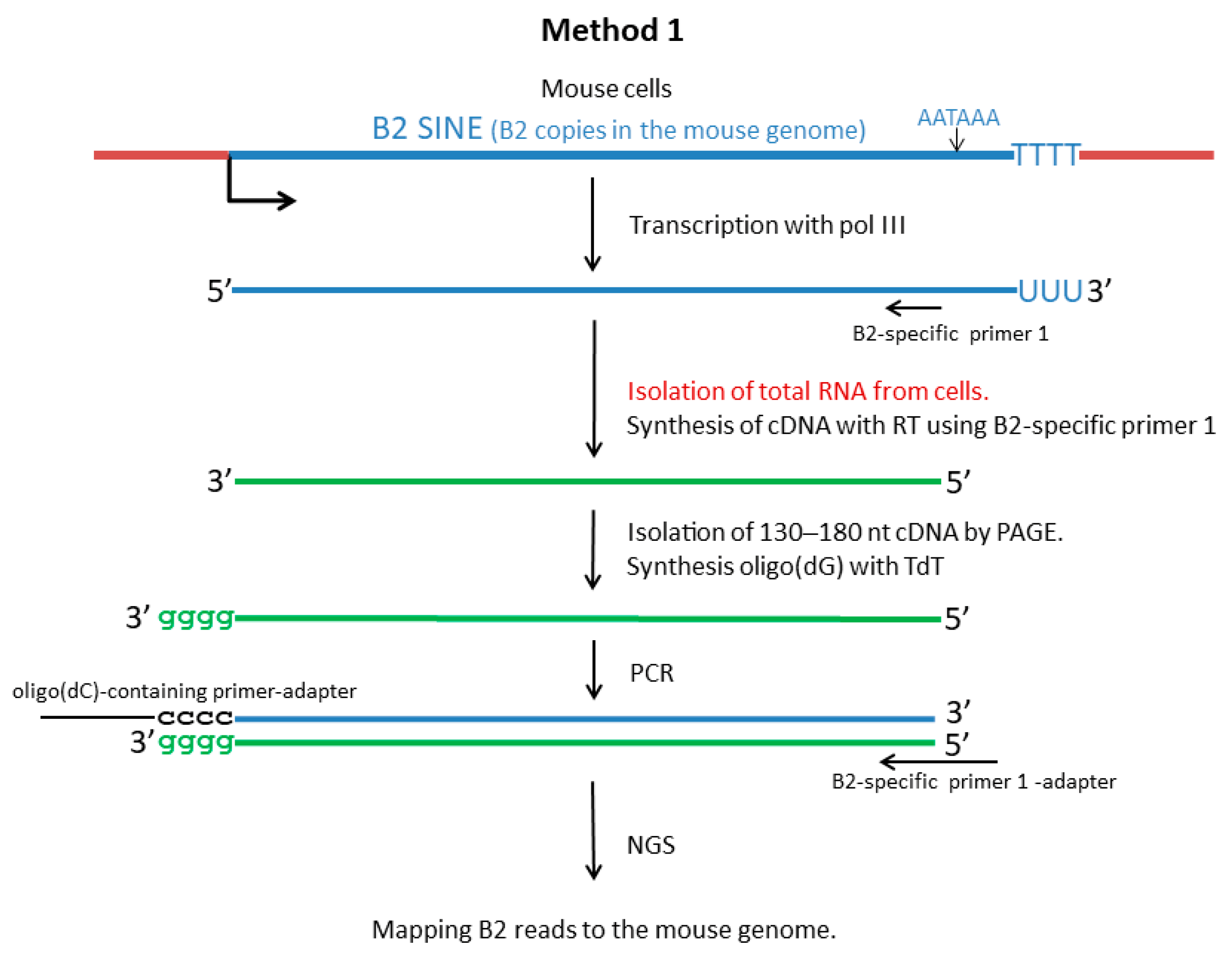
2.2. NGS of B2 Transcripts (Method 1)
2.3. NGS of B2 Transcripts (Method 2)
2.3.1. The Method Concept and Initial Analysis of NGS Data
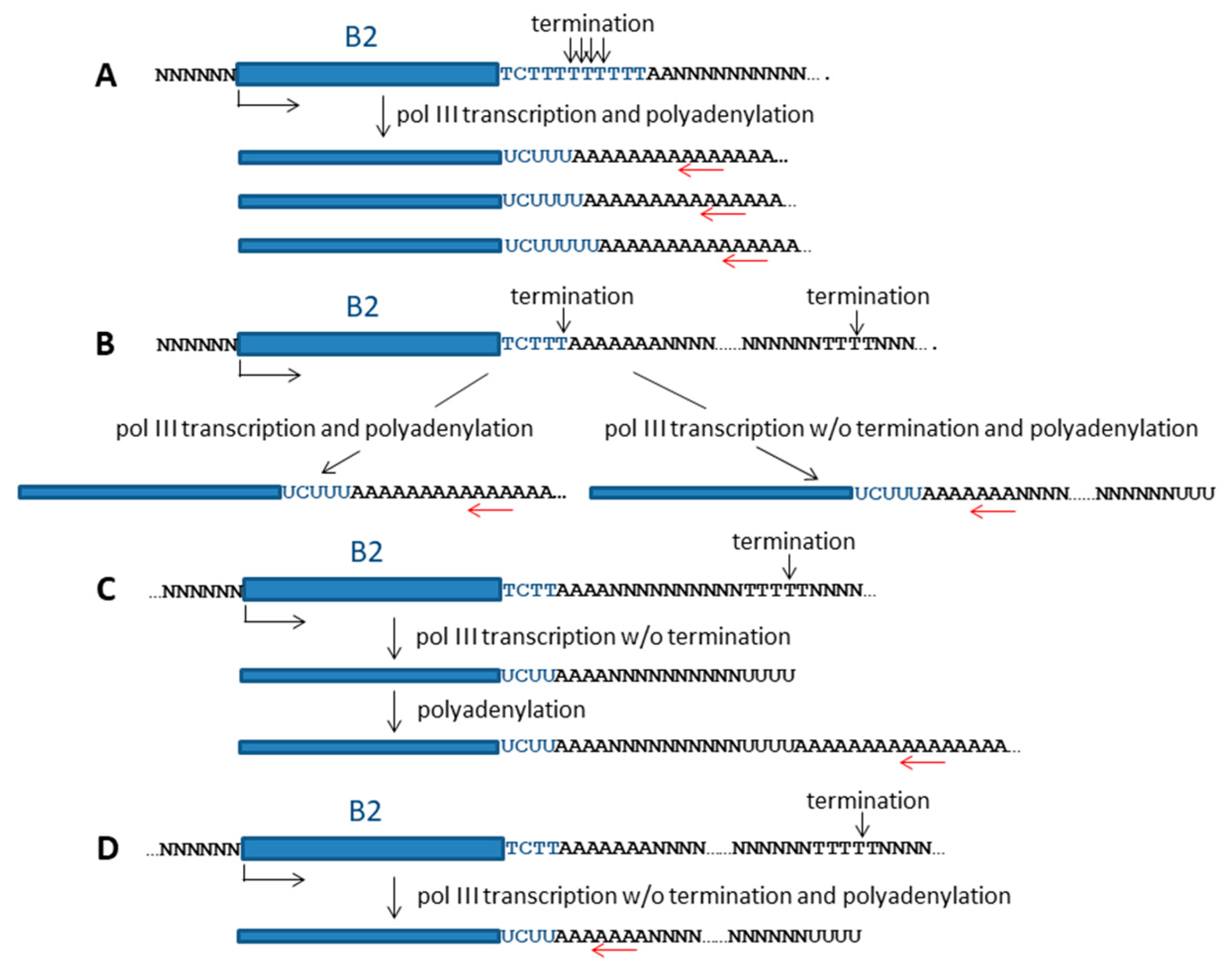
2.3.2. Evidence for the Polyadenylation Capacity of Pol III Transcripts of B2 Copies in Mouse Cells
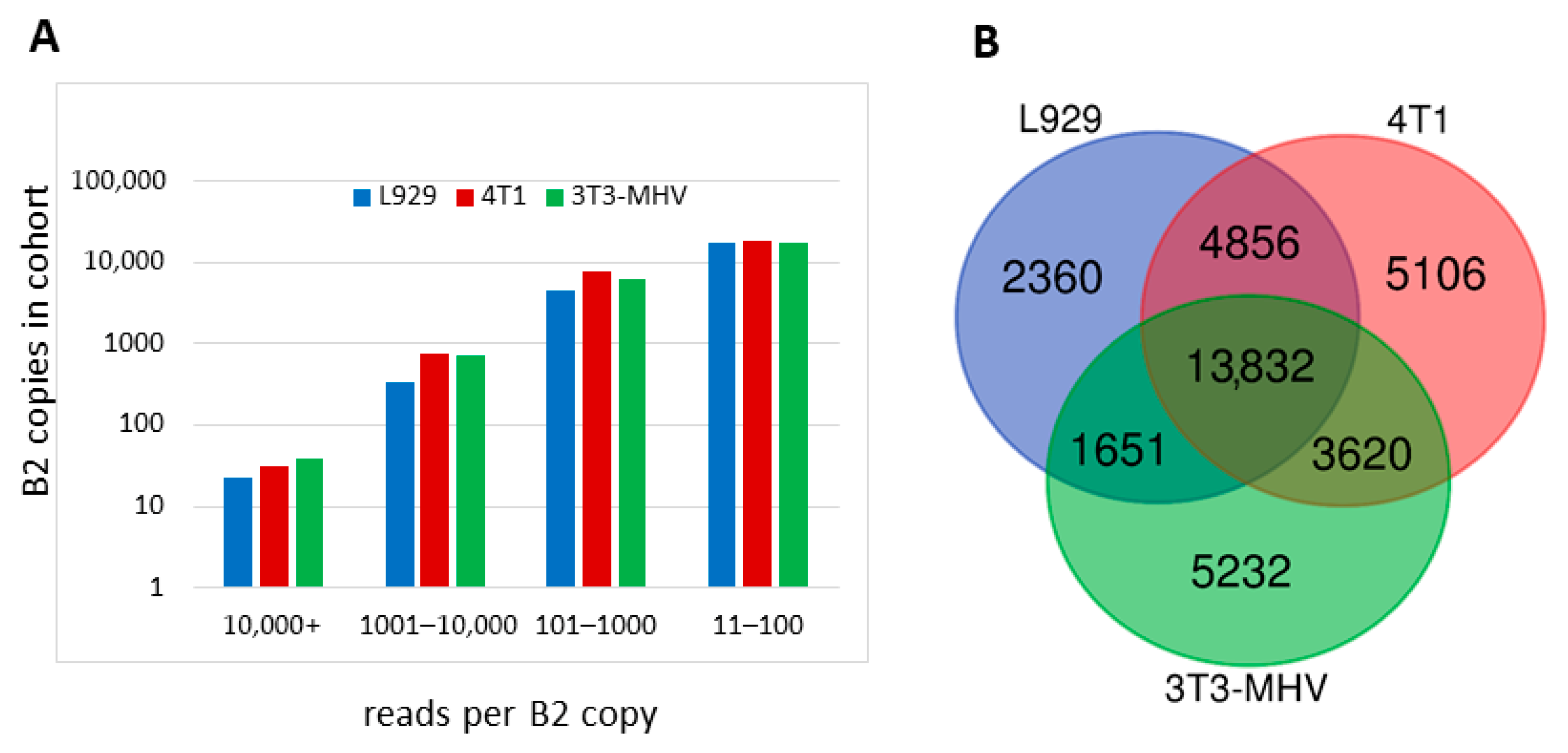
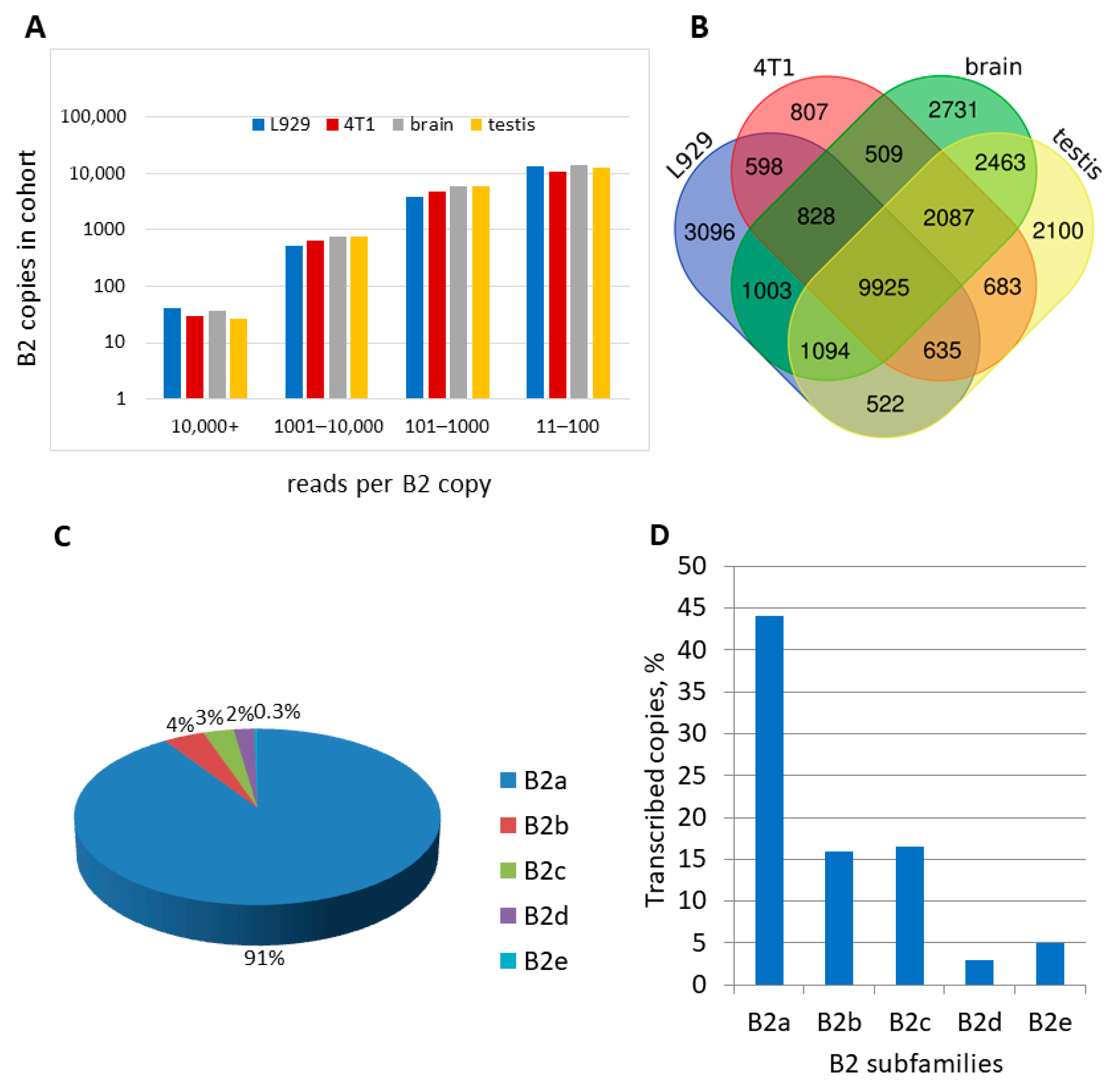
2.3.3. Comparison of B2 Transcription Termination Profiles in Different Murine Cells
2.3.4. Analysis of the Number and Types of B2 Copies Whose Pol III Transcripts Can Be Polyadenylated in Mouse Cells
3. Discussion
3.1. Comparison of Approaches to Sequencing B2 Transcripts
3.2. Termination of Transcription and Polyadenylation of B2 Transcripts
4. Materials and Methods
4.1. Cell Cultures and Mouse Organs
4.2. RNA Isolation and Northern-Blot Analysis
4.3. Preparation of cDNA Libraries and NGS
4.4. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next-Generation Sequencing |
| SINE | Short Interspersed Element |
| LINE | Long Interspersed Element |
| pol III | RNA polymerase III |
| pol II | RNA polymerase II |
| PA | Polyadenylation |
| PAS | Polyadenylation signal |
| mPSF | mammalian Polyadenylation Specificity Factor |
| CFIm | Cleavage Factor I mammalian |
References
- Kramerov, D.A.; Vassetzky, N.S. SINEs. Wiley Interdiscip. Rev. RNA 2011, 2, 772–786. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Vassetzky, N.S.; Kramerov, D.A. SINEBase: A database and tool for SINE analysis. Nucleic Acids Res. 2013, 41, D83–D89. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Ferec, C.; Cooper, D.N. LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease: Mutation detection bias and multiple mechanisms of target gene disruption. J. Biomed. Biotechnol. 2006, 2006, 56182. [Google Scholar] [CrossRef]
- Ferrigno, O.; Virolle, T.; Djabari, Z.; Ortonne, J.P.; White, R.J.; Aberdam, D. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat. Genet. 2001, 28, 77–81. [Google Scholar] [CrossRef]
- Su, M.; Han, D.; Boyd-Kirkup, J.; Yu, X.; Han, J.J. Evolution of Alu elements toward enhancers. Cell Rep. 2014, 7, 376–385. [Google Scholar] [CrossRef]
- Policarpi, C.; Crepaldi, L.; Brookes, E.; Nitarska, J.; French, S.M.; Coatti, A.; Riccio, A. Enhancer SINEs Link Pol III to Pol II Transcription in Neurons. Cell Rep. 2017, 21, 2879–2894. [Google Scholar] [CrossRef]
- Krull, M.; Brosius, J.; Schmitz, J. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 2005, 22, 1702–1711. [Google Scholar] [CrossRef]
- Wang, W.; Kirkness, E.F. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005, 15, 1798–1808. [Google Scholar] [CrossRef]
- Chen, C.; Ara, T.; Gautheret, D. Using Alu elements as polyadenylation sites: A case of retroposon exaptation. Mol. Biol. Evol. 2009, 26, 327–334. [Google Scholar] [CrossRef]
- Choi, J.D.; Del Pinto, L.A.; Sutter, N.B. SINE retrotransposons import polyadenylation signals to 3′UTRs in dog (Canis familiaris). Mob. DNA 2025, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Zovoilis, A.; Cifuentes-Rojas, C.; Chu, H.P.; Hernandez, A.J.; Lee, J.T. Destabilization of B2 RNA by EZH2 Activates the Stress Response. Cell 2016, 167, 1788–1802.E13. [Google Scholar] [CrossRef] [PubMed]
- Ponicsan, S.L.; Kugel, J.F.; Goodrich, J.A. Genomic gems: SINE RNAs regulate mRNA production. Curr. Opin. Genet. Dev. 2010, 20, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, A.A.; Guffanti, G.; Hunter, R.G. B2 SINE RNA as a novel regulator of glucocorticoid receptor transcriptional activity. Neurobiol. Stress 2023, 23, 100522. [Google Scholar] [CrossRef]
- Haynes, S.R.; Jelinek, W.R. Low molecular weight RNAs transcribed in vitro by RNA polymerase III from Alu-type dispersed repeats in Chinese hamster DNA are also found in vivo. Proc. Natl. Acad. Sci. USA 1981, 78, 6130–6134. [Google Scholar] [CrossRef]
- Krayev, A.S.; Markusheva, T.V.; Kramerov, D.A.; Ryskov, A.P.; Skryabin, K.G.; Bayev, A.A.; Georgiev, G.P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982, 10, 7461–7475. [Google Scholar] [CrossRef]
- Daniels, G.R.; Deininger, P.L. Repeat sequence families derived from mammalian tRNA genes. Nature 1985, 317, 819–822. [Google Scholar] [CrossRef]
- Sakamoto, K.; Okada, N. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J. Mol. Evol. 1985, 22, 134–140. [Google Scholar] [CrossRef]
- Vassetzky, N.S.; Borodulina, O.R.; Ustyantsev, I.G.; Kosushkin, S.A.; Kramerov, D.A. Analysis of SINE Families B2, Dip, and Ves with Special Reference to Polyadenylation Signals and Transcription Terminators. Int. J. Mol. Sci. 2021, 22, 9897. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Ichiyanagi, T.; Katoh, H.; Mori, Y.; Hirafuku, K.; Boyboy, B.A.; Kawase, M.; Ichiyanagi, K. B2 SINE Copies Serve as a Transposable Boundary of DNA Methylation and Histone Modifications in the Mouse. Mol. Biol. Evol. 2021, 38, 2380–2395. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Kominami, R.; Mishima, Y.; Okada, N. The 6S RNA transcribed from rodent total DNA in vitro is the transcript of the type 2 Alu family. Mol. Gen. Genet. 1984, 194, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kramerov, D.A.; Lekakh, I.V.; Samarina, O.P.; Ryskov, A.P. The sequences homologous to major interspersed repeats B1 and B2 of mouse genome are present in mRNA and small cytoplasmic poly(A) + RNA. Nucleic Acids Res. 1982, 10, 7477–7491. [Google Scholar] [CrossRef]
- Bladon, T.S.; Fregeau, C.J.; McBurney, M.W. Synthesis and processing of small B2 transcripts in mouse embryonal carcinoma cells. Mol. Cell. Biol. 1990, 10, 4058–4067. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Tillib, S.V.; Shumyatsky, G.P.; Georgiev, G.P. The most abundant nascent poly(A) + RNAs are transcribed by RNA polymerase III in murine tumor cells. Nucleic Acids Res. 1990, 18, 4499–4506. [Google Scholar] [CrossRef]
- Borodulina, O.R.; Kramerov, D.A. Short interspersed elements (SINEs) from insectivores. Two classes of mammalian SINEs distinguished by A-rich tail structure. Mamm. Genome 2001, 12, 779–786. [Google Scholar] [CrossRef]
- Borodulina, O.R.; Golubchikova, J.S.; Ustyantsev, I.G.; Kramerov, D.A. Polyadenylation of RNA transcribed from mammalian SINEs by RNA polymerase III: Complex requirements for nucleotide sequences. Biochim. Biophys. Acta 2016, 1859, 355–365. [Google Scholar] [CrossRef]
- Ustyantsev, I.G.; Kosushkin, S.A.; Borodulina, O.R.; Vassetzky, N.S.; Kramerov, D.A. Ere, a Family of Short Interspersed Elements in the Genomes of Odd-Toed Ungulates (Perissodactyla). Animals 2024, 14, 1982. [Google Scholar] [CrossRef]
- Ustyantsev, I.G.; Borodulina, O.R.; Kramerov, D.A. Identification of nucleotide sequences and some proteins involved in polyadenylation of RNA transcribed by Pol III from SINEs. RNA Biol. 2021, 18, 1475–1488. [Google Scholar] [CrossRef]
- Ustyantsev, I.G.; Borodulina, O.R.; Kramerov, D.A. Participation of Proteins of the CPSF Complex in Polyadenylation of Transcripts Read by RNA Polymerase III from SINEs. Mol. Biol. 2024, 58, 494–503. [Google Scholar] [CrossRef]
- Ustyantsev, I.G.; Tatosyan, K.A.; Stasenko, D.V.; Kochanova, N.Y.; Borodulina, O.R.; Kramerov, D.A. [Polyadenylation of Sine Transcripts Generated by RNA Polymerase III Dramatically Prolongs Their Lifetime in Cells]. Mol. Biol. 2020, 54, 78–86. [Google Scholar] [CrossRef]
- Dewannieux, M.; Heidmann, T. Role of poly(A) tail length in Alu retrotransposition. Genomics 2005, 86, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Roy-Engel, A.M.; Salem, A.H.; Oyeniran, O.O.; Deininger, L.; Hedges, D.J.; Kilroy, G.E.; Batzer, M.A.; Deininger, P.L. Active Alu element “A-tails”: Size does matter. Genome Res. 2002, 12, 1333–1344. [Google Scholar] [CrossRef]
- Roy-Engel, A.M. LINEs, SINEs and other retroelements: Do birds of a feather flock together? Front. Biosci. (Landmark Ed.) 2012, 17, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, B.J.; Hedges, D.J.; Derbes, R.S.; Campos Sanchez, R.; Chiaromonte, F.; Makova, K.D.; Roy-Engel, A.M. Rescuing Alu: Recovery of new inserts shows LINE-1 preserves Alu activity through A-tail expansion. PLoS Genet. 2012, 8, e1002842. [Google Scholar] [CrossRef]
- Gogolevskaya, I.K.; Kramerov, D.A. Evolutionary history of 4.5SI RNA and indication that it is functional. J. Mol. Evol. 2002, 54, 354–364. [Google Scholar] [CrossRef]
- Karijolich, J.; Zhao, Y.; Alla, R.; Glaunsinger, B. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017, 45, 6194–6208. [Google Scholar] [CrossRef]
- Orioli, A.; Pascali, C.; Quartararo, J.; Diebel, K.W.; Praz, V.; Romascano, D.; Percudani, R.; van Dyk, L.F.; Hernandez, N.; Teichmann, M.; et al. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res. 2011, 39, 5499–5512. [Google Scholar] [CrossRef]
- Linker, S.B.; Randolph-Moore, L.; Kottilil, K.; Qiu, F.; Jaeger, B.N.; Barron, J.; Gage, F.H. Identification of bona fide B2 SINE retrotransposon transcription through single-nucleus RNA-seq of the mouse hippocampus. Genome Res. 2020, 30, 1643–1654. [Google Scholar] [CrossRef]
- Oomen, M.E.; Rodriguez-Terrones, D.; Kurome, M.; Zakhartchenko, V.; Mottes, L.; Simmet, K.; Noll, C.; Nakatani, T.; Mourra-Diaz, C.M.; Aksoy, I.; et al. An atlas of transcription initiation reveals regulatory principles of gene and transposable element expression in early mammalian development. Cell 2025, 188, 1156–1174.e1120. [Google Scholar] [CrossRef]
- Proudfoot, N.J. Ending the message: Poly(A) signals then and now. Genes Dev. 2011, 25, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hamilton, K.; Tong, L. Recent molecular insights into canonical pre-mRNA 3′-end processing. Transcription 2020, 11, 83–96. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Forouzmand, E.; Jeong, J.; Qiao, F.; Sowd, G.A.; Engelman, A.N.; Xie, X.; Hertel, K.J.; Shi, Y. Molecular Mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation. Mol. Cell 2018, 69, 62–74.E4. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Hu, J.; Zhang, H.; Lutz, C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005, 33, 201–212. [Google Scholar] [CrossRef]
- Borodulina, O.R.; Ustyantsev, I.G.; Kramerov, D.A. SINEs as Potential Expression Cassettes: Impact of Deletions and Insertions on Polyadenylation and Lifetime of B2 and Ves SINE Transcripts Generated by RNA Polymerase III. Int. J. Mol. Sci. 2023, 24, 4600. [Google Scholar] [CrossRef]
- Pulaski, B.A.; Ostrand-Rosenberg, S. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 2001, 39, 20–22. [Google Scholar] [CrossRef]
- Senin, V.M.; Buntsevich, A.M.; Afanasyeva, A.V.; Kiseleva, N.S. A new line of murine carcinosarcoma. Exp. Oncol. USSR 1983, 5, 35–39. [Google Scholar]
- Ebralidze, A.; Tulchinsky, E.; Grigorian, M.; Afanasyeva, A.; Senin, V.; Revazova, E.; Lukanidin, E. Isolation and characterization of a gene specifically expressed in different metastatic cells and whose deduced gene product has a high degree of homology to a Ca2+-binding protein family. Genes Dev. 1989, 3, 1086–1093. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303, 3997 v3991. [Google Scholar]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.D.; Tomii, K.; Katoh, K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 2016, 32, 3246–3251. [Google Scholar] [CrossRef]


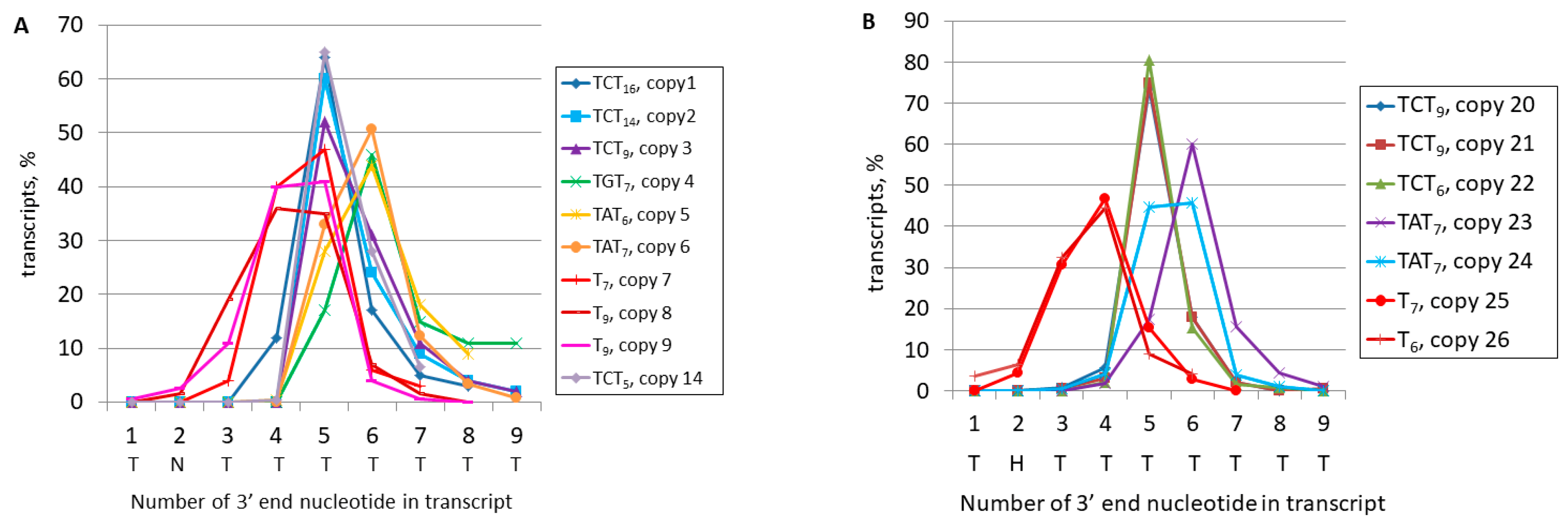
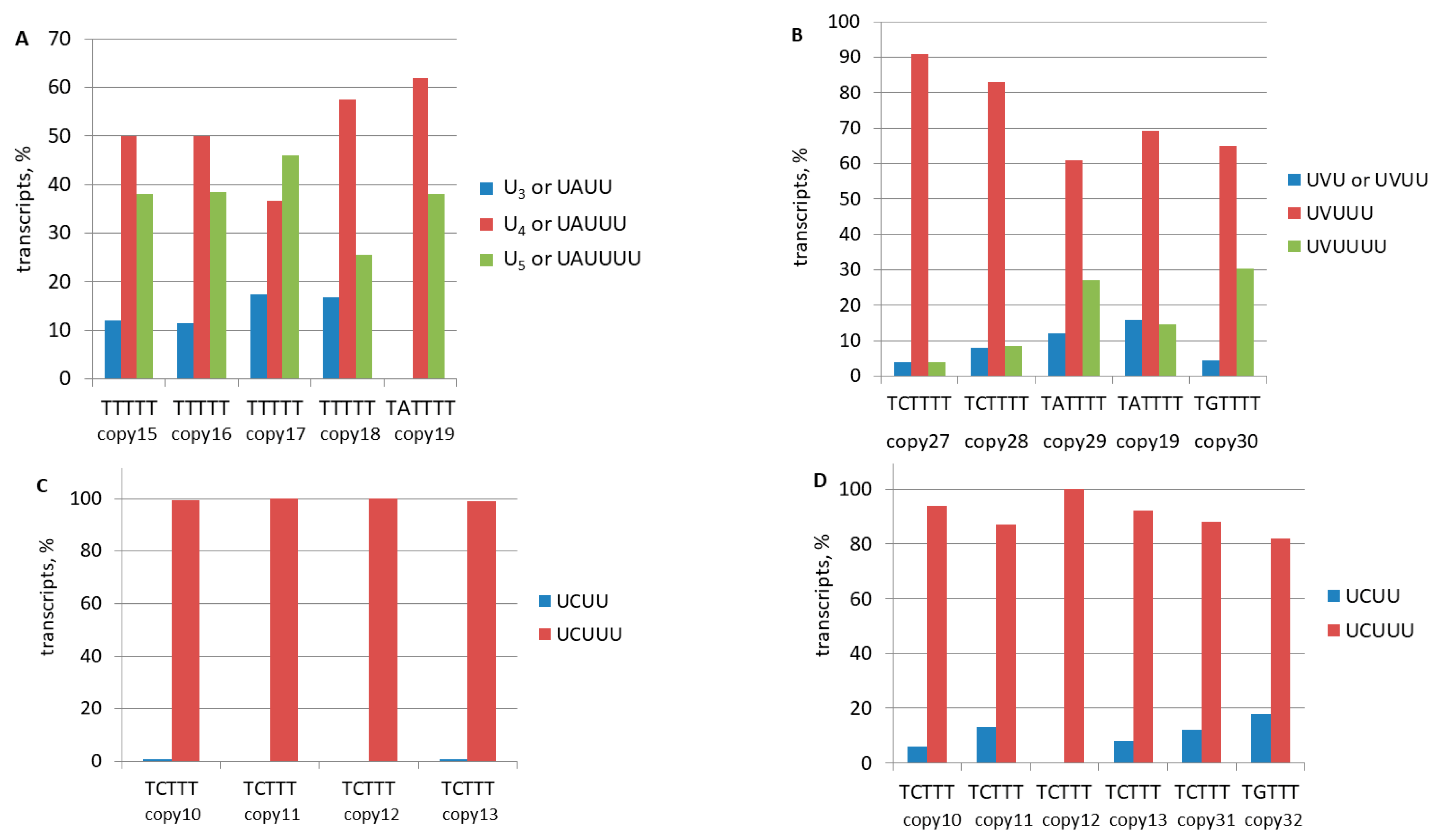
| Copy Number | Coordinates of B2 Copy in the Mouse Genome | Terminator (Uppercase) and Its Flanking Sequences (Lowercase) | Reads in the Library (Number of Reads Analyzed) |
|---|---|---|---|
| copy 1 | chr1:33718543–33718720 | taaaTCTTTTTTTTTTTTTTTTaagttctctgact | 8733 (300) |
| copy 2 | chr7:43662585–43662767 | taaaTCTTTTTTTTTTTTTTaaatctgtgt | 1880 (300) |
| copy 3 | chr1:39288250–39288427 | taaaTCTTTTTTTTTaagagtctggg | 648 (300) |
| copy 4 | chr7:79747308–79747485 | taaaTGTTTTTTTaaaagtatgtatgtc | 17,195 (300) |
| copy 5 | chr13:24776694–24776875 | taaaTATTTTTTaaaagaccatgccata | 6303 (300) |
| copy 6 | chr11:120514490–120514667 | taaaTATTTTTTTaaaaaatgactgaag | 7358 (300) |
| copy 7 | chr11:87432668–87432843 | caaaTTTTTTTcttttttttaaattaggtatttt | 1996 (300) |
| copy 8 * | chr8:111186123–111186298 | aaaaTTTTTTTTTgtacagtctataacaattt | 791 (131) |
| copy 9 | chr15:102348646–102348820 | taaaTTTTTTTTTaaaaaagatatcagtaagg | 4797 (300) |
| copy 10 | chr6:73015125–73015298 | taaaTCTTTaaaaaaatttcatagtctg | 12,839 (284) |
| copy 11 | chr5:121594718–121594889 | taaaTCTTTagactgaagccagcgggg | 254 (126) |
| copy 12 | chr3:107861564–107861740 | taaaTCTTTgggccggagccagcag | 330 (45) |
| copy 13 * | chr8:111186123–111186298 | taaaTCTTTaaaaaatttttttttgtac | 791 (169) |
| copy 14 | chr12:40255314–40255487 | taaaTCTTTTTaaaaaatatgtgcacca | 393 (336) |
| copy 15 | chr2:25257465–25257635 | taaaTTTTTaaaaattagggtgttatgca | 351 (300) |
| copy 16 | chr10:22009277–22009448 | aaaaTTTTTagtgaatatcttgtttgtatt | 35 (26) |
| copy 17 | chr9:60734676–60734853 | taaaTTTTTaaaaatcccctcccatgatg | 1704 (300) |
| copy 18 | chr9:49043798–49043971 | ttaaTTTTTaaaaaagcctgtgagaaaa | 232 (184) |
| copy 19 | chr2:32853715–32853892 | taaaTATTTTaaaaaaaccatccagag | 4115 (300) |
| Copy Number | Coordinates of B2 Copy in the Mouse Genome | Terminator (Uppercase) and Its Flanking Sequences (Lowercase) | Reads in the Library (Number of Reads Analyzed) |
|---|---|---|---|
| copy 20 | chr4:134947766–134947944 | taaaTCTTTTTTTTTaagaaagaaaagg | 7024 (1000) |
| copy 21 | chr17:32112666–32112839 | taaaTCTTTTTTTTTaaaagaaatgtttagc | 4393 (188) |
| copy 22 | chr11:87503376–87503546 | taaaTCTTTTTTaaaaaggaggtaatgaactg | 680 (420) |
| copy 23 | chr1:58466632–58466809 | taaaTATTTTTTTaagaaaaataagtgctag | 923 (600) |
| copy 24 | chr11:97993976–97994153 | taaaTATTTTTTTaaaaatgtcttaccagatg | 67,195 (190) |
| copy 25 | chr9:65300382–65300568 | aaataaTTTTTTTaaagtacaatgagtatat | 183 (183) |
| copy 26 | chr11:89376866–89377041 | taaataaTTTTTTaaaaaaatagataatc | 735 (200) |
| copy 27 | chr13:99830672–99830886 | taaaTCTTTTaaaaaaagtactaaa | 730 (188) |
| copy 28 | chr4:129762801–129762978 | taaaTCTTTTaaaaaaaattttttaaaaagaaaga | 268 (200) |
| copy 29 | chr1:133036101–133036282 | ataaTATTTTaaaaaaacaaacaaacaa | 20,212 (193) |
| copy 30 | chr15:36647144–36647318 | taaaTGTTTTaaagactgaatagttagcaaatg | 1579 (200) |
| copy 31 | chr15:82218406–82218583 | taaaTCTTTgtgtgccctctgtgga | 739 (156) |
| copy 32 | chr4:44554747–44554923 | taaaTGTTTgcttcaagcagctgttggaatcac | 2087 (147) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borodulina, O.R.; Kosushkin, S.A.; Ustyantsev, I.G.; Vassetzky, N.S.; Kramerov, D.A. Analysis of RNA Transcribed by RNA Polymerase III from B2 SINEs in Mouse Cells. Non-Coding RNA 2025, 11, 39. https://doi.org/10.3390/ncrna11030039
Borodulina OR, Kosushkin SA, Ustyantsev IG, Vassetzky NS, Kramerov DA. Analysis of RNA Transcribed by RNA Polymerase III from B2 SINEs in Mouse Cells. Non-Coding RNA. 2025; 11(3):39. https://doi.org/10.3390/ncrna11030039
Chicago/Turabian StyleBorodulina, Olga R., Sergey A. Kosushkin, Ilia G. Ustyantsev, Nikita S. Vassetzky, and Dmitri A. Kramerov. 2025. "Analysis of RNA Transcribed by RNA Polymerase III from B2 SINEs in Mouse Cells" Non-Coding RNA 11, no. 3: 39. https://doi.org/10.3390/ncrna11030039
APA StyleBorodulina, O. R., Kosushkin, S. A., Ustyantsev, I. G., Vassetzky, N. S., & Kramerov, D. A. (2025). Analysis of RNA Transcribed by RNA Polymerase III from B2 SINEs in Mouse Cells. Non-Coding RNA, 11(3), 39. https://doi.org/10.3390/ncrna11030039






