In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact
Abstract
1. Introduction
2. Results
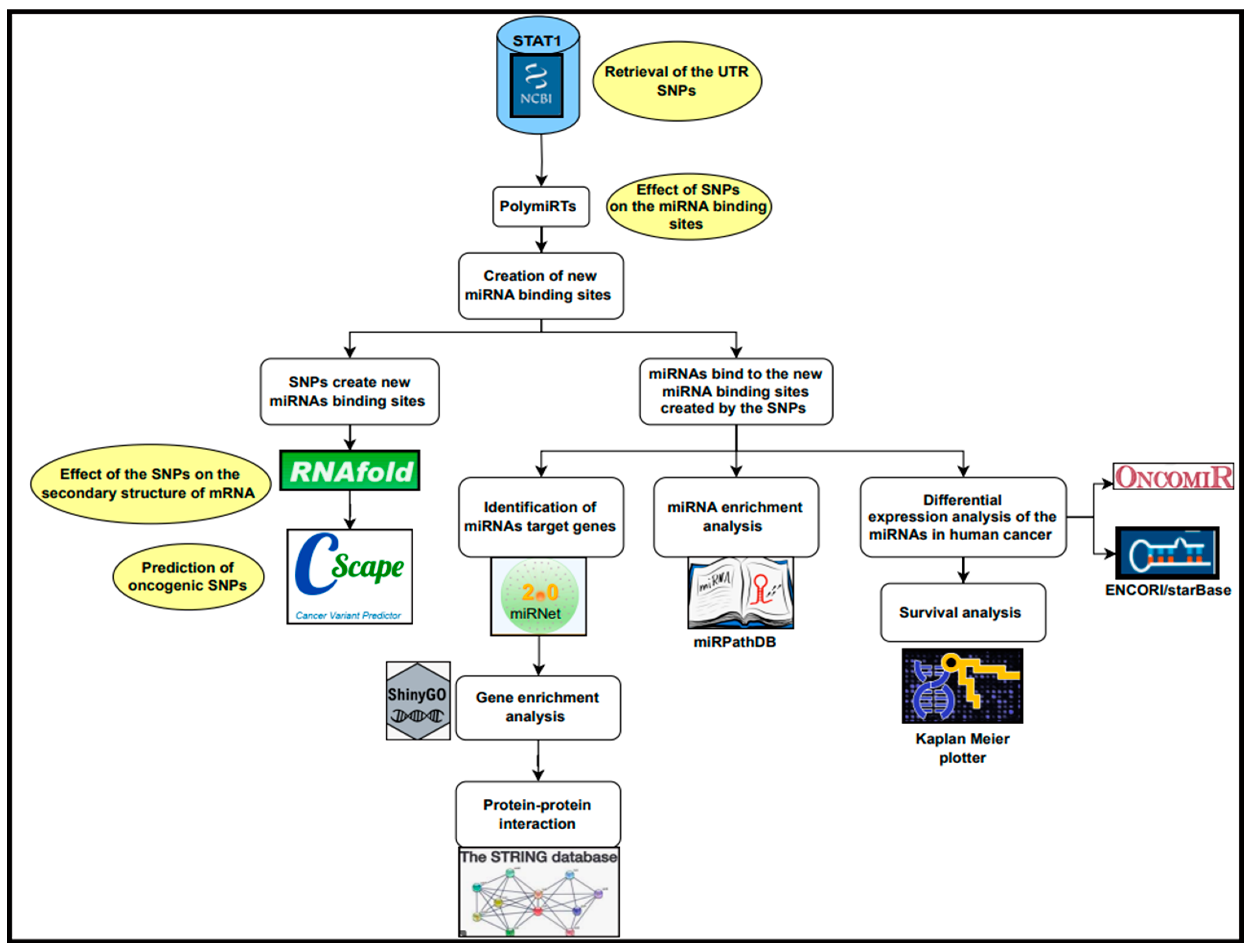
2.1. Retrieval of 3′ UTR SNPs of STAT1 from NCBI
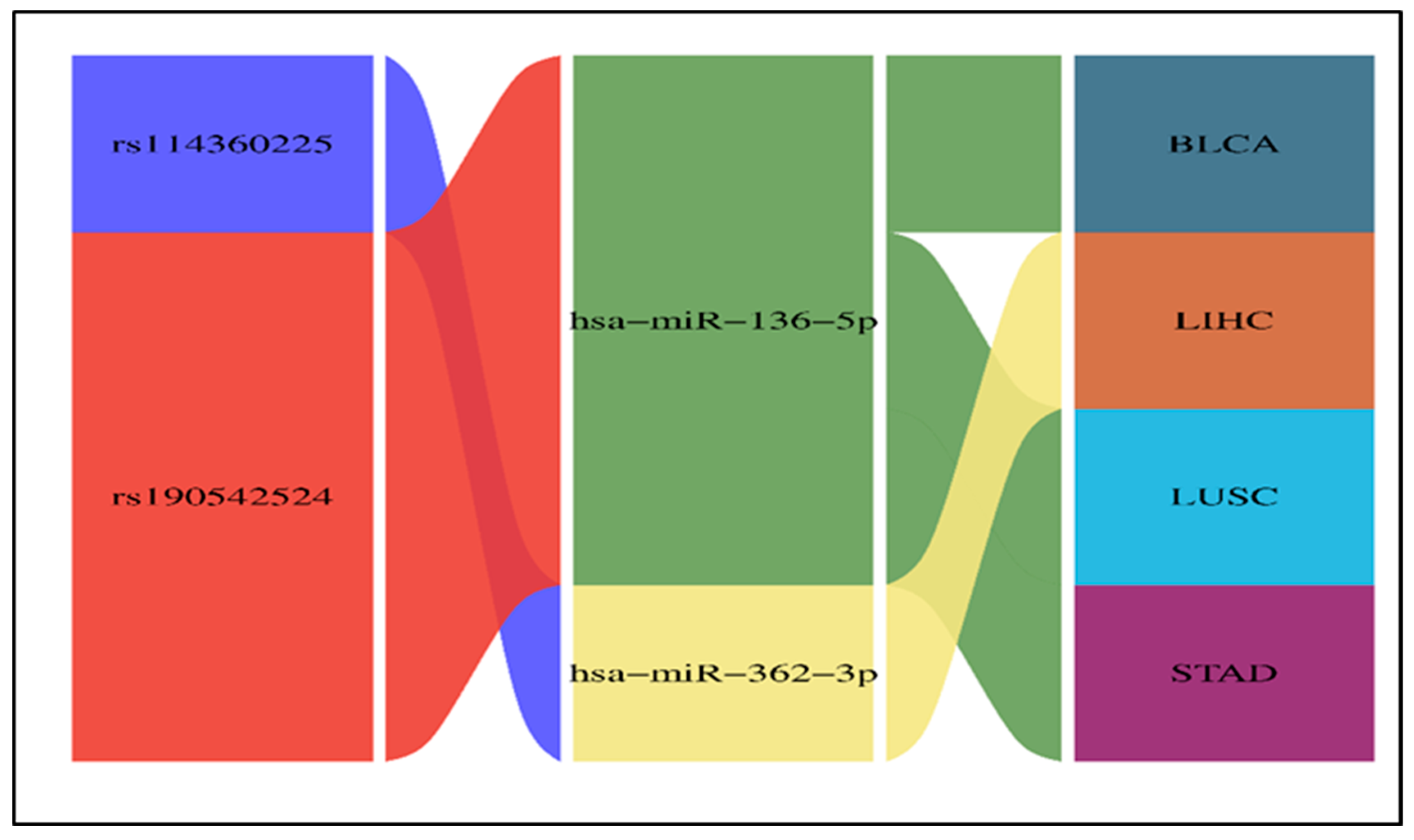
2.2. Results of the Impact of 3′ UTR SNPs on miRNA Binding Sites
2.3. The Effect of 3′ UTR SNPs on the Secondary Structure of mRNA
2.4. Cscape Results of Cancer-Associated 3′ UTR SNPs
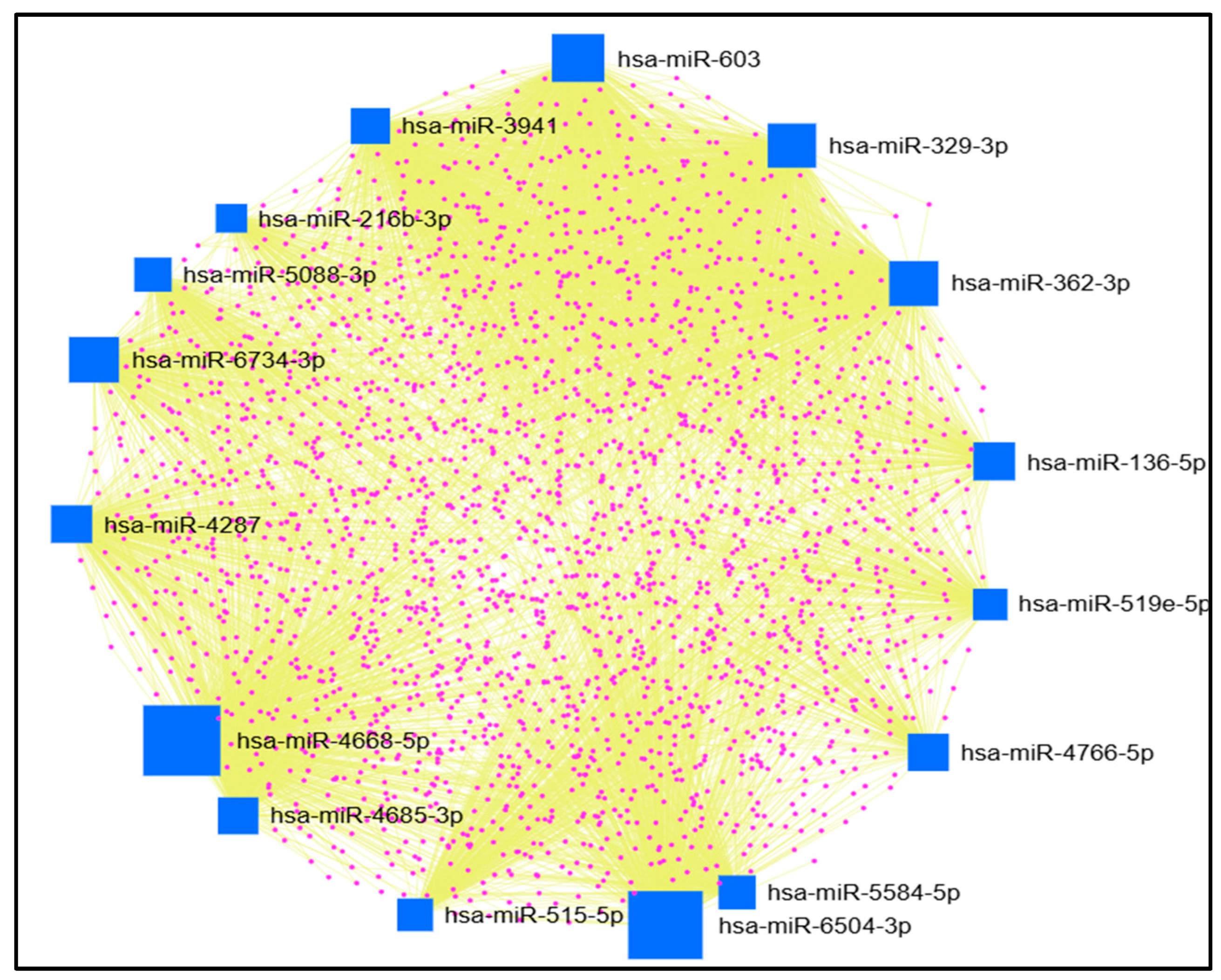
2.5. Results of miRNet Identification of miRNAs’ Target Genes
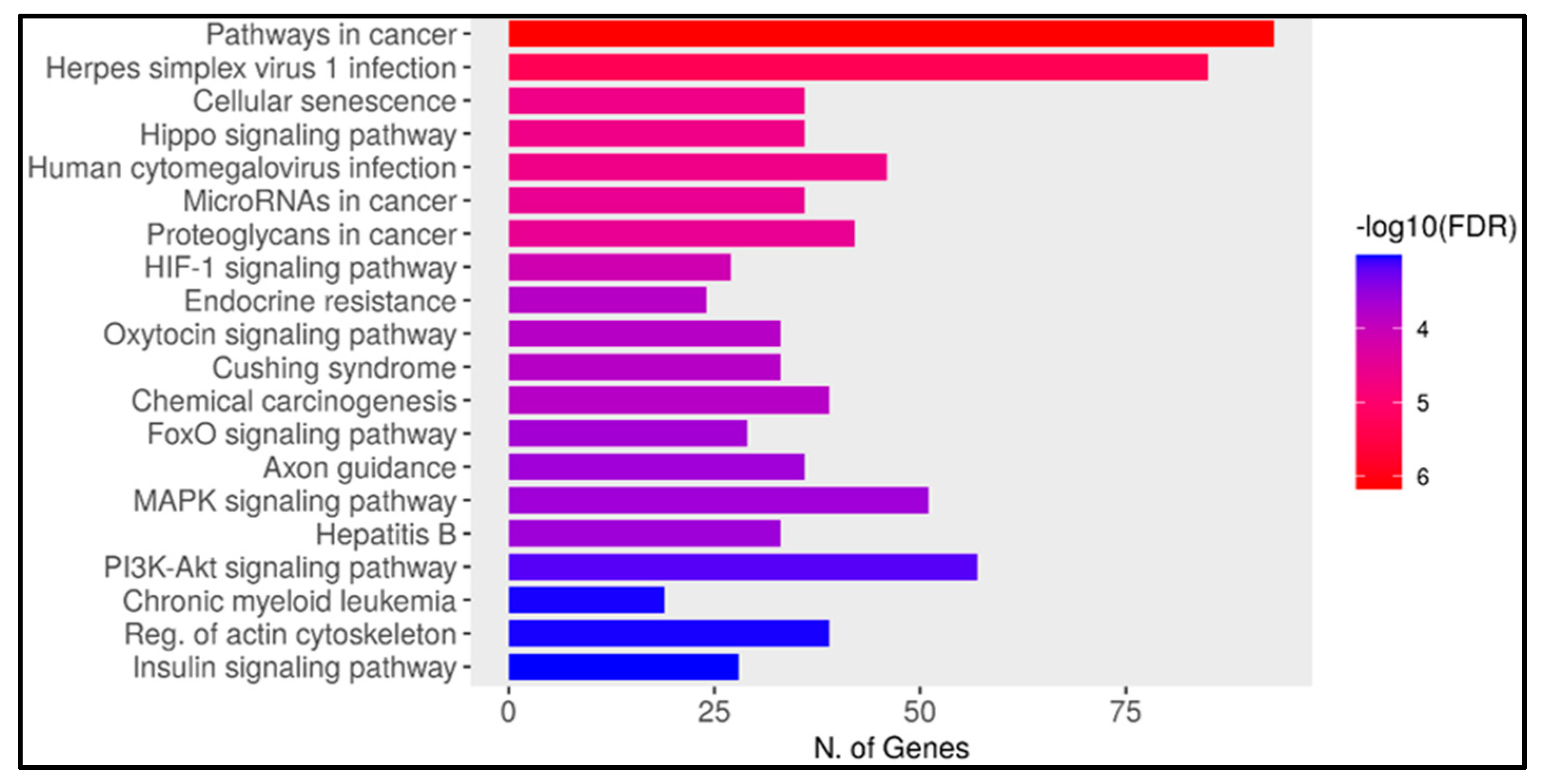
2.6. Result of Gene Enrichment Analysis
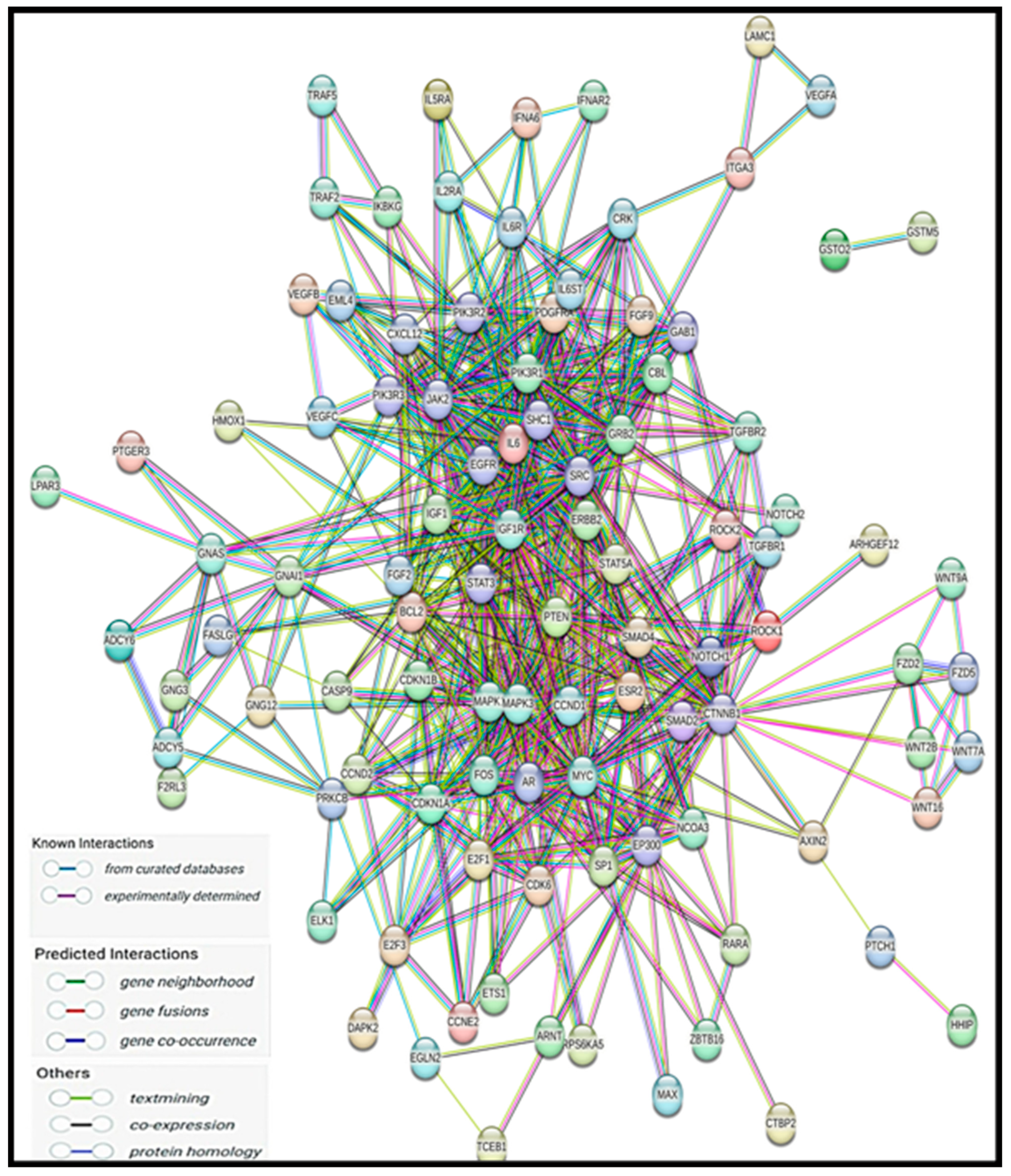
2.7. Protein–Protein Interaction and Disease–Gene Association Enrichment
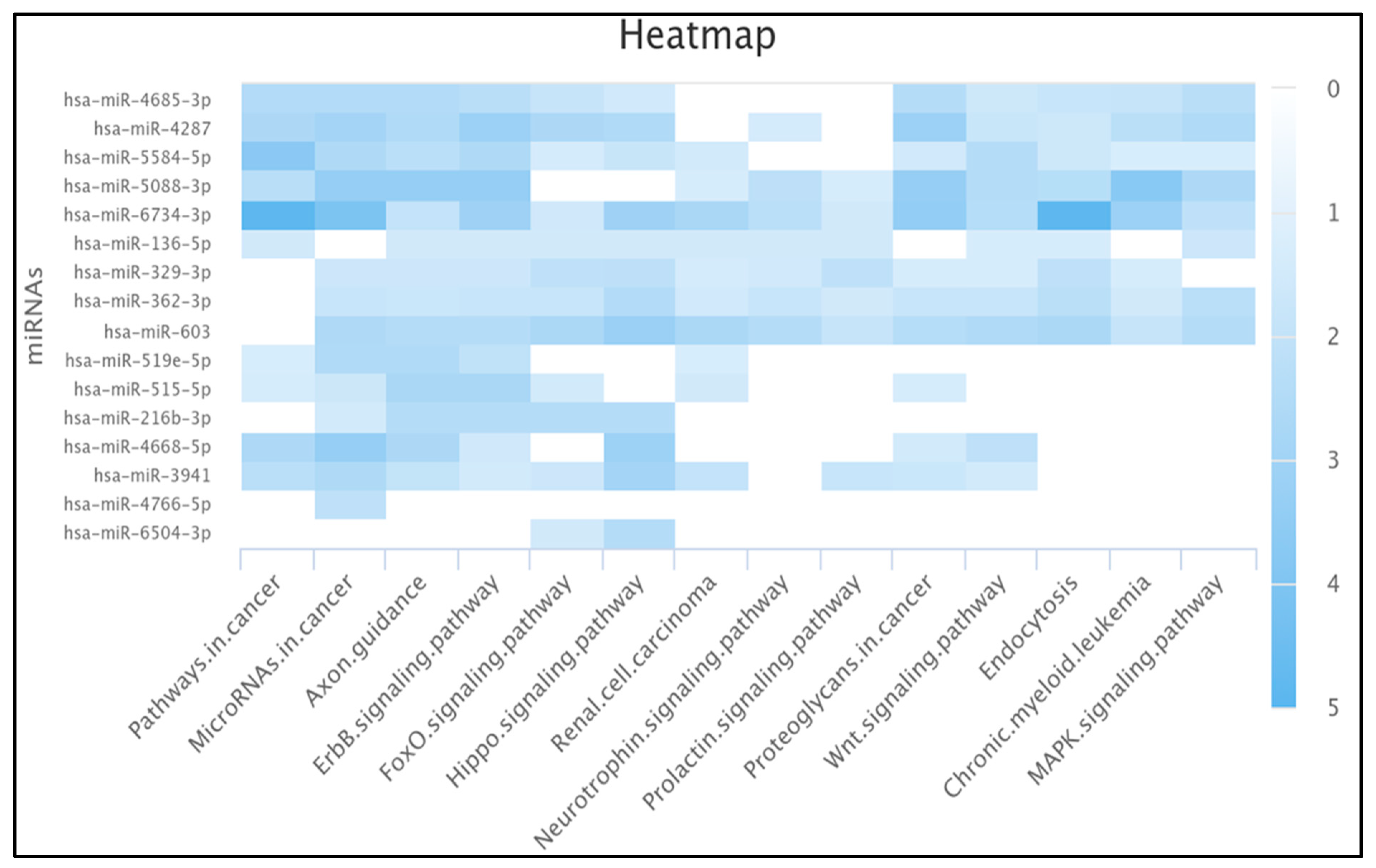
2.8. Ten miRNAs Enriched in the Pathway of Cancer
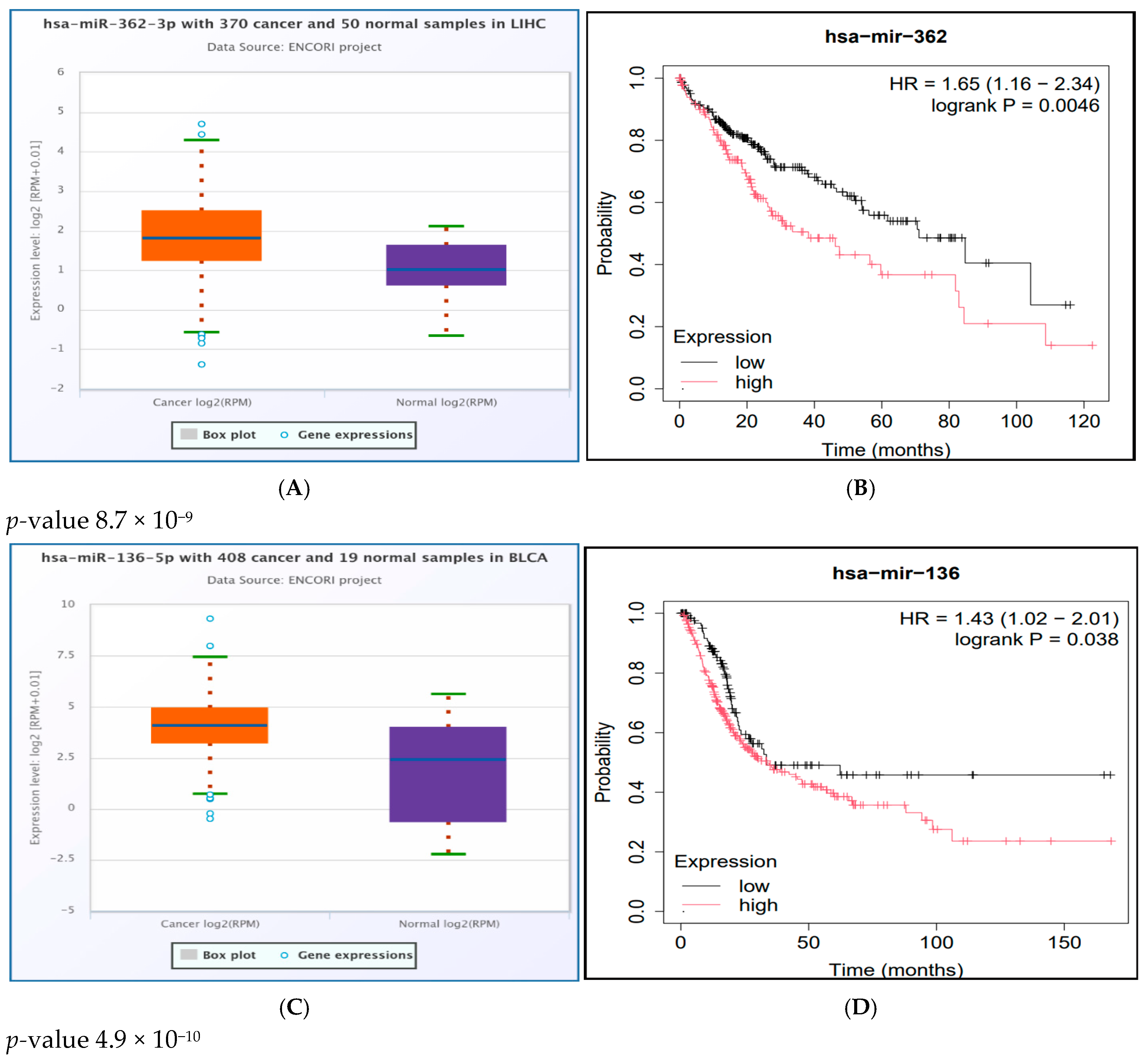
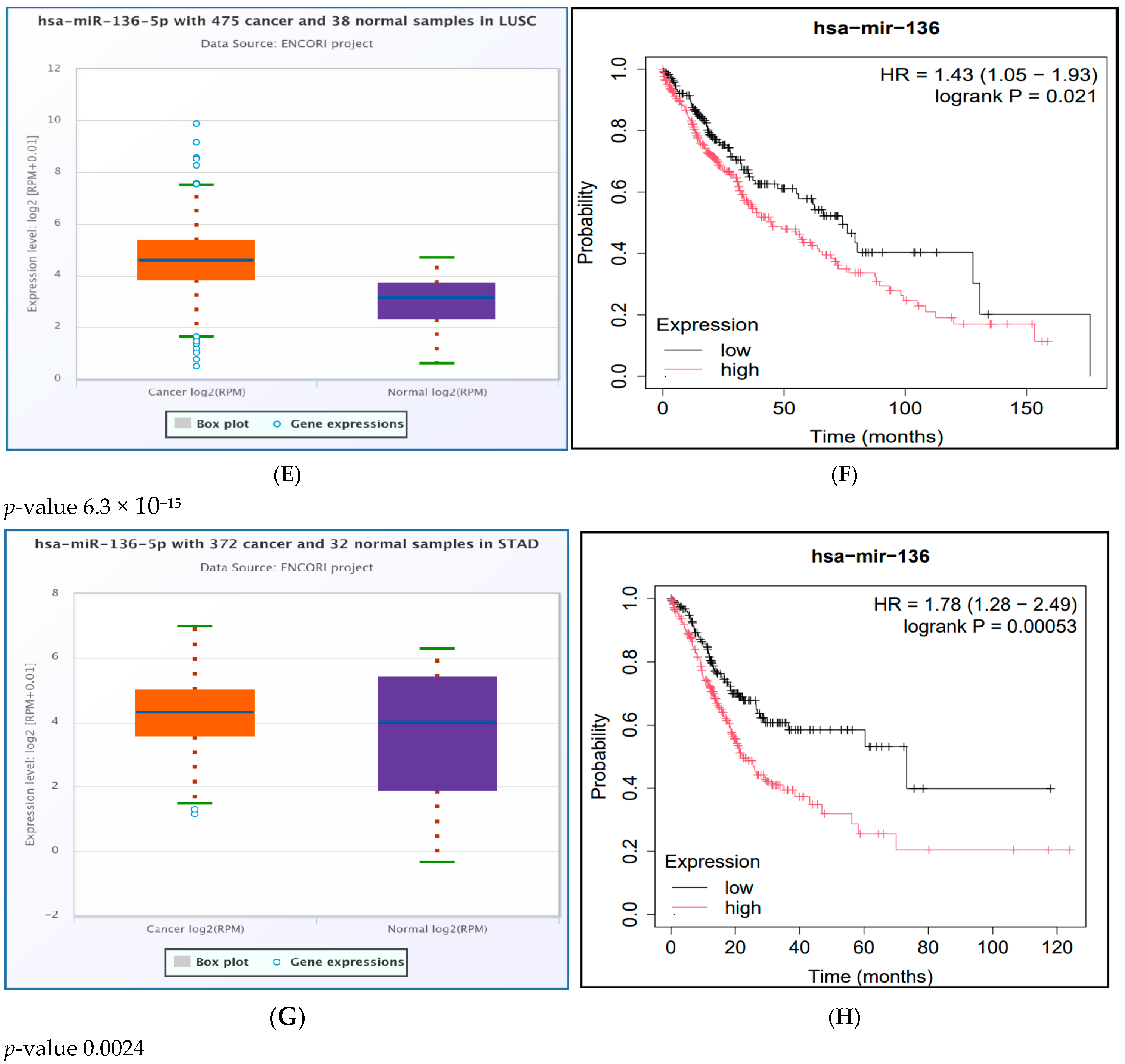
2.9. Results of miRNA Differential Expression Analysis in Human Cancer
2.10. Survival Analysis Study of the Significantly UpRegulated miRNAs in Cancer
3. Materials and Methods
3.1. Retrieval of 3′ UTR SNPs
3.2. Evaluation of the Impact of 3′ UTR SNPs on miRNA Binding Sites
3.3. Determination of the Effect of SNPs on the Secondary Structure of mRNA
3.4. Prediction of Cancer-Associated SNPs
3.5. Identification of miRNAs’ Target Genes
3.6. Gene Enrichment Analysis
3.7. Protein–Protein Interaction Using the STRING Database
3.8. MiRNAs Enrichment Analysis
3.9. Pan-Cancer miRNA Differential Expression Analysis
3.10. MiRNAs Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gongora, C.; Mechti, N. Interferon signaling pathways. Bull. Cancer 1999, 86, 911–919. [Google Scholar] [PubMed]
- Horvath, C.M. The Jak-STAT pathway stimulated by interferon gamma. Sci. STKE 2004, 2004, tr8. [Google Scholar] [CrossRef] [PubMed]
- Abroun, S.; Saki, N.; Ahmadvand, M.; Asghari, F.; Salari, F.; Rahim, F. Stats: An old story, yet mesmerizing. Cell J. 2015, 17, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Benekli, M.; Baer, M.R.; Baumann, H.; Wetzler, M. Signal transducer and activator of transcription proteins in leukemias. Blood 2003, 101, 2940–2954. [Google Scholar] [CrossRef]
- Shuai, K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene 2000, 19, 2638–2644. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Kamal, E.; Kaddam, L.A.; Ahmed, M.; Alabdulkarim, A. Integrating artificial intelligence and bioinformatics methods to identify disruptive STAT1 variants impacting protein stability and function. Genes 2025, 16, 303. [Google Scholar] [CrossRef]
- Lee, H.-C.; Md Yusof, H.H.; Leong, M.P.-Y.; Zainal Abidin, S.; Seth, E.A.; Hewitt, C.A.; Vidyadaran, S.; Nordin, N.; Scott, H.S.; Cheah, P.-S.; et al. Gene and protein expression profiles of JAK-STAT signalling pathway in the developing brain of the Ts1Cje down syndrome mouse model. Int. J. Neurosci. 2019, 129, 871–881. [Google Scholar] [CrossRef]
- Heim, M.H. The Jak-STAT pathway: Cytokine signalling from the receptor to the nucleus. J. Recept. Signal Transduct. Res. 1999, 19, 75–120. [Google Scholar] [CrossRef]
- Gałecka, M.; Szemraj, J.; Su, K.-P.; Halaris, A.; Maes, M.; Skiba, A.; Gałecki, P.; Bliźniewska-Kowalska, K. Is the JAK-STAT Signaling Pathway Involved in the Pathogenesis of Depression? J. Clin. Med. 2022, 11, 2056. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29. [Google Scholar] [PubMed]
- Koromilas, A.E.; Sexl, V. The tumor suppressor function of STAT1 in breast cancer. JAK-STAT 2013, 2, e23353. [Google Scholar] [CrossRef] [PubMed]
- Crnčec, I.; Modak, M.; Gordziel, C.; Svinka, J.; Scharf, I.; Moritsch, S.; Pathria, P.; Schlederer, M.; Kenner, L.; Timelthaler, G.; et al. STAT1 is a sex-specific tumor suppressor in colitis-associated colorectal cancer. Mol. Oncol. 2018, 12, 514–528. [Google Scholar] [CrossRef]
- Zhang, Y.; Molavi, O.; Su, M.; Lai, R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer 2014, 14, 791. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, B.; Stoiber, D.; Moriggl, R.; Weisz, E.; Ott, R.G.; Kreibich, R.; Levy, D.E.; Beug, H.; Freissmuth, M.; Sexl, V. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell 2006, 10, 77–87. [Google Scholar] [CrossRef]
- Zellmer, V.R.; Schnepp, P.M.; Fracci, S.L.; Tan, X.; Howe, E.N.; Zhang, S. Tumor-induced Stromal STAT1 Accelerates Breast Cancer via Deregulating Tissue Homeostasis. Mol. Cancer Res. 2017, 15, 585–597. [Google Scholar] [CrossRef]
- Li, X.; Wang, F.; Xu, X.; Zhang, J.; Xu, G. The dual role of STAT1 in ovarian cancer: Insight into molecular mechanisms and application potentials. Front. Cell Dev. Biol. 2021, 9, 636595. [Google Scholar] [CrossRef]
- Russo, F.; Fiscon, G.; Conte, F.; Rizzo, M.; Paci, P.; Pellegrini, M. Interplay between long noncoding rnas and micrornas in cancer. Methods Mol. Biol. 2018, 1819, 75–92. [Google Scholar] [CrossRef]
- Li, X.; Peng, J.; Yi, C. The epitranscriptome of small non-coding RNAs. Noncoding RNA Res. 2021, 6, 167–173. [Google Scholar] [CrossRef]
- Grosshans, H.; Slack, F.J. Micro-RNAs: Small is plentiful. J. Cell Biol. 2002, 156, 17–21. [Google Scholar] [CrossRef]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef]
- Anglicheau, D.; Muthukumar, T.; Suthanthiran, M. MicroRNAs: Small RNAs with big effects. Transplantation 2010, 90, 105–112. [Google Scholar] [CrossRef]
- Fang, Z.; Rajewsky, N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS ONE 2011, 6, e18067. [Google Scholar] [CrossRef] [PubMed]
- Våge, J.; Lingaas, F. Single nucleotide polymorphisms (SNPs) in coding regions of canine dopamine- and serotonin-related genes. BMC Genet. 2008, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.G.; Farnham, P.J. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 2015, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Ryczek, N.; Łyś, A.; Makałowska, I. The Functional Meaning of 5′UTR in Protein-Coding Genes. Int. J. Mol. Sci. 2023, 24, 2976. [Google Scholar] [CrossRef]
- Pan, X.; Zhao, J.; Zhou, Z.; Chen, J.; Yang, Z.; Wu, Y.; Bai, M.; Jiao, Y.; Yang, Y.; Hu, X.; et al. 5′-UTR SNP of FGF13 causes translational defect and intellectual disability. eLife 2021, 10, e63021. [Google Scholar] [CrossRef]
- Soukarieh, O.; Meguerditchian, C.; Proust, C.; Aïssi, D.; Eyries, M.; Goyenvalle, A.; Trégouët, D.-A. Common and rare 5′UTR variants altering upstream open reading frames in cardiovascular genomics. Front. Cardiovasc. Med. 2022, 9, 841032. [Google Scholar] [CrossRef]
- Guo, M.; Xu, C.; Chen, Y.-Z.; Sun, Q.-W.; Zhao, X.-Y.; Liu, X.; Yang, Y.; Hu, Y.-Y.; Li, F.-F.; Liu, S.-L. Associations of CXCL1 gene 5′UTR variations with ovarian cancer. J. Ovarian Res. 2020, 13, 43. [Google Scholar] [CrossRef]
- Lim, Y.; Arora, S.; Schuster, S.L.; Corey, L.; Fitzgibbon, M.; Wladyka, C.L.; Wu, X.; Coleman, I.M.; Delrow, J.J.; Corey, E.; et al. Multiplexed functional genomic analysis of 5′ untranslated region mutations across the spectrum of prostate cancer. Nat. Commun. 2021, 12, 4217. [Google Scholar] [CrossRef]
- Mendell, J.T.; Dietz, H.C. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell 2001, 107, 411–414. [Google Scholar] [CrossRef]
- Song, C.-Q.; Zhang, J.-H.; Shi, J.-C.; Cao, X.-Q.; Song, C.-H.; Hassan, A.; Wang, P.; Dai, L.-P.; Zhang, J.-Y.; Wang, K.-J. Bioinformatic Prediction of SNPs within miRNA Binding Sites of Inflammatory Genes Associated with Gastric Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 937–943. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ergun, S.; Oztuzcu, S. Sequence-based analysis of 5′UTR and coding regions of CASP3 in terms of miRSNPs and SNPs in targetting miRNAs. Comput. Biol. Chem. 2016, 62, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Tong, Y.; Zhang, H.-M.; Wang, K.; Hu, T.; Shan, G.; Sun, J.; Guo, A.-Y. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 2012, 33, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.; Munirajan, A.K. Single nucleotide polymorphisms in microRNA binding sites of oncogenes: Implications in cancer and pharmacogenomics. OMICS 2014, 18, 142–154. [Google Scholar] [CrossRef]
- Ergun, S.; Oztuzcu, S. Computational analysis of 3′UTR region of CASP3 with respect to miRSNPs and SNPs in targetting miRNAs. Comput. Biol. Chem. 2014, 53PB, 235–241. [Google Scholar] [CrossRef]
- Preskill, C.; Weidhaas, J.B. SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Crit. Rev. Oncog. 2013, 18, 327–340. [Google Scholar] [CrossRef]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Noncoding RNA 2021, 7, 47. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42, D86–D91. [Google Scholar] [CrossRef]
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA web services. Methods Mol. Biol. 2015, 1269, 307–326. [Google Scholar] [CrossRef]
- Rogers, M.F.; Shihab, H.A.; Gaunt, T.R.; Campbell, C. CScape: A tool for predicting oncogenic single-point mutations in the cancer genome. Sci. Rep. 2017, 7, 11597. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to “The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets”. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.-P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- Wong, N.W.; Chen, Y.; Chen, S.; Wang, X. OncomiR: An online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018, 34, 713–715. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Bartoszewski, R.A.; Jablonsky, M.; Bartoszewska, S.; Stevenson, L.; Dai, Q.; Kappes, J.; Collawn, J.F.; Bebok, Z. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 2010, 285, 28741–28748. [Google Scholar] [CrossRef]
- Naslavsky, M.S.; Crovella, S.; Lima Filho, J.L.; Rocha, C.R.C. The sound of silence: Human beta-defensin-1 gene untranslated SNPs change the predicted mRNA secondary structure in a length-dependent manner. Immunol. Lett. 2010, 129, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Karthi, S.; Rajeshwari, M.; Francis, A.; Saravanan, M.; Varalakshmi, P.; Houlden, H.; Thangaraj, K.; Ashokkumar, B. 3′-UTR SNP rs2229611 in G6PC1 affects mRNA stability, expression and Glycogen Storage Disease type-Ia risk. Clin. Chim. Acta 2017, 471, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A comprehensive review on MAPK: A promising therapeutic target in cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef]
- Braga, E.A.; Fridman, M.V.; Loginov, V.I.; Dmitriev, A.A.; Morozov, S.G. Molecular Mechanisms in Clear Cell Renal Cell Carcinoma: Role of miRNAs and Hypermethylated miRNA Genes in Crucial Oncogenic Pathways and Processes. Front. Genet. 2019, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Wong, H.K.; Gu, W.; Yu, M.; To, K.-F.; Wang, C.C.; Wong, Y.F.; Cheung, T.H.; Chung, T.K.H.; Choy, K.W. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol. Oncol. 2013, 129, 199–208. [Google Scholar] [CrossRef]
- Wu, X.; Shen, J.; Xiao, Z.; Li, J.; Zhao, Y.; Zhao, Q.; Cho, C.H.; Li, M. An overview of the multifaceted roles of miRNAs in gastric cancer: Spotlight on novel biomarkers and therapeutic targets. Biochem. Pharmacol. 2019, 163, 425–439. [Google Scholar] [CrossRef]
- Ren, R.; Du, Y.; Niu, X.; Zang, R. ZFPM2-AS1 transcriptionally mediated by STAT1 regulates thyroid cancer cell growth, migration and invasion via miR-515-5p/TUSC3. J. Cancer 2021, 12, 3393–3406. [Google Scholar] [CrossRef]
- Li, T.-T.; Gao, X.; Gao, L.; Gan, B.-L.; Xie, Z.-C.; Zeng, J.-J.; Chen, G. Role of upregulated miR-136-5p in lung adenocarcinoma: A study of 1242 samples utilizing bioinformatics analysis. Pathol. Res. Pract. 2018, 214, 750–766. [Google Scholar] [CrossRef]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.-G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef]
- Assiri, A.A.; Mourad, N.; Shao, M.; Kiel, P.; Liu, W.; Skaar, T.C.; Overholser, B.R. MicroRNA 362-3p Reduces hERG-related Current and Inhibits Breast Cancer Cells Proliferation. Cancer Genom. Proteom. 2019, 16, 433–442. [Google Scholar] [CrossRef]
- Kang, H.; Kim, C.; Lee, H.; Rho, J.G.; Seo, J.W.; Nam, J.W.; Song, W.K.; Nam, S.W.; Kim, W.; Lee, E.K. Downregulation of microRNA-362-3p and microRNA-329 promotes tumor progression in human breast cancer. Cell Death Differ. 2016, 23, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.L.; Tobiasen, H.; Holm, A.; Schepeler, T.; Ostenfeld, M.S.; Thorsen, K.; Rasmussen, M.H.; Birkenkamp-Demtroeder, K.; Sieber, O.M.; Gibbs, P.; et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int. J. Cancer 2013, 133, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, X.; Sun, Y.; Shi, J.; Jiang, D.; Wang, J.; Liu, Y.; Hu, C.; Pan, J.; Zheng, L.; et al. MicroRNA-362-3p Inhibits Migration and Invasion via Targeting BCAP31 in Cervical Cancer. Front. Mol. Biosci. 2020, 7, 107. [Google Scholar] [CrossRef]
- Yang, S.; Sun, Y.; Jiang, D.; Wang, J.; Dang, E.; Li, Z.; Zhou, J.; Lu, Y.; Shi, J.; Tao, L.; et al. MiR-362 suppresses cervical cancer progression via directly targeting BAP31 and activating TGFβ/Smad pathway. Cancer Med. 2021, 10, 305–316. [Google Scholar] [CrossRef]
- Yuan, J.; Li, T.; Yi, K.; Hou, M. The suppressive role of miR-362-3p in epithelial ovarian cancer. Heliyon 2020, 6, e04258. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, Y.; Wang, H.; Song, Y.; Huang, H. miR-362-3p suppresses ovarian cancer by inhibiting LRP8. Transl. Oncol. 2022, 15, 101284. [Google Scholar] [CrossRef]
- Cao, S.; Li, N.; Liao, X. miR-362-3p acts as a tumor suppressor by targeting SERBP1 in ovarian cancer. J. Ovarian Res. 2021, 14, 23. [Google Scholar] [CrossRef]
| SNP ID | miRNAs |
|---|---|
| rs11305 | hsa-miR-6504-3p |
| rs184180073 | hsa-miR-4287, hsa-miR-4685-3p, hsa-miR-5088-3p, hsa-miR-6734-3p |
| rs41363648 | hsa-miR-134-3p, hsa-miR-4778-5p, hsa-miR-7114-5p |
| rs188557905 | hsa-miR-4699-3p |
| rs186033487 | hsa-miR-122-3p |
| rs79073086 | hsa-miR-4766-5p |
| rs114360225 | hsa-miR-216b-3p, hsa-miR-329-3p, hsa-miR-362-3p, hsa-miR-3941, hsa-miR-603 |
| rs139958571 | hsa-miR-6814-3p, hsa-miR-6872-5p |
| rs182394503 | hsa-miR-202-5p, hsa-miR-337-3p |
| rs190542524 | hsa-miR-136-5p, hsa-miR-515-5p, hsa-miR-519e-5p |
| rs182725919 | hsa-miR-6741-5p |
| rs41481847 | hsa-miR-3148, hsa-miR-3162-5p, hsa-miR-4668-5p, hsa-miR-5584-5p, hsa-miR-6750-5p, hsa-miR-6822-5p |
| SNP ID | Minimum Free Energy of Wild Type kcal/mol | Wild mRNA | Minimum Free Energy of Mutant Type kcal/mol | Mutant mRNA | Interpretation |
|---|---|---|---|---|---|
| rs114360225 | −16.70 |  | −17.20 | 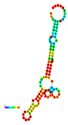 | A reduction in MFE in the mutant mRNA induces structural alterations in the mRNA, hence stabilizing its structure. |
| rs139958571 | −10.40 |  | −10.40 |  | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs41363648 | −10.30 | 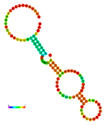 | −10.30 | 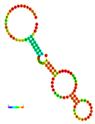 | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs41481847 | −21.40 |  | −26.00 | 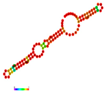 | A reduction in MFE in the mutant mRNA induces structural alterations in the mRNA, hence stabilizing its structure. |
| rs11305 | −19.40 | 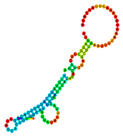 | −19.40 |  | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs184180073 | −10.40 | 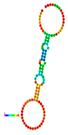 | −10.40 | 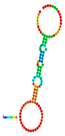 | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs79073086 | −11.50 | 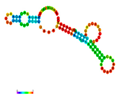 | −12.50 | 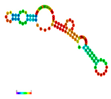 | A reduction in MFE in the mutant mRNA induces structural alterations in the mRNA, hence stabilizing its structure. |
| rs41363648 | −10.30 | 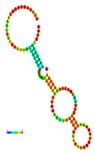 | −10.30 | 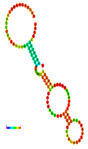 | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs186033487 | −10.70 |  | −10.90 | 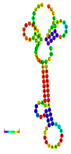 | A reduction in MFE in the mutant mRNA induces structural alterations in the mRNA, hence stabilizing its structure. |
| rs188557905 | −35.80 | 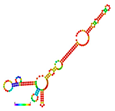 | −13.90 | 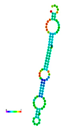 | The energy elevation destabilizes the mRNA structure. |
| rs139958571 | −10.40 |  | −10.40 |  | No alteration in energy, accompanied by no alteration in mRNA structure. |
| rs190542524 | −23.80 | 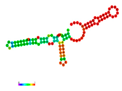 | −22.30 |  | The energy elevation destabilizes the mRNA structure. |
| SNP ID | Chromosomal Location | Cscape Score | Interpretation |
|---|---|---|---|
| rs114360225 | 2,190970130,T,C | 0.468060 | Benign |
| rs139958571 | 2,191835001,C,G | 0.589398 | Oncogenic |
| rs41363648 | 2,191834487,T,C | 0.686973 | Oncogenic |
| rs41481847 | 2,191835275,A,G | 0.527544 | Oncogenic |
| rs11305 | 2,191834030,T,A 2,191834030,T,C | 0.707913 0.516490 | Oncogenic Oncogenic |
| rs184180073 | 2,191834477,T,C | 0.667051 | Oncogenic |
| rs79073086 | 2,191834832,G,A 2,191834832,G,C | 0.666260 0.596381 | Oncogenic Oncogenic |
| rs41363648 | 2,191834487,T,C | 0.686973 | Oncogenic |
| rs186033487 | 2,191834759,A,C 2,191834759,A,G | 0.717679 0.672660 | Oncogenic Oncogenic |
| rs188557905 | 2,191834574,C,T | 0.753580 | Oncogenic |
| rs139958571 | 2,191835001,C,G | 0.589398 | Oncogenic |
| rs190542524 | 2,191835125,T,A 2,191835125,T,C 2,191835125,T,G | 0.802671 0.534609 0.746269 | Oncogenic Oncogenic Oncogenic |
| miRNAs | Cancer Type | p-Value | Upregulated in |
|---|---|---|---|
| hsa-miR-362-3p | BLCA | 6.86 × 10−3 | Tumor |
| BRCA | 3.61 × 10−3 | Tumor | |
| HNSC | 3.07 × 10−6 | Normal | |
| KIRC | 4.77 × 10−8 | Normal | |
| KIRP | 2.08 × 10−5 | Normal | |
| LIHC | 7.37 × 10−3 | Tumor | |
| LUSC | 4.55 × 10−7 | Normal | |
| STAD | 2.12 × 10−7 | Tumor | |
| THCA | 1.17 × 10−5 | Normal | |
| UCEC | 2.49 × 10−2 | Tumor | |
| hsa-miR-136-5p | BLCA | 4.01 × 10−2 | Tumor |
| BRCA | 6.39 × 10−4 | Tumor | |
| HNSC | 3.36 × 10−6 | Normal | |
| KIRC | 1.51 × 10−14 | Normal | |
| KIRP | 1.09× 10−11 | Normal | |
| LIHC | 2.47× 10−11 | Normal | |
| LUAD | 4.05 × 10−6 | Tumor | |
| LUSC | 2.51 × 10−6 | Tumor | |
| STAD | 2.14 × 10−2 | Tumor | |
| THCA | 6.54 × 10−7 | Normal | |
| hsa-miR-515-5p | LUAD | 4.89 × 10−2 | Normal |
| hsa-miR-329-3p | BRCA | 7.22 × 10−9 | Normal |
| HNSC | 2.42 × 10−3 | Normal | |
| STAD | 4.45 × 10−3 | Tumor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamal, E. In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact. Non-Coding RNA 2025, 11, 32. https://doi.org/10.3390/ncrna11030032
Kamal E. In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact. Non-Coding RNA. 2025; 11(3):32. https://doi.org/10.3390/ncrna11030032
Chicago/Turabian StyleKamal, Ebtihal. 2025. "In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact" Non-Coding RNA 11, no. 3: 32. https://doi.org/10.3390/ncrna11030032
APA StyleKamal, E. (2025). In Silico Prioritization of STAT1 3′ UTR SNPs Identifies rs190542524 as a miRNA-Linked Variant with Potential Oncogenic Impact. Non-Coding RNA, 11(3), 32. https://doi.org/10.3390/ncrna11030032






