The Role of Non-Coding RNAs in MYC-Mediated Metabolic Regulation: Feedback Loops and Interactions
Abstract
1. Introduction
2. Overview of MYC’s Metabolic Function
3. Non-Coding RNA and MYC
3.1. Overview of ncRNA
3.2. ncRNA and MYC Regulation
4. Interaction Between ncRNAs and MYC in Cancer Metabolism
4.1. ncRNAs That Affect Metabolism by Regulating MYC Stability
4.2. ncRNAs with Direct Metabolic Functions and Regulated by MYC
4.3. ncRNAs That Regulate MYC and Are Targets of MYC
5. Clinical Relevance of the ncRNAs Discussed
5.1. Prognostic Value and Therapeutic Potential
5.2. Clinical Relevance to MYC Regulation and Synthetic Lethality
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ncRNA | Non-coding RNA |
| GLUT1 | Glucose transporter 1 |
| HK2 | Hexokinase 2 |
| LDHA | Lactate dehydrogenase A |
| Eno1 | Alpha enolase |
| MCT | Monocarboxylate transporter |
| PFKM | Phosphofructokinase muscle isoform |
| SCLC | Small-cell lung carcinoma |
| NSCLC | Non-small-cell lung cancer |
| LncRNA | Long non-coding RNA |
| CircRNA | Circular RNA |
| MiRNA | MicroRNA |
| TNBC | Triple-negative breast cancer |
| IGFBP | Insulin-like growth factor mRNA-binding protein |
| M6A | N6-methyladenosine |
| CRC | Colorectal cancer |
| HCC | Hepatocellular carcinoma |
| PCa | Prostate cancer |
| BPTF | Bromodomain and PHD finger-containing transcription factor |
| HIF1α | Hypoxia-inducible factor 1 alpha |
| FBXW7 | F-box and WD repeat domain-containing 7 |
References
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Das, S.K.; Lewis, B.A.; Levens, D. MYC: A complex problem. Trends Cell Biol. 2023, 33, 235–246. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of cancer cell metabolism: Oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef]
- Dejure, F.R.; Eilers, M. MYC and tumor metabolism: Chicken and egg. Embo J. 2017, 36, 3409–3420. [Google Scholar] [CrossRef]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Coan, M.; Haefliger, S.; Ounzain, S.; Johnson, R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat. Rev. Genet. 2024, 25, 578–595. [Google Scholar] [CrossRef] [PubMed]
- Stasevich, E.M.; Murashko, M.M.; Zinevich, L.S.; Demin, D.E.; Schwartz, A.M. The Role of Non-Coding RNAs in the Regulation of the Proto-Oncogene MYC in Different Types of Cancer. Biomedicines 2021, 9, 921. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L.; Pucci, P.; Ishola, T.; Trigg, R.M.; Williams, J.A.; Pereira, J.; Cavanagh, M.L.; Turner, S.D.; Gkoutos, G.V.; Tsaprouni, L. The Contribution of Autophagy and LncRNAs to MYC-Driven Gene Regulatory Networks in Cancers. Int. J. Mol. Sci. 2021, 22, 8527. [Google Scholar] [CrossRef] [PubMed]
- Cargill, K.R.; Stewart, C.A.; Park, E.M.; Ramkumar, K.; Gay, C.M.; Cardnell, R.J.; Wang, Q.; Diao, L.; Shen, L.; Fan, Y.H.; et al. Targeting MYC-enhanced glycolysis for the treatment of small cell lung cancer. Cancer Metab. 2021, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Balakrishnan, A.; Bok, R.A.; Anderton, B.; Larson, P.E.; Nelson, S.J.; Kurhanewicz, J.; Vigneron, D.B.; Goga, A. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011, 14, 131–142. [Google Scholar] [CrossRef]
- Shin, P.J.; Zhu, Z.; Camarda, R.; Bok, R.A.; Zhou, A.Y.; Kurhanewicz, J.; Goga, A.; Vigneron, D.B. Cancer recurrence monitoring using hyperpolarized [1-(13)C]pyruvate metabolic imaging in murine breast cancer model. Magn. Reson. Imaging 2017, 43, 105–109. [Google Scholar] [CrossRef]
- Doherty, J.R.; Yang, C.; Scott, K.E.; Cameron, M.D.; Fallahi, M.; Li, W.; Hall, M.A.; Amelio, A.L.; Mishra, J.K.; Li, F.; et al. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res. 2014, 74, 908–920. [Google Scholar] [CrossRef]
- Gan, L.; Xiu, R.; Ren, P.; Yue, M.; Su, H.; Guo, G.; Xiao, D.; Yu, J.; Jiang, H.; Liu, H.; et al. Metabolic targeting of oncogene MYC by selective activation of the proton-coupled monocarboxylate family of transporters. Oncogene 2016, 35, 3037–3048. [Google Scholar] [CrossRef]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef]
- Yu, L.; Kim, J.; Jiang, L.; Feng, B.; Ying, Y.; Ji, K.Y.; Tang, Q.; Chen, W.; Mai, T.; Dou, W.; et al. MTR4 drives liver tumorigenesis by promoting cancer metabolic switch through alternative splicing. Nat. Commun. 2020, 11, 708. [Google Scholar] [CrossRef]
- Uppaluri, K.R.; Challa, H.J.; Gaur, A.; Jain, R.; Krishna Vardhani, K.; Geddam, A.; Natya, K.; Aswini, K.; Palasamudram, K. Unlocking the potential of non-coding RNAs in cancer research and therapy. Transl. Oncol. 2023, 35, 101730. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cao, L.; Fan, P.; Mei, Y.; Wu, M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016, 17, 1204–1220. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xue, X.; Zheng, L.; Bi, J.; Zhou, Y.; Zhi, K.; Gu, Y.; Fang, G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. Febs J. 2014, 281, 802–813. [Google Scholar] [CrossRef]
- Yu, Y.; Chang, Z.; Han, C.; Zhuang, L.; Zhou, C.; Qi, X.; Peng, Z. Long non-coding RNA MINCR aggravates colon cancer via regulating miR-708-5p-mediated Wnt/β-catenin pathway. Biomed. Pharmacother. 2020, 129, 110292. [Google Scholar] [CrossRef]
- Taheri, M.; Askari, A.; Hussen, B.M.; Eghbali, A.; Ghafouri-Fard, S. A review on the role of MYC-induced long non-coding RNA in human disorders. Pathol. Res. Pract. 2023, 248, 154568. [Google Scholar] [CrossRef]
- Swier, L.; Dzikiewicz-Krawczyk, A.; Winkle, M.; van den Berg, A.; Kluiver, J. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol. Oncol. 2019, 13, 26–45. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, H.; Liu, M.; Wu, Q.; Li, W.; Zhang, J. Corrigendum to “MicroRNA-24-1 suppresses mouse hepatoma cell invasion and metastasis via directly targeting O-GlcNAc transferase” [Biomed. Pharmacother., vol. 91 (July 2017) Pages 731-738/BIOPHA 5337]. Biomed. Pharmacother. 2024, 177, 116946. [Google Scholar] [CrossRef]
- Shams, R.; Asadzadeh Aghdaei, H.; Behmanesh, A.; Sadeghi, A.; Zali, M.; Salari, S.; Padrón, J.M. MicroRNAs Targeting MYC Expression: Trace of Hope for Pancreatic Cancer Therapy. A Systematic Review. Cancer Manag. Res. 2020, 12, 2393–2404. [Google Scholar] [CrossRef] [PubMed]
- Arndt, G.M.; Dossey, L.; Cullen, L.M.; Lai, A.; Druker, R.; Eisbacher, M.; Zhang, C.; Tran, N.; Fan, H.; Retzlaff, K.; et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Capizzi, M.; Strappazzon, F.; Cianfanelli, V.; Papaleo, E.; Cecconi, F. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy 2017, 13, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Mori, S.; Nevins, J.R. Myc-induced microRNAs integrate Myc-mediated cell proliferation and cell fate. Cancer Res. 2010, 70, 4820–4828. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Prochownik, E.V. MicroRNA-based screens for synthetic lethal interactions with c-Myc. RNA Dis. 2016, 3, e1330. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Wang, Y.; Li, Q.; Zeng, K.; Li, X.; Feng, X. Myc derived circRNA promotes triple-negative breast cancer progression via reprogramming fatty acid metabolism. Discov. Oncol. 2023, 14, 67. [Google Scholar] [CrossRef]
- Yang, X.; Tao, L.; Xu, Y.; Li, S.; Yang, W.; Wang, L.; Zhu, J. CircMYC promotes proliferation, migration, invasion and inhibits apoptosis of small cell lung cancer by targeting miR-145/ Matrix Metallopeptidase 2 axis. Bioengineered 2022, 13, 10552–10563. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, W.; Tao, G.; Wang, W. Circular RNA circ-LRP6 facilitates Myc-driven tumorigenesis in esophageal squamous cell cancer. Bioengineered 2020, 11, 932–938. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.H.; Wu, Q.N.; Jin, Y.; Wang, D.S.; Chen, Y.X.; Liu, J.; Luo, X.J.; Meng, Q.; Pu, H.Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174. [Google Scholar] [CrossRef]
- Zhu, Y.; Ma, K.; Ye, Y.; Tang, J.; Zhu, J. Long non-coding RNA LINRIS is upregulated in non-small cell lung cancer and its silencing inhibits cell proliferation by suppressing microRNA-10a maturation. Bioengineered 2022, 13, 4340–4346. [Google Scholar] [CrossRef]
- Zhai, S.; Xu, Z.; Xie, J.; Zhang, J.; Wang, X.; Peng, C.; Li, H.; Chen, H.; Shen, B.; Deng, X. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene 2021, 40, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Guo, S.; Wang, S.; Zhang, Y.; Chen, H.; Wang, Y.; Liu, R.; Niu, Y.; Xu, Y. EIF4A3-Induced circARHGAP29 Promotes Aerobic Glycolysis in Docetaxel-Resistant Prostate Cancer through IGF2BP2/c-Myc/LDHA Signaling. Cancer Res. 2022, 82, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yan, T.; Bao, Y.; Shen, C.; Yu, C.; Zhu, X.; Tian, X.; Guo, F.; Liang, Q.; Liu, Q.; et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019, 10, 3499. [Google Scholar] [CrossRef]
- Lin, J.; Wang, X.; Zhai, S.; Shi, M.; Peng, C.; Deng, X.; Fu, D.; Wang, J.; Shen, B. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 2022, 15, 128. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, T.; Liu, H. circPDK1 competitively binds miR-4731-5p to mediate GIGYF1 expression and increase paclitaxel sensitivity in non-small cell lung cancer. Discov. Oncol. 2024, 15, 157. [Google Scholar] [CrossRef] [PubMed]
- Ebron, J.S.; Shankar, E.; Singh, J.; Sikand, K.; Weyman, C.M.; Gupta, S.; Lindner, D.J.; Liu, X.; Campbell, M.J.; Shukla, G.C. MiR-644a Disrupts Oncogenic Transformation and Warburg Effect by Direct Modulation of Multiple Genes of Tumor-Promoting Pathways. Cancer Res. 2019, 79, 1844–1856. [Google Scholar] [CrossRef]
- Wu, Z.; Jiang, H.; Fu, H.; Zhang, Y. A circGLIS3/miR-644a/PTBP1 positive feedback loop promotes the malignant biological progressions of non-small cell lung cancer. Am. J. Cancer Res. 2021, 11, 108–122. [Google Scholar]
- Kim, S.; Lee, E.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.J.; Lee, H.J.; Chae, S.Y.; Jeon, S.M.; et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Yao, H.; Li, H.; Meng, F.; Li, Q.; Lin, X.; Liu, L. MNX1-AS1, a c-Myc induced lncRNA, promotes the Warburg effect by regulating PKM2 nuclear translocation. J. Exp. Clin. Cancer Res. 2022, 41, 337. [Google Scholar] [CrossRef]
- Xiang, S.; Gu, H.; Jin, L.; Thorne, R.F.; Zhang, X.D.; Wu, M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc. Natl. Acad. Sci. USA 2018, 115, E1465–E1474. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jin, L.; Shi, R.; Li, J.; Wang, Y.; Zhang, L.; Liang, C.Z.; Narayana, V.K.; De Souza, D.P.; Thorne, R.F.; et al. The long noncoding RNA glycoLINC assembles a lower glycolytic metabolon to promote glycolysis. Mol. Cell 2022, 82, 542–554.e546. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, X.; Zhou, S.; Li, W.; Liu, C.; Chadwick, M.; Qian, C. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019, 450, 63–75. [Google Scholar] [CrossRef]
- Wang, F.; Hu, Y.; Wang, H.; Hu, P.; Xiong, H.; Zeng, Z.; Han, S.; Wang, D.; Wang, J.; Zhao, Y.; et al. LncRNA FTO-IT1 promotes glycolysis and progression of hepatocellular carcinoma through modulating FTO-mediated N6-methyladenosine modification on GLUT1 and PKM2. J. Exp. Clin. Cancer Res. 2023, 42, 267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, J.; Sun, R.; Sheng, H.; Yin, K.; Pan, Y.; Jimenez, R.; Chen, S.; Cui, X.L.; Zou, Z.; et al. A lncRNA from the FTO locus acts as a suppressor of the m(6)A writer complex and p53 tumor suppression signaling. Mol. Cell 2023, 83, 2692–2708.e2697. [Google Scholar] [CrossRef]
- Hua, Q.; Jin, M.; Mi, B.; Xu, F.; Li, T.; Zhao, L.; Liu, J.; Huang, G. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J. Hematol. Oncol. 2019, 12, 91. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, L.; Gu, Y.; Huang, Y.; Liang, X.; Kong, L.; Sun, Y. Long non-coding RNA LINC01123 promotes cell proliferation, migration and invasion via interacting with SRSF7 in colorectal cancer. Pathol. Res. Pract. 2022, 232, 153843. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Samuels, T.J.; Järvelin, A.I.; Ish-Horowicz, D.; Davis, I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife 2020, 9, e51529. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, M.; Guo, F.; Liu, X.; Zhang, Q.; Yang, S.; Yeung, Y.T.; Yang, R.; Wang, K.; Wu, Q.; et al. A novel lncRNA MTAR1 promotes cancer development through IGF2BPs mediated post-transcriptional regulation of c-MYC. Oncogene 2022, 41, 4736–4753. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef] [PubMed]
- Richart, L.; Carrillo-de Santa Pau, E.; Río-Machín, A.; de Andrés, M.P.; Cigudosa, J.C.; Lobo, V.J.S.; Real, F.X. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat. Commun. 2016, 7, 10153. [Google Scholar] [CrossRef]
- Li, B.; He, L.; Zuo, D.; He, W.; Wang, Y.; Zhang, Y.; Liu, W.; Yuan, Y. Mutual Regulation of MiR-199a-5p and HIF-1α Modulates the Warburg Effect in Hepatocellular Carcinoma. J. Cancer 2017, 8, 940–949. [Google Scholar] [CrossRef]
- Ji, S.; Qin, Y.; Liang, C.; Huang, R.; Shi, S.; Liu, J.; Jin, K.; Liang, D.; Xu, W.; Zhang, B.; et al. FBW7 (F-box and WD Repeat Domain-Containing 7) Negatively Regulates Glucose Metabolism by Targeting the c-Myc/TXNIP (Thioredoxin-Binding Protein) Axis in Pancreatic Cancer. Clin. Cancer Res. 2016, 22, 3950–3960. [Google Scholar] [CrossRef]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical significance of FBXW7 loss of function in human cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef]
- Kim, T.; Croce, C.M. MicroRNA: Trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 2023, 55, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhou, S.; Yang, Y.; Xu, Y.; Gong, X.; Cheng, Y.; Wang, Y. LncRNA MNX1-AS1: A novel oncogenic propellant in cancers. Biomed. Pharmacother. 2022, 149, 112801. [Google Scholar] [CrossRef]
- Qin, H.; Wang, C.; Hua, Y. LINC01123 is associated with prognosis of oral squamous cell carcinoma and involved in tumor progression by sponging miR-34a-5p. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2022, 133, 50–59. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Yin, J.; Gan, Y.; Xu, S.; Gu, Y.; Huang, W. Alternative approaches to target Myc for cancer treatment. Signal Transduct. Target. Ther. 2021, 6, 117. [Google Scholar] [CrossRef]
- Donati, G.; Amati, B. MYC and therapy resistance in cancer: Risks and opportunities. Mol. Oncol. 2022, 16, 3828–3854. [Google Scholar] [CrossRef]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Shahabi, S.; Kumaran, V.; Castillo, J.; Cong, Z.; Nandagopal, G.; Mullen, D.J.; Alvarado, A.; Correa, M.R.; Saizan, A.; Goel, R.; et al. LINC00261 Is an Epigenetically Regulated Tumor Suppressor Essential for Activation of the DNA Damage Response. Cancer Res. 2019, 79, 3050–3062. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Zhang, D.; Yang, Y.T.; Li, X.Y.; Li, H.N.; Zhang, X.P.; Long, J.Y.; Lu, Y.Q.; Liu, L.; Yang, G.; et al. Suppression of microRNA-222-3p ameliorates ulcerative colitis and colitis-associated colorectal cancer to protect against oxidative stress via targeting BRG1 to activate Nrf2/HO-1 signaling pathway. Front. Immunol. 2023, 14, 1089809. [Google Scholar] [CrossRef] [PubMed]
- Dauch, D.; Rudalska, R.; Cossa, G.; Nault, J.C.; Kang, T.W.; Wuestefeld, T.; Hohmeyer, A.; Imbeaud, S.; Yevsa, T.; Hoenicke, L.; et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat. Med. 2016, 22, 744–753. [Google Scholar] [CrossRef]
- Tayyar, Y.; Jubair, L.; Fallaha, S.; McMillan, N.A.J. Critical risk-benefit assessment of the novel anti-cancer aurora a kinase inhibitor alisertib (MLN8237): A comprehensive review of the clinical data. Crit. Rev. Oncol. Hematol. 2017, 119, 59–65. [Google Scholar] [CrossRef]
- Lei, Z.; Zhu, Z.; Yao, Z.; Dai, X.; Dong, Y.; Chen, B.; Wang, S.; Wang, S.; Bentum-Ennin, L.; Jin, L.; et al. Reciprocal interactions between lncRNAs and MYC in colorectal cancer: Partners in crime. Cell Death Dis. 2024, 15, 539. [Google Scholar] [CrossRef]
- He, T.L.; Zhang, Y.J.; Jiang, H.; Li, X.H.; Zhu, H.; Zheng, K.L. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med. Oncol. 2015, 32, 187. [Google Scholar] [CrossRef]
- Tateishi, K.; Iafrate, A.J.; Ho, Q.; Curry, W.T.; Batchelor, T.T.; Flaherty, K.T.; Onozato, M.L.; Lelic, N.; Sundaram, S.; Cahill, D.P.; et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin. Cancer Res. 2016, 22, 4452–4465. [Google Scholar] [CrossRef]

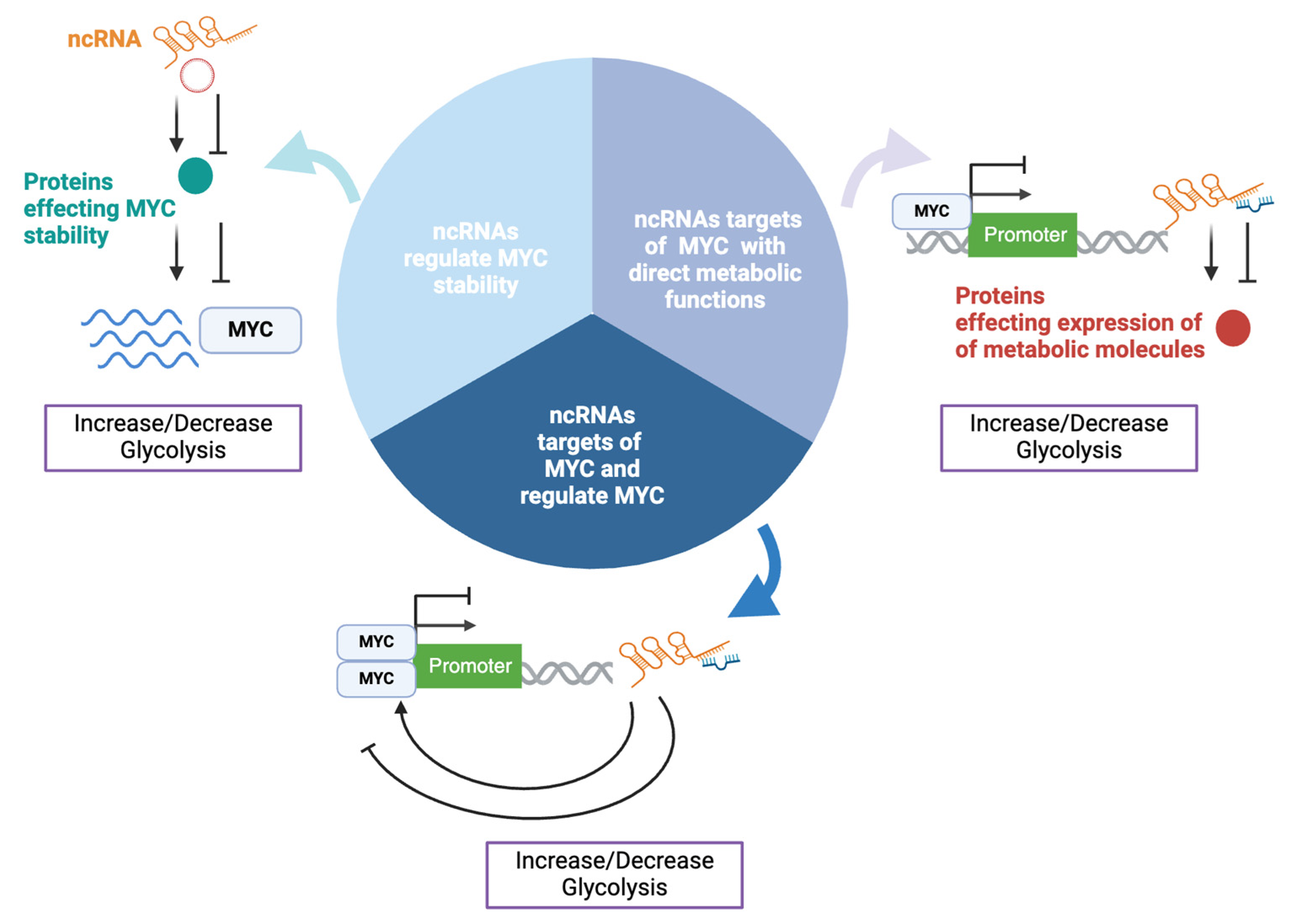
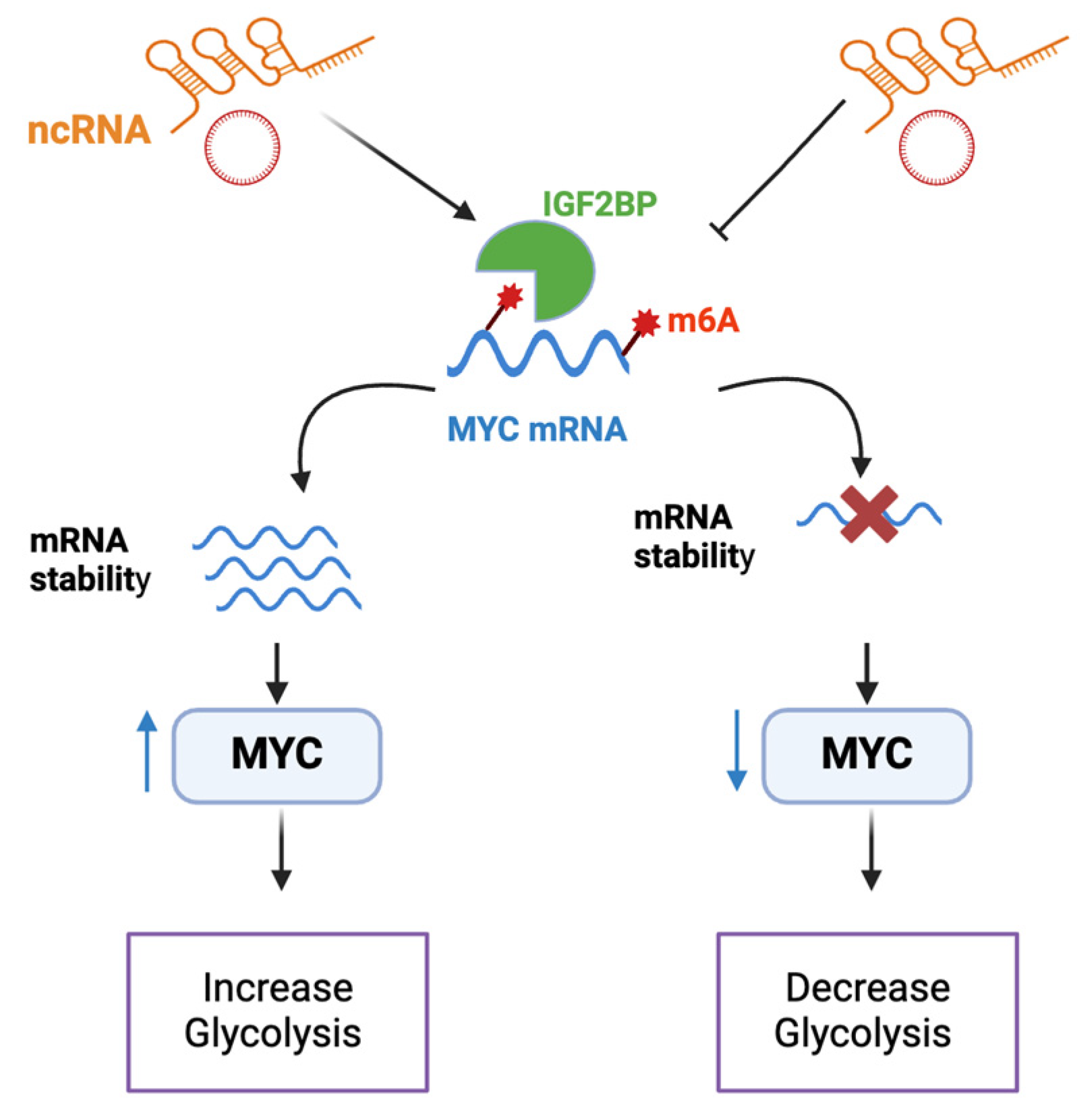

| ncRNA | Relation to MYC | Effect on Metabolism/Tumorigenesis | Clinical Relevance | Refs. |
|---|---|---|---|---|
| ncRNAs that affect metabolism by regulating MYC Stability | ||||
| LINRIS | Stabilizes MYC via IGFBP2 interaction | Increases glycolysis and promotes cancer proliferation | Poor prognosis in NSCLC patients | [39,40] |
| LINC00261 | Reduces MYC via IGF2BP2 interaction and miRNA sponging | Decreases glycolysis | Decreased—poor prognosis in Pancreatic cancer | [41] |
| circARHGAP2 | Stabilizes MYC and LDHA via IGFBP2 interaction | Increases glycolysis and promotes docetaxel resistance | [42] | |
| GLCC1 | Stabilizes MYC via HSP90 interaction | Increases glycolysis and correlates with CRC progression | Poor prognosis in CRC | [43] |
| circPDK1 | Increases MYC activity via miRNA sponging | Increases glycolysis and promotes cancer proliferation | Poor prognosis in pancreatic cancer | [44,45] |
| miR-644a | Inhibits MYC directly | Decreases glycolysis | Poor prognosis in NSCLC patients | [46,47] |
| miR-155 | Inhibits MYC indirectly via FOXO3a | Decreases glycolysis | Decreased—poor prognosis in breast cancer | [48,49] |
| ncRNAs with direct metabolic functions and regulated by MYC | ||||
| MNX1-AS1 | Induced by MYC | Increases glycolysis and cancer progression | [50] | |
| IDH1-AS1 | Repressed by MYC | Decreases glycolysis and suppresses HIF1α | [51] | |
| ncRNAs that regulate MYC and are targets of MYC | ||||
| gLINC | Induced by MYC and activates MYC | Increases glycolysis and supports cell survival under serine deprivation conditions | [52] | |
| FGF13-AS1 | Reduces MYC via IGF2BP2 interaction | Decreases glycolysis and stemness | [53] | |
| FTO-IT1 | Induced by MYC and stabilizes MYC via m6A modification | Increases glycolysis | Poor prognosis in HCC and PCa | [54,55] |
| LINC01123 | Induced by MYC and increases MYC expression via miRNA sponging | Increases glycolysis | Poor prognosis in CRC and NSCLC | [56,57] |
| LncRNA-MIF | Induced by MYC and inhibits MYC via miRNA sponging and FBXW7 interaction | Decreases glycolysis | [25] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamoudi, A.A. The Role of Non-Coding RNAs in MYC-Mediated Metabolic Regulation: Feedback Loops and Interactions. Non-Coding RNA 2025, 11, 27. https://doi.org/10.3390/ncrna11020027
Alamoudi AA. The Role of Non-Coding RNAs in MYC-Mediated Metabolic Regulation: Feedback Loops and Interactions. Non-Coding RNA. 2025; 11(2):27. https://doi.org/10.3390/ncrna11020027
Chicago/Turabian StyleAlamoudi, Aliaa Amr. 2025. "The Role of Non-Coding RNAs in MYC-Mediated Metabolic Regulation: Feedback Loops and Interactions" Non-Coding RNA 11, no. 2: 27. https://doi.org/10.3390/ncrna11020027
APA StyleAlamoudi, A. A. (2025). The Role of Non-Coding RNAs in MYC-Mediated Metabolic Regulation: Feedback Loops and Interactions. Non-Coding RNA, 11(2), 27. https://doi.org/10.3390/ncrna11020027






