Interplay of microRNAs and circRNAs in Epithelial Ovarian Cancer
Abstract
1. Introduction
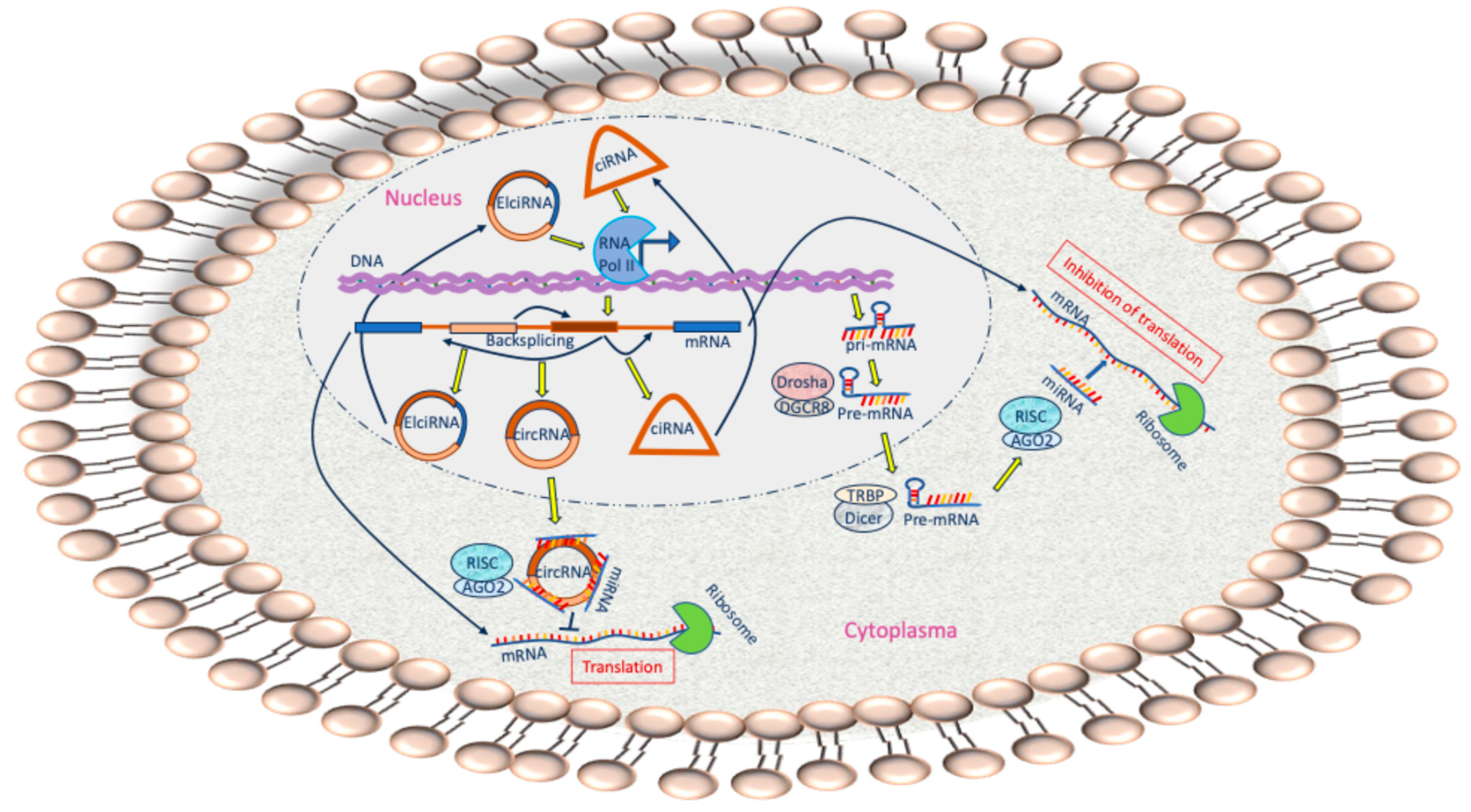
2. ncRNAs
3. Biogenesis and Characteristics of miRNAs
4. Biogenesis and Characteristics of circRNA
5. circRNAs Serve as Sponge for miRNAs
6. Sponging of a miRNA by Several circRNAs
7. EOC Therapies
8. circRNAs as Predictive Biomarkers
9. circRNAs as Therapeutic Agents and Targets
10. Conclusions
Funding
Conflicts of Interest
Abbreviations
| ABCC | ATP-Binding Cassette Subfamily C Member |
| ANXA | Annexin A |
| ATG | Autophagy-related |
| BRD4 | Bromodomain |
| Capn | Calpain |
| CBX | Chromobox |
| CDC5L/PEAK1 | Cell Division Cycle 5 Like |
| CEBPG | CCAAT Enhancer-Binding Protein Gamma |
| CLU | Clusterin |
| CUL | Cullin |
| DUSP | Dual Specificity Phosphatase |
| EIF | Eukaryotic Translation Initiation Factor |
| ELK | ETS Transcription Factor |
| EZH | Enhancer of Zeste Homolog |
| FBX | F-box protein |
| FMNL | Formin-like |
| FOX | Forkhead-box |
| FZD | Frizzled Class Receptor |
| GAS | Growth Arrest-Specific |
| hTERT | Human Telomerase Reverse Transcriptase |
| IGFBP | Insulin-like growth factor binding protein |
| JAK1 | Janus Kinase |
| KLF | Krüppel-like factor |
| KLK | Kallikrein-Related Peptidase |
| LARP | La Ribonucleoprotein |
| LETM1 | Leucine Zipper-EF-Hand-Containing Transmembrane Protein |
| LIF | Leukemia Inhibitory Factor |
| LSM | U6 snRNA-Associated Sm-like Protein |
| MAP3K | Mitogen-Activated Protein Kinase |
| MDM | Mouse double minute |
| MTM | Myotubular Myopathy |
| MTSS | Metastasis suppressor |
| MUC1 | Mucin |
| NF | Nuclear Factor |
| p-GP | P-Glycoprotein |
| PIK3R | Phosphoinositide-3-Kinase Regulatory Subunit |
| PLXN | Plexin |
| PPA | Inorganic Pyrophosphatase |
| PSAT | Phosphoserine Aminotransferase |
| PTEN | Phosphatase and Tensin homolog |
| RAB | Ras-related protein |
| RACGAP | Rac GTPase Activating Protein |
| RASSF | Ras Association Domain Family Member |
| ROCK | Rho-Kinase |
| S100B | Calcium Binding Protein B |
| SFRP | Secreted Frizzled Related Protein |
| SIK | Salt Inducible Kinase |
| SLC | Solute Carrier Family |
| SMG | Serine/threonine-protein kinase |
| SOCS | Suppressor of Cytokine Signaling |
| SOX | SRY-Box Transcription Factor |
| SREBF | Sterol regulatory element-binding transcription factor |
| STAT | Signal transducer and activator of transcription |
| TGFβR | Transforming Growth Factor β receptor |
| TUBB | Tubulin Beta |
| VAMP | Vesicle-Associated Membrane Protein |
| VEGF | Vascular Endothelial Growth Factor |
References
- Sambasivan, S. Epithelial Ovarian Cancer: Review Article. Cancer Treat Res. Commun. 2022, 33, 100629. [Google Scholar] [CrossRef] [PubMed]
- Kandukuri, S.R.; Rao, J. FIGO 2013 Staging System for Ovarian Cancer: What Is New in Comparison to the 1988 Staging System? Curr. Opin. Obstet. Gynecol. 2015, 27, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Organs 2022, 211, 157–182. [Google Scholar] [CrossRef] [PubMed]
- Morand, S.; Devanaboyina, M.; Staats, H.; Stanbery, L.; Nemunaitis, J. Ovarian Cancer Immunotherapy and Personalized Medicine. Int. J. Mol. Sci. 2021, 22, 6532. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liang, X.; Wang, L.; Zhang, X. The Role of MiRNA in Ovarian Cancer: An Overview. Reprod. Sci. 2022, 29, 561–575. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, M.; Zhang, X.; Huang, F.; Wu, K.; Zhang, J.; Liu, J.; Huang, Z.; Luo, H.; Tao, L.; et al. Cellular MicroRNAs Up-Regulate Transcription via Interaction with Promoter TATA-Box Motifs. RNA 2014, 20, 1878–1889. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Wu, J.; Yang, J.; Cho, W.C.; Zheng, Y. Argonaute Proteins: Structural Features, Functions and Emerging Roles. J. Adv. Res. 2020, 24, 317–324. [Google Scholar] [CrossRef]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking Back Three Decades and into the Future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef]
- Hu, G.; Drescher, K.M.; Chen, X.-M. Exosomal MiRNAs: Biological Properties and Therapeutic Potential. Front. Genet. 2012, 3, 56. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28. [Google Scholar] [CrossRef]
- Chen, L.; Shan, G. CircRNA in Cancer: Fundamental Mechanism and Clinical Potential. Cancer Lett. 2021, 505, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jha, A.; Panda, A.C.; Dixit, A. Cancer-Associated CircRNA–MiRNA–MRNA Regulatory Networks: A Meta-Analysis. Front. Mol. Biosci. 2021, 8, 671309. [Google Scholar] [CrossRef] [PubMed]
- Park, E.G.; Ha, H.; Lee, D.H.; Kim, W.R.; Lee, Y.J.; Bae, W.H.; Kim, H.S. Genomic Analyses of Non-Coding RNAs Overlapping Transposable Elements and Its Implication to Human Diseases. Int. J. Mol. Sci. 2022, 23, 8950. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-Coding RNA Networks in Cancer. Nat. Rev. Cancer 2017, 18, 5–18. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J. Therapeutic Potentials of Noncoding RNAs: Targeted Delivery of NcRNAs in Cancer Cells. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2016; Volume 927, pp. 429–458. [Google Scholar]
- Chan, J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- Bortolin-Cavaille, M.-L.; Dance, M.; Weber, M.; Cavaille, J. C19MC MicroRNAs Are Processed from Introns of Large Pol-II, Non-Protein-Coding Transcripts. Nucleic Acids Res. 2009, 37, 3464–3473. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA Genes Are Transcribed by RNA Polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP Recruits the Dicer Complex to Ago2 for MicroRNA Processing and Gene Silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Updated Overview. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2023; Volume 2595, pp. 1–12. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical Relevance of Circulating Cell-Free MicroRNAs in Cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Staicu, C.E.; Predescu, D.V.; Rusu, C.M.; Radu, B.M.; Cretoiu, D.; Suciu, N.; Crețoiu, S.M.; Voinea, S.C. Role of MicroRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells 2020, 9, 169. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P.B. Circulating Non-Coding RNAs in Recurrent and Metastatic Ovarian Cancer. Cancer Drug Resist. 2019, 2, 399. [Google Scholar] [CrossRef]
- Liu, H.Y.; Zhang, Y.Y.; Zhu, B.L.; Feng, F.Z.; Yan, H.; Zhang, H.Y.; Zhou, B. MiR-21 Regulates the Proliferation and Apoptosis of Ovarian Cancer Cells through PTEN/PI3K/AKT. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4149–4155. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA Expression Profiling in Human Ovarian Cancer: MiR-214 Induces Cell Survival and Cisplatin Resistance by Targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. MiRNA Profile in Ovarian Cancer. Exp. Mol. Pathol. 2020, 113, 104381. [Google Scholar] [CrossRef]
- Li, N.; Yang, L.; Wang, H.; Yi, T.; Jia, X.; Chen, C.; Xu, P. MiR-130a and MiR-374a Function as Novel Regulators of Cisplatin Resistance in Human Ovarian Cancer A2780 Cells. PLoS ONE 2015, 10, e0128886. [Google Scholar] [CrossRef]
- Meng, X.; Müller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and Prognostic Relevance of Circulating Exosomal MiR-373, MiR-200a, MiR-200b and MiR-200c in Patients with Epithelial Ovarian Cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef]
- Pan, C.; Stevic, I.; Müller, V.; Ni, Q.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Exosomal MicroRNAs as Tumor Markers in Epithelial Ovarian Cancer. Mol. Oncol. 2018, 12, 1935–1948. [Google Scholar] [CrossRef]
- Yu, R.; Cai, L.; Chi, Y.; Ding, X.; Wu, X. MiR-377 Targets CUL4A and Regulates Metastatic Capability in Ovarian Cancer. Int. J. Mol. Med. 2018, 41, 3147–3156. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, Q.; Luo, K.; Zhang, Q.; Geng, J.; Zhou, X.; Xu, Y.; Qian, M.; Zhang, J.-a.; Ji, L.; et al. MiR-340-FHL2 Axis Inhibits Cell Growth and Metastasis in Ovarian Cancer. Cell Death Dis. 2019, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, Y.; Zhang, Y. CircRNA in Ocular Neovascular Diseases: Fundamental Mechanism and Clinical Potential. Pharmacol. Res. 2023, 197, 106946. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Hu, Q.; Zhou, T. EIciRNA-Mediated Gene Expression: Tunability and Bimodality. FEBS Lett. 2018, 592, 3460–3471. [Google Scholar] [CrossRef]
- Li, J.; Zhang, G.; Liu, C.-G.; Xiang, X.; Le, M.T.N.; Sethi, G.; Wang, L.; Goh, B.-C.; Ma, Z. The Potential Role of Exosomal CircRNAs in the Tumor Microenvironment: Insights into Cancer Diagnosis and Therapy. Theranostics 2022, 12, 87–104. [Google Scholar] [CrossRef]
- Chen, J.; Gu, J.; Tang, M.; Liao, Z.; Tang, R.; Zhou, L.; Su, M.; Jiang, J.; Hu, Y.; Chen, Y.; et al. Regulation of Cancer Progression by CircRNA and Functional Proteins. J. Cell Physiol. 2022, 237, 373–388. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond Transcriptional Regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct. Target Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/Beta-Catenin Signaling: ComponentMacDonald BT; Tamai, K.; He, X. Wnt/Beta-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, J.; Yu, L.; Wu, S.; Qiu, X. Circular RNA Hsa_circ_0000144 Aggravates Ovarian Cancer Progression by Regulating ELK3 via Sponging MiR-610. J. Ovarian Res. 2022, 15, 113. [Google Scholar] [CrossRef]
- Yu, S.; Yu, M.; Chen, J.; Tang, H.; Gong, W.; Tan, H. Circ_0000471 Suppresses the Progression of Ovarian Cancer through Mediating Mir-135b-5p/Dusp5 Axis. Am. J. Reprod. Immunol. 2023, 89, e13651. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.; Li, Y.; Shi, X.; Shen, F.; Chen, M.; Chen, Y.; Wang, J. Hsa_circ_0001445 Works as a Cancer Suppressor via MiR-576-5p/SFRP1 Axis Regulation in Ovarian Cancer. Cancer Med. 2023, 12, 5736–5750. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, J.; Guo, S. Hsa_circ_0004712 Downregulation Attenuates Ovarian Cancer Malignant Development by Targeting the MiR-331-3p/FZD4 Pathway. J. Ovarian Res. 2021, 14, 118. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, Y.; Xu, H.; Shi, Y.; Shen, R.; Teng, F.; Xu, J.; Jia, X. Circular Rna Hsa_circ_0007444 Inhibits Ovarian Cancer Progression through Mir-23a-3p/Dicer1 Axis. Acta Biochim. Biophys. Sin. 2023, 55, 574–586. [Google Scholar] [CrossRef]
- Wei, W.; Wang, N.; Lin, L. Prognostic Value of Hsa_circ_0007615 in Epithelial Ovarian Cancer and Its Regulatory Effect on Tumor Progression. Horm. Metab. Res. 2023, 55, 801–808. [Google Scholar] [CrossRef]
- Li, L.; Yu, P.; Zhang, P.; Wu, H.; Chen, Q.; Li, S.; Wang, Y. Upregulation of Hsa_circ_0007874 Suppresses the Progression of Ovarian Cancer by Regulating the MiR-760/SOCS3 Pathway. Cancer Med. 2020, 9, 2491–2499. [Google Scholar] [CrossRef]
- Li, Y.; Lin, S.; An, N. Hsa_circ_0009910: Oncogenic Circular RNA Targets MicroRNA-145 in Ovarian Cancer Cells. Cell Cycle 2020, 19, 1857–1868. [Google Scholar] [CrossRef]
- Pan, Y.; Huang, Q.; Peng, X.; Yu, S.; Liu, N. Circ_0015756 Promotes Ovarian Cancer Progression via the MiR-145–5p/PSAT1 Axis. Reprod. Biol. 2022, 22, 100702. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Han, B.; Liu, L.; Cui, H.; Liu, H.; Jia, R.; Zhang, X.; Lu, X. Circ_0021573 Acts as a Competing Endogenous RNA to Promote the Malignant Phenotypes of Human Ovarian Cancer Cells. Reprod. Biol. 2023, 23, 100704. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.; Li, X. Circ_0072995 Promotes Ovarian Cancer Progression Through Regulating MiR-122-5p/SLC1A5 Axis. Biochem. Genet. 2022, 60, 153–172. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, H. Circ_0078607 Inhibits the Progression of Ovarian Cancer via Regulating the MiR-32-5p/SIK1 Network. J. Ovarian Res. 2022, 15, 3. [Google Scholar] [CrossRef]
- Hou, W.; Zhang, Y. Circ_0025033 Promotes the Progression of Ovarian Cancer by Activating the Expression of LSM4 via Targeting MiR-184. Pathol. Res. Pract. 2021, 217, 153275. [Google Scholar] [CrossRef]
- Zhang, Y.; Di, Q.; Chen, J.; Chang, M.; Ma, Y.; Yu, J. Circ_0061140 Contributes to the Malignant Progression in Ovarian Cancer Cells by Mediating the RAB1A Level Through Sponging MiR-361-5p. Biochem. Genet. 2022, 60, 1946–1962. [Google Scholar] [CrossRef]
- Ma, L.; Liu, W.; Li, M. Circ_0061140 Contributes to Ovarian Cancer Progression by Targeting MiR-761/LETM1 Signaling. Biochem. Genet. 2023, 61, 628–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Jin, Y.; Hu, Q.; Cheng, S.; Wang, C.; Yang, Z.; Wang, Y. Circular RNA Hsa_circ_0078607 Suppresses Ovarian Cancer Progression by Regulating MiR-518a-5p/Fas Signaling Pathway. J. Ovarian Res. 2020, 13, 64. [Google Scholar] [CrossRef]
- Lyu, M.; Li, X.; Shen, Y.; Lu, J.; Zhang, L.; Zhong, S.; Wang, J. CircATRNL1 and CircZNF608 Inhibit Ovarian Cancer by Sequestering MiR-152-5p and Encoding Protein. Front. Genet. 2022, 13, 784089. [Google Scholar] [CrossRef]
- Lu, M.; Gong, B.; Wang, Y.; Li, J. CircBNC2 Affects Epithelial Ovarian Cancer Progression through the MiR-223-3p/LARP4 Axis. Anticancer Drugs 2023, 34, 384–394. [Google Scholar] [CrossRef]
- Liu, T.; Yuan, L.; Zou, X. Circular RNA Circ-BNC2 (Hsa_circ_0008732) Inhibits the Progression of Ovarian Cancer through MicroRNA-223-3p/FBXW7 Axis. J. Ovarian Res. 2022, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- He, S.L.; Zhao, X.; Yi, S.J. CircAHNAK Upregulates EIF2B5 Expression to Inhibit the Progression of Ovarian Cancer by Modulating the JAK2/STAT3 Signaling Pathway. Carcinogenesis 2022, 43, 941–955. [Google Scholar] [CrossRef]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as Acts as a Sponge of MiR-135b-5p to Suppress Ovarian Cancer Progression. Onco. Targets Ther. 2019, 12, 3869–3879. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, X.; Zhang, S.; Chen, S.; Guan, X.; Li, Q.; Chen, X.; Zhao, Y. CircCRIM1 Promotes Ovarian Cancer Progression by Working as CeRNAs of CRIM1 and Targeting MiR-383-5p/ZEB2 Axis. Reprod. Biol. Endocrinol. 2021, 19, 176. [Google Scholar] [CrossRef]
- Xie, J.; Wang, S.; Li, G.; Zhao, X.; Jiang, F.; Liu, J.; Tan, W. CircEPSTI1 Regulates Ovarian Cancer Progression via Decoying MiR-942. J. Cell Mol. Med. 2019, 23, 3597–3602. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, W.; Fang, J.; Xie, P.; Miao, M.; Yang, H. Circular Rna Circexoc6b Inhibits the Progression of Ovarian Cancer by Sponging MiR-421 and Regulating Rus1 Expression. Onco. Targets Ther. 2020, 13, 8233–8243. [Google Scholar] [CrossRef]
- Sun, D.; Liu, J.; Zhou, L. Upregulation of Circular RNA Circ-FAM53B Predicts Adverse Prognosis and Accelerates the Progression of Ovarian Cancer via the MiR-646/VAMP2 and MiR-647/MDM2 Signaling Pathways. Oncol. Rep. 2019, 42, 2728–2737. [Google Scholar] [CrossRef]
- Wu, M.; Qiu, Q.; Zhou, Q.; Li, J.; Yang, J.; Zheng, C.; Luo, A.; Li, X.; Zhang, H.; Cheng, X.; et al. CircFBXO7/MiR-96-5p/MTSS1 Axis Is an Important Regulator in the Wnt Signaling Pathway in Ovarian Cancer. Mol. Cancer 2022, 21, 137. [Google Scholar] [CrossRef]
- Hu, J.; Wang, L.; Chen, J.; Gao, H.; Zhao, W.; Huang, Y.; Jiang, T.; Zhou, J.; Chen, Y. The Circular RNA Circ-ITCH Suppresses Ovarian Carcinoma Progression through Targeting MiR-145/RASA1 Signaling. Biochem. Biophys. Res. Commun. 2018, 505, 222–228. [Google Scholar] [CrossRef]
- Song, R.; Chai, T.; Liu, J.; Chu, A.; Sun, C.; Liu, Z. Knockdown of CircMFN2 Inhibits Cell Progression and Glycolysis by MiR-198/CUL4B Pathway in Ovarian Cancer. J. Biochem. Mol. Toxicol. 2023, 37, e23383. [Google Scholar] [CrossRef]
- Yang, H.; Guo, Y.; Zhang, Y.; Wang, D.; Zhang, G.; Hou, J.; Yang, J. Circ_MUC16 Attenuates the Effects of Propofol to Promote the Aggressive Behaviors of Ovarian Cancer by Mediating the MiR-1182/S100B Signaling Pathway. BMC Anesthesiol. 2021, 21, 297. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ye, H.; Yang, B.; Ao, M.; Yu, X.; Wu, Y.; Xi, M.; Hou, M. M6A-Modified CircNFIX Promotes Ovarian Cancer Progression and Immune Escape via Activating IL-6R/JAK1/STAT3 Signaling by Sponging MiR-647. Int. Immunopharmacol. 2023, 124, 110879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, Y.; Zhao, W.; Liu, G.; Yang, Q. Circ-PGAM1 Promotes Malignant Progression of Epithelial Ovarian Cancer through Regulation of the MiR-542-3p/CDC5L/PEAK1 Pathway. Cancer Med. 2020, 9, 3500–3521. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lan, S.; Wang, L.; Zhao, J.; Jia, X.; Xu, J.; Sun, G.; Liu, L.; Gong, S.; Wang, N.; et al. Expression of Circ-PHC3 Enhances Ovarian Cancer Progression via Regulation of the MiR-497-5p/SOX9 Pathway. J. Ovarian Res. 2023, 16, 142. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, X.; Chen, A.; Shi, W.; Wang, L.; Yi, R.; Qiu, J. CircPIP5K1A Serves as a Competitive Endogenous RNA Contributing to Ovarian Cancer Progression via Regulation of MiR-661/IGFBP5 Signaling. J. Cell. Biochem. 2019, 120, 19406–19414. [Google Scholar] [CrossRef]
- Wu, S.G.; Zhou, P.; Chen, J.X.; Lei, J.; Hua, L.; Dong, Y.; Hu, M.; Lian, C.L.; Yang, L.C.; Zhou, J. Circ-PTK2 (Hsa_circ_0008305) Regulates the Pathogenic Processes of Ovarian Cancer via MiR-639 and FOXC1 Regulatory Cascade. Cancer Cell Int. 2021, 21, 277. [Google Scholar] [CrossRef]
- Song, W.; Zeng, Z.; Zhang, Y.; Li, H.; Cheng, H.; Wang, J.; Wu, F. CircRNF144B/MiR-342-3p/FBXL11 Axis Reduced Autophagy and Promoted the Progression of Ovarian Cancer by Increasing the Ubiquitination of Beclin-1. Cell Death Dis. 2022, 13, 857. [Google Scholar] [CrossRef]
- Wang, L.L.; Zong, Z.H.; Liu, Y.; Guan, X.; Chen, S.; Zhao, Y. CircRhoC Promotes Tumorigenicity and Progression in Ovarian Cancer by Functioning as a MiR-302e Sponge to Positively Regulate VEGFA. J. Cell Mol. Med. 2019, 23, 8472–8481. [Google Scholar] [CrossRef]
- Li, B.; Zhang, L. CircSETDB1 Knockdown Inhibits the Malignant Progression of Serous Ovarian Cancer through MiR-129-3p-Dependent Regulation of MAP3K3. J. Ovarian Res. 2021, 14, 160. [Google Scholar] [CrossRef]
- Zong, Z.H.; Du, Y.P.; Guan, X.; Chen, S.; Zhao, Y. CircWHSC1 Promotes Ovarian Cancer Progression by Regulating MUC1 and HTERT through Sponging MiR-145 and MiR-1182. J. Exp. Clin. Cancer Res. 2019, 38, 437. [Google Scholar] [CrossRef]
- Sheng, M.; Wei, N.; Yang, H.Y.; Yan, M.; Zhao, Q.X.; Jing, L.J. CircRNA UBAP2 Promotes the Progression of Ovarian Cancer by Sponging MicroRNA-144. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7283–7294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, Q.; Qiu, Q.; Hou, L.; Wu, M.; Li, J.; Li, X.; Lu, B.; Cheng, X.; Liu, P.; et al. CircPLEKHM3 Acts as a Tumor Suppressor through Regulation of the MiR-9/BRCA1/DNAJB6/KLF4/AKT1 Axis in Ovarian Cancer. Mol. Cancer 2019, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Fang, H. Curcumin Inhibits Ovarian Cancer Progression by Regulating Circ-PLEKHM3/MiR-320a/SMG1 Axis. J. Ovarian Res. 2021, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, Y.; Shen, Q.; Chen, Q.; Zhu, X.J.; Jiang, S.S.; Zhang, Q. CircRNA-MYLK Promotes Malignant Progression of Ovarian Cancer through Regulating MicroRNA-652. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5281–5291. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, C.; Lin, J.; Dong, Y.; Wang, Y.; Xia, L. Hsa_circ_0001741 Suppresses Ovarian Cancer Cell Proliferations Through Adsorption of MiR-188-5p and Promotion of FOXN2 Expression. Mol. Biotechnol. 2024, 66, 1477–1483. [Google Scholar] [CrossRef]
- Xu, F.; Ni, M.; Li, J.; Cheng, J.; Zhao, H.; Zhao, J.; Huang, S.; Wu, X. Circ0004390 Promotes Cell Proliferation through Sponging MiR-198 in Ovarian Cancer. Biochem. Biophys. Res. Commun. 2020, 526, 14–20. [Google Scholar] [CrossRef]
- Luo, L.; Gao, Y.Q.; Sun, X.F. Circular RNA ITCH Suppresses Proliferation and Promotes Apoptosis in Human Epithelial Ovarian Cancer Cells by Sponging MiR-10a-α. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8119–8126. [Google Scholar]
- Sun, X.; Luo, L.; Gao, Y. Circular RNA PVT1 Enhances Cell Proliferation but Inhibits Apoptosis through Sponging MicroRNA-149 in Epithelial Ovarian Cancer. J. Obstet. Gynaecol. Res. 2020, 46, 625–635. [Google Scholar] [CrossRef]
- Xu, Q.; Deng, B.; Li, M.; Chen, Y.; Zhuan, L. CircRNA-UBAP2 Promotes the Proliferation and Inhibits Apoptosis of Ovarian Cancer Though MiR-382-5p/PRPF8 Axis. J. Ovarian Res. 2020, 13, 81. [Google Scholar] [CrossRef]
- Tian, Q.; Mu, Q.; Liu, S.; Huang, K.; Tang, Y.; Zhang, P.; Zhao, J.; Shu, C. M6A-Modified CircASXL1 Promotes Proliferation and Migration of Ovarian Cancer through the MiR-320d/RACGAP1 Axis. Carcinogenesis 2023, 44, 859–870. [Google Scholar] [CrossRef]
- Qu, B.; Sun, L.; Xiao, P.; Shen, H.; Ren, Y.; Zhang, J. CircCDK17 Promotes the Proliferation and Metastasis of Ovarian Cancer Cells by Sponging MiR-22-3p to Regulate CD147 Expression. Carcinogenesis 2024, 45, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zheng, Y.; You, J.; Han, Y.; Lu, X.; Wang, X.; Shi, C.; Zhu, W. Hsa_circ_0001535 Inhibits the Proliferation and Migration of Ovarian Cancer by Sponging MiR-593-3p, Upregulating PTEN Expression. Clin. Transl. Oncol. 2023, 25, 2901–2910. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, X.; Qiao, L.; Wang, H. CircRNA Circ_0000554 Promotes Ovarian Cancer Invasion and Proliferation by Regulating MiR-567. Environ. Sci. Pollut. Res. 2022, 29, 19072–19080. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Meng, K.; Qiu, R. Circular RNA Circ_0013958 Functions as a Tumor Promoter in Ovarian Cancer by Regulating MiR-637/PLXNB2 Axis. Front. Genet. 2021, 12, 644451. [Google Scholar] [CrossRef]
- Lin, X.; Chen, Y.; Ye, X.; Xia, X. Circular RNA ABCB10 Promotes Cell Proliferation and Invasion, but Inhibits Apoptosis via Regulating the MicroRNA-1271-Mediated Capn4/Wnt/β-Catenin Signaling Pathway in Epithelial Ovarian Cancer. Mol. Med. Rep. 2021, 23, 387. [Google Scholar] [CrossRef]
- Wang, N.; Cao, Q.X.; Tian, J.; Ren, L.; Cheng, H.L.; Yang, S.Q. Circular RNA MTO1 Inhibits the Proliferation and Invasion of Ovarian Cancer Cells Through the MiR-182-5p/KLF15 Axis. Cell Transplant. 2020, 29, 963689720943613. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sun, H. The Molecular Mechanism of CircRHOBTB3 Inhibits the Proliferation and Invasion of Epithelial Ovarian Cancer by Serving as the CeRNA of MiR-23a-3p. J. Ovarian Res. 2022, 15, 66. [Google Scholar] [CrossRef]
- Gong, J.; Xu, X.; Zhang, X.; Zhou, Y. Circular RNA-9119 Suppresses in Ovarian Cancer Cell Viability via Targeting the MicroRNA-21-5p-PTEN-Akt Pathway. Aging 2020, 12, 14314–14328. [Google Scholar] [CrossRef]
- Lu, H.; Zheng, G.; Gao, X.; Chen, C.; Zhou, M.; Zhang, L. Propofol Suppresses Cell Viability, Cell Cycle Progression and Motility and Induces Cell Apoptosis of Ovarian Cancer Cells through Suppressing MEK/ERK Signaling via Targeting CircVPS13C/MiR-145 Axis. J. Ovarian Res. 2021, 14, 30. [Google Scholar] [CrossRef]
- Wei, X.; Lv, H.; Yang, S.; Yang, X. CircRNA PLOD2 Enhances Ovarian Cancer Propagation by Controlling MiR-378. Saudi J. Biol. Sci. 2021, 28, 6260–6265. [Google Scholar] [CrossRef]
- Zhang, M.; Xia, B.; Xu, Y.; Zhang, Y.; Xu, J.; Lou, G. Circular RNA (Hsa:Circ_0051240) Promotes Cell Proliferation, Migration and Invasion in Ovarian Cancer through MiR-637/KLK4 Axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 224–1233. [Google Scholar] [CrossRef]
- Li, Q.-h.; Liu, Y.; Chen, S.; Zong, Z.-h.; Du, Y.-p; Sheng, X.-j.; Zhao, Y. Circ-CSPP1 Promotes Proliferation, Invasion and Migration of Ovarian Cancer Cells by Acting as a MiR-1236-3p Sponge. Biomed. Pharmacother. 2019, 114, 108832. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wang, R.Y.; Yu, X.Z.; Wu, Y.K.; Yang, B.W.; Ao, M.Y.; Xi, M.R.; Hou, M.M. Exosomal CircNFIX Promotes Angiogenesis in Ovarian Cancer via MiR-518a-3p/TRIM44 Axis. Kaohsiung J. Med. Sci. 2023, 39, 26–39. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Li, H.; Sun, Y.; Tong, X. Downregulation of Hsa_circ_0026123 Suppresses Ovarian Cancer Cell Metastasis and Proliferation through the MiR-124-3p/EZH2 Signaling Pathway. Int. J. Mol. Med. 2021, 47, 668–676. [Google Scholar] [CrossRef]
- Zeng, X.Y.; Yuan, J.; Wang, C.; Zeng, D.; Yong, J.H.; Jiang, X.Y.; Lan, H.; Xiao, S.S. CircCELSR1 Facilitates Ovarian Cancer Proliferation and Metastasis by Sponging MiR-598 to Activate BRD4 Signals. Mol. Med. 2020, 26, 70. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Zhou, J.H.; Shen, F.R.; Shi, X.; Chen, Y.G. CircATRNL1 Activates Smad4 Signaling to Inhibit Angiogenesis and Ovarian Cancer Metastasis via MiR-378. Mol. Oncol. 2021, 15, 1217–1233. [Google Scholar] [CrossRef]
- Wang, F.; Niu, Y.; Chen, K.; Yuan, X.; Qin, Y.; Zheng, F.; Cui, Z.; Lu, W.; Wu, Y.; Xia, D. Extracellular Vesicle-Packaged CircATP2B4 Mediates M2 Macrophage Polarization via MiR-532-3p/SREBF1 Axis to Promote Epithelial Ovarian Cancer Metastasis. Cancer Immunol. Res. 2023, 11, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ren, X.; Jiaoting, E.; Zhou, Y.; Bian, R. Exosome-Transmitted CircIFNGR2 Modulates Ovarian Cancer Metastasis via MiR-378/ST5 Axis. Mol. Cell Biol. 2023, 43, 22–42. [Google Scholar] [CrossRef]
- Xie, W.; Liu, L.; He, C.; Zhao, M.; Ni, R.; Zhang, Z.; Shui, C. Circ_0002711 Knockdown Suppresses Cell Growth and Aerobic Glycolysis by Modulating MiR-1244/ROCK1 Axis in Ovarian Cancer. J. Biosci. 2021, 46, 21. [Google Scholar] [CrossRef]
- Deng, G.; Zhou, X.; Chen, L.; Yao, Y.; Li, J.; Zhang, Y.; Luo, C.; Sun, L.; Tang, J. High Expression of ESRP1 Regulated by Circ-0005585 Promotes Cell Colonization in Ovarian Cancer. Cancer Cell Int. 2020, 20, 174. [Google Scholar] [CrossRef]
- Tang, Q.; Wen, H.; Hu, H.; Chen, X.; Xu, S.; Fan, L.; Liu, L.; Li, J. Circ_0070203 Promotes Epithelial-Mesenchymal Transition in Ovarian Serous Cystadenocarcinoma through MiR-370-3p/TGFβR2 Axis. Recent Pat. Anticancer Drug Discov. 2023, 19, 233–246. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, Y.; Yang, M.; Qiao, L.; Wang, H.; Jiang, H.; Fu, M.; Qin, J.; Xu, S. Hsa_circ_0013561 Promotes Epithelial-Mesenchymal Transition and Tumor Progression by Regulating ANXA2 via MiR-23b-3p in Ovarian Cancer. Cancer Gene Ther. 2024, 31, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, Z.N.; Qiu, B.Q.; Hu, M.; Liang, X.Q.; Dai, X.; Hong, D.; Sun, Y.F. CircRNA FGFR3 Induces Epithelial-Mesenchymal Transition of Ovarian Cancer by Regulating MiR-29a-3p/E2F1 Axis. Aging 2020, 12, 14080–14091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, Y.; Ye, W.; Jiang, J.; Wu, C. Circular RNA S-7 Promotes Ovarian Cancer EMT via Sponging MiR-641 to up-Regulate ZEB1 and MDM2. Biosci. Rep. 2020, 40, BSR20200825. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.; Zhu, G.; Hong, L.; Hu, C.; Wang, K.; Cui, K.; Hao, C. RNA-Binding Protein IGF2BP2 Enhances Circ_0000745 Abundancy and Promotes Aggressiveness and Stemness of Ovarian Cancer Cells via the MicroRNA-3187-3p/ERBB4/PI3K/AKT Axis. J. Ovarian Res. 2021, 14, 154. [Google Scholar] [CrossRef]
- Huang, K.; Liu, D.; Su, C. Circ_0007841 Accelerates Ovarian Cancer Development through Facilitating MEX3C Expression by Restraining MiR-151-3p Activity. Aging 2021, 13, 12058–12066. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Qu, S.; Zhai, Y.; Yang, X. Circ_0025033 Promotes Ovarian Cancer Development via Regulating the Hsa_miR-370-3p/SLC1A5 Axis. Cell. Mol. Biol. Lett. 2022, 27, 94. [Google Scholar] [CrossRef]
- Li, X.; Jiang, X.; Lu, J.; Lin, Y.; Jiang, L.; Li, Y.; Wan, F.; Wang, C. CircCERS6 Suppresses the Development of Epithelial Ovarian Cancer Through Mediating MiR-630/RASSF8. Biochem. Genet. 2022, 60, 2611–2629. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, M.; Feng, Z.; Wu, H.; Wu, J.; Ha, X.; Wu, Y.; Chen, S.; Xu, F.; Wen, H.; et al. AUF1-Induced Circular RNA Hsa_circ_0010467 Promotes Platinum Resistance of Ovarian Cancer through MiR-637/LIF/STAT3 Axis. Cell. Mol. Life Sci. 2023, 80, 256. [Google Scholar] [CrossRef]
- Dai, C.; Dai, S.Y.; Gao, Y.; Yan, T.; Zhou, Q.Y.; Liu, S.-j.; Liu, X.; Deng, D.N.; Wang, D.H.; Qin, Q.F.; et al. Circ_0078607 Increases Platinum Drug Sensitivity via MiR-196b-5p/GAS7 Axis in Ovarian Cancer. Epigenetics 2023, 18, 2175565. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, Y. Circ_0007841 Knockdown Confers Cisplatin Sensitivity to Ovarian Cancer Cells by Down-Regulation of NFIB Expression in a MiR-532-5p-Dependent Manner. J. Chemother. 2023, 35, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; He, W.; Zhao, H.; Zhao, P. Circ_0026123 Promotes Cisplatin Resistance and Progression of Ovarian Cancer by Upregulating RAB1A through Sequestering MiR-543. Anticancer Drugs 2022, 33, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Han, Y.; Qiao, H.; Han, Y.; Lu, X.; Lu, Y.; Wang, X.; Kai, H.; Zheng, Y. Hsa_circ_0063804 Enhances Ovarian Cancer Cells Proliferation and Resistance to Cisplatin by Targeting MiR-1276/CLU Axis. Aging 2022, 14, 4699–4713. [Google Scholar] [CrossRef]
- Yin, Y.; Li, J.; Rong, J.; Zhang, B.; Wang, X.; Han, H. Circ_0067934 Reduces JNK Phosphorylation through a MicroRNA-545-3p/PPA1 Axis to Enhance Tumorigenesis and Cisplatin Resistance in Ovarian Cancer. Immunopharmacol. Immunotoxicol. 2022, 44, 261–274. [Google Scholar] [CrossRef]
- Luo, Y.; Gui, R. Circulating Exosomal Circfoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J. Gynecol. Oncol. 2020, 31, e75. [Google Scholar] [CrossRef]
- Sheng, H.; Wang, X. Knockdown of Circ-PIP5K1A Overcomes Resistance to Cisplatin in Ovarian Cancer by MiR-942-5p/NFIB Axis. Anticancer Drugs 2023, 34, 214–226. [Google Scholar] [CrossRef]
- Li, H.; Lin, R.; Zhang, Y.; Zhu, Y.; Huang, S.; Lan, J.; Lu, N.; Xie, C.; He, S.; Zhang, W. N6-Methyladenosine-Modified CircPLPP4 Sustains Cisplatin Resistance in Ovarian Cancer Cells via PIK3R1 Upregulation. Mol. Cancer 2024, 23, 5. [Google Scholar] [CrossRef]
- Guo, M.; Li, S.; Zhao, X.; Yuan, Y.; Zhang, B.; Guan, Y. Knockdown of Circular Rna Hsa_circ_0000714 Can Regulate Rab17 by Sponging Mir-370-3p to Reduce Paclitaxel Resistance of Ovarian Cancer through Cdk6/Rb Pathway. Onco. Targets Ther. 2020, 13, 13211–13224. [Google Scholar] [CrossRef]
- Huang, H.; Yan, L.; Zhong, J.; Hong, L.; Zhang, N.; Luo, X. Circ_0025033 Deficiency Suppresses Paclitaxel Resistance and Malignant Development of Paclitaxel-Resistant Ovarian Cancer Cells by Modulating the MiR-532-3p/FOXM1 Network. Immunopharmacol. Immunotoxicol. 2022, 44, 275–286. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, J.-e.; Chen, Y.; Wu, Q. Circ_0061140 Knockdown Inhibits Tumorigenesis and Improves PTX Sensitivity by Regulating MiR-136/CBX2 Axis in Ovarian Cancer. J. Ovarian Res. 2021, 14, 136. [Google Scholar] [CrossRef]
- Ying, H.; Zhao, R.; Yu, Q.; Zhang, K.; Deng, Q. CircATL2 Enhances Paclitaxel Resistance of Ovarian Cancer via Impacting MiR-506-3p/NFIB Axis. Drug Dev. Res. 2022, 83, 512–524. [Google Scholar] [CrossRef]
- Wei, S.; Qi, L.; Wang, L. Overexpression of Circ-CELSR1 Facilitates Paclitaxel Resistance of Ovarian Cancer by Regulating MiR-149-5p/SIK2 Axis. Anticancer Drugs 2021, 32, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, J.; Quan, C.; Wen, H.; Feng, Z.; Hu, Q.; Zhu, J.; Huang, Y.; Wu, X. CircCELSR1 (Hsa_circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via MiR-1252. Mol. Ther. Nucleic Acids 2020, 19, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, Z.; Yang, S.; Wang, Y.; Luan, Z. CircEXOC6B Suppresses the Proliferation and Motility and Sensitizes Ovarian Cancer Cells to Paclitaxel Through MiR-376c-3p/FOXO3 Axis. Cancer Biother. Radiopharm. 2022, 37, 802–814. [Google Scholar] [CrossRef]
- Huang, C.; Qin, L.; Chen, S.; Huang, Q. CircSETDB1 Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Sponging MiR-508-3p and Regulating ABCC1 Expression. Anticancer Drugs 2023, 34, 395–404. [Google Scholar] [CrossRef]
- Chen, Y.-y.; Tai, Y. chun Hsa_circ_0006404 and Hsa_circ_0000735 Regulated Ovarian Cancer Response to Docetaxel Treatment via Regulating P-GP Expression. Biochem. Genet. 2022, 60, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Guan, R.; Gong, K.; Xie, H.; Shi, L. Circ_FURIN Knockdown Assuages Testosterone-Induced Human Ovarian Granulosa-like Tumor Cell Disorders by Sponging MiR-423-5p to Reduce MTM1 Expression in Polycystic Ovary Syndrome. Reprod. Biol. Endocrinol. 2022, 20, 32. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 Promotes Autophagy of Epithelial Ovarian Cancer via Interaction with ATG13 and MiR-199a. Mol. Cancer 2020, 19, 45. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, H.; Hu, J. CircRAB11FIP1 Promoted Autophagy Flux of Ovarian Cancer through DSC1 and MiR-129. Cell Death Dis. 2021, 12, 219. [Google Scholar] [CrossRef]
- Qu, D.D.; Zou, X.; Liu, Z.L. Propofol Modulates Glycolysis Reprogramming of Ovarian Tumor via Restraining Circular RNA-Zinc Finger RNA-Binding Protein/MicroRNA-212-5p/Superoxide Dismutase 2 Axis. Bioengineered 2022, 13, 11881–11892. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Liu, Y.; Liu, C.; Wang, Y.; He, L.; Cheng, X.; Peng, Y.; Xia, L.; Wu, X.; et al. NFIB Facilitates Replication Licensing by Acting as a Genome Organizer. Nat. Commun. 2023, 14, 5076. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.C.; Park, S.J.; Nam, M.; Kang, J.; Kim, K.; Yeo, J.H.; Kim, J.K.; Heo, Y.; Lee, H.S.; Lee, M.Y.; et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab. 2020, 31, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.-B.; Li, X.Z.; Zeng, S.; Liu, C.; Yang, S.M.; Yang, L.; Hu, C.J.; Bai, J.Y. Regulation of the Master Regulator FOXM1 in Cancer. Cell Commun. Signal. 2018, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liao, J.; Liu, J.; Meng, C.; Liu, B.; Shao, C. Analysis of Correlation between Rab1A Expression and Its Prognosis in Cancers: A Meta-Analysis. Clin. Lab. 2024, 70, 1. [Google Scholar] [CrossRef]
- Bertorello, A.M.; Zhu, J.K. SIK1/SOS2 Networks: Decoding Sodium Signals via Calcium-Responsive Protein Kinase Pathways. Pflug. Arch. 2009, 458, 613–619. [Google Scholar] [CrossRef]
- Green, D.R.; Llambi, F. Cell Death Signaling. Cold Spring Harb. Perspect. Biol. 2015, 7, a006080. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Du, J.H.; Xing, Y.J.; Cheng, Y.M.; He, R.F.; Liang, X.L.; Li, H.L.; Yang, Y.X. SIK2: A Critical Glucolipid Metabolic Reprogramming Regulator and Potential Target in Ovarian Cancer. J. Obstet. Gynaecol. Res. 2023, 49, 2000–2009. [Google Scholar] [CrossRef]
- Karin, M.; Baud, V. Is NF-KappaB a Good Target for Cancer Therapy? Hopes and Pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar]
- Brzozowa-Zasada, M.; Piecuch, A.; Michalski, M.; Segiet, O.; Kurek, J.; Harabin-Słowińska, M.; Wojnicz, R. Notch and Its Oncogenic Activity in Human Malignancies. Eur. Surg. 2017, 49, 199–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, Q.; Su, B.; Xu, H.; Sun, Y.; Sun, P.; Li, R.; Peng, X.; Cai, J. Role of RASA1 in Cancer: A Review and Update (Review). Oncol. Rep. 2020, 44, 2386–2396. [Google Scholar] [CrossRef]
- Damia, G.; Broggini, M. Platinum Resistance in Ovarian Cancer: Role of DNA Repair. Cancers 2019, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Chen, L. Progress in Research on Paclitaxel and Tumor Immunotherapy. Cell. Mol. Biol. Lett. 2019, 24, 40. [Google Scholar] [CrossRef] [PubMed]
- Yélamos, J.; Moreno-Lama, L.; Jimeno, J.; Ali, S.O. Immunomodulatory Roles of PARP-1 and PARP-2: Impact on PARP-Centered Cancer Therapies. Cancers 2020, 12, 392. [Google Scholar] [CrossRef]
- Irusta, G. Roads to the Strategic Targeting of Ovarian Cancer Treatment. Reproduction 2021, 161, R1–R11. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Beilerli, A.; Begliarzade, S.; Sufianov, A.; Ilyasova, T.; Liang, Y.; Beylerli, O. Circulating CiRS-7 as a Potential Non-Invasive Biomarker for Epithelial Ovarian Cancer: An Investigative Study. Noncoding RNA Res. 2022, 7, 197–204. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Zhang, L.Y.; Wang, L. CircHIPK3 Is Upregulated and Predicts a Poor Prognosis in Epithelial Ovarian Cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3713–3718. [Google Scholar] [CrossRef]
- Ning, L.; Long, B.; Zhang, W.; Yu, M.; Wang, S.; Cao, D.; Yang, J.; Shen, K.; Huang, Y.; Lang, J. Circular RNA Profiling Reveals CircEXOC6B and CircN4BP2L2 as Novel Prognostic Biomarkers in Epithelial Ovarian Cancer. Int. J. Oncol. 2018, 53, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Lang, J.; Long, B.; Wu, L. Diagnostic Value of CircN4BP2L2 in Type I and Type II Epithelial Ovarian Cancer. BMC Cancer 2022, 22, 1210. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Liu, Y.; Wang, M. Circular RNA Profiling Reveals CircRNA1656 as a Novel Biomarker in High Grade Serous Ovarian Cancer. Biosci. Trends 2019, 13, 204–211. [Google Scholar] [CrossRef]
- Ge, L.; Sun, Y.; Shi, Y.; Liu, G.; Teng, F.; Geng, Z.; Chen, X.; Xu, H.; Xu, J.; Jia, X. Plasma CircRNA Microarray Profiling Identifies Novel CircRNA Biomarkers for the Diagnosis of Ovarian Cancer. J. Ovarian Res. 2022, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, Z.; Jin, Y.; Cheng, S.; Yang, J.; Wang, Y. Low Expression of Circular RNA Hsa_Circ_0078607 Predicts Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancer Manag. Res. 2021, 13, 2877–2883. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Wang, P.L.; Gao, Y.; Liang, W.T. Circular RNA-LARP4 Is Lower Expressed and Serves as a Potential Biomarker of Ovarian Cancer Prognosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7178–7182. [Google Scholar]
- Bao, L.; Zhong, J.; Pang, L. Upregulation of Circular RNA VPS13C-Has-Circ-001567 Promotes Ovarian Cancer Cell Proliferation and Invasion. Cancer Biother. Radiopharm. 2019, 34, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, W.; Li, Q.H.; Xie, B.M.; Shen, F.; Du, Y.P.; Zong, Z.H.; Wang, L.L.; Wei, X.Q.; Zhao, Y. Circ-NOLC1 Promotes Epithelial Ovarian Cancer Tumorigenesis and Progression by Binding ESRP1 and Modulating CDK1 and RhoA Expression. Cell Death Discov. 2021, 7, 22. [Google Scholar] [CrossRef]
- Pei, C.; Wang, H.; Shi, C.; Zhang, C.; Wang, M. CircRNA Hsa_circ_0013958 May Contribute to the Development of Ovarian Cancer by Affecting Epithelial-Mesenchymal Transition and Apoptotic Signaling Pathways. J. Clin. Lab. Anal. 2020, 34, e23292. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Y.; Zhang, W.; Lang, J.; Ning, L. Utility of Plasma CircBNC2 as a Diagnostic Biomarker in Epithelial Ovarian Cancer. Onco. Targets Ther. 2019, 12, 9715–9723. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Zhang, X.; Liu, G. Serum CircSETDB1 Is a Promising Biomarker for Predicting Response to Platinum-Taxane-Combined Chemotherapy and Relapse in High-Grade Serous Ovarian Cancer. Onco Targets Ther. 2019, 12, 7451–7457. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, X.; Xia, X.; Lin, X. Circular RNA ABCB10 Correlates with Advanced Clinicopathological Features and Unfavorable Survival, and Promotes Cell Proliferation While Reduces Cell Apoptosis in Epithelial Ovarian Cancer. Cancer Biomark. 2019, 26, 151–161. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Xing, Y.; Wang, Z.; Lu, Z.; Xia, J.; Xie, Z.; Jiao, M.; Liu, R.; Chu, Y. MicroRNAs: Immune Modulators in Cancer Immunotherapy. Immunother. Adv. 2021, 1, ltab006. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. Micrornas: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Circular RNAs as Therapeutic Agents and Targets. Front. Physiol. 2018, 9, 1262. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering Circular RNA for Potent and Stable Translation in Eukaryotic Cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef] [PubMed]
- Rossbach, O. Artificial Circular RNA Sponges Targeting MicroRNAs as a Novel Tool in Molecular Biology. Mol. Ther. Nucleic Acids 2019, 17, 452–454. [Google Scholar] [CrossRef]
- Yang, Q.; Li, F.; He, A.T.; Yang, B.B. Circular RNAs: Expression, Localization, and Therapeutic Potentials. Mol. Ther. 2021, 29, 1683–1702. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.Z.; Cao, S.M.; Lei, Y.N.; Liu, C.X.; Guo, S.K.; Shan, L.; et al. Screening for Functional Circular RNAs Using the CRISPR–Cas13 System. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef]
- Meganck, R.M.; Borchardt, E.K.; Castellanos Rivera, R.M.; Scalabrino, M.L.; Wilusz, J.E.; Marzluff, W.F.; Asokan, A. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol. Ther. Nucleic Acids 2018, 13, 89–98. [Google Scholar] [CrossRef]
- Ma, J.; Du, W.W.; Zeng, K.; Wu, N.; Fang, L.; Lyu, J.; Yee, A.J.; Yang, B.B. An Antisense Circular RNA CircSCRIB Enhances Cancer Progression by Suppressing Parental Gene Splicing and Translation. Mol. Ther. 2021, 29, 2754–2768. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering Exosomes as Refined Biological Nanoplatforms for Drug Delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 Family: A Potential Tumor Suppressor and Therapeutic Candidate in Cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I Study of MRX34, a Liposomal MiR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]

| circRNAs | miRNAs | Targets | Function | Ref. | |
|---|---|---|---|---|---|
| * Increasing | ** Decreasing | ||||
| circ_0000144 | miR-610 | ELK3 | progression | - | [45] |
| circ_0000471 | miR-135b | dusp5 | progression | [46] | |
| circ_0001445 | miR-576 | SFRP1 | - | progression | [47] |
| circ_0004712 | miR-331 | FZD4 | progression | - | [48] |
| circ_0007444 | miR-23a | Dicer1 | - | progression | [49] |
| circ_0007615 | miR-874 | TUBB3 | progression | - | [50] |
| circ_0007874 | miR-760 | SOCS3 | - | progression | [51] |
| circ_0009910 | miR-145 | - | progression | - | [52] |
| circ_0015756 | miR-145 | PSAT1 | progression | - | [53] |
| circ_0021573 | miR-936 | CUL4B | progression | - | [54] |
| circ_0072995 | miR-122 | SLC1A5 | progression | - | [55] |
| circ_0078607 | miR-32 | SIK1 | - | progression | [56] |
| circ_0025033 | miR-184 | LSM4 | progression | - | [57] |
| circ_0061140 | miR-361 | RAB1A | progression | - | [58] |
| miR-761 | LETM1 | progression | - | [59] | |
| circ_0070203 | miR-518a | Fas | - | progression | [60] |
| circ_ATRNL1 | miR-152 | - | progression | - | [61] |
| circ_BNC2 | miR-223 | LARP4 | - | progression | [62] |
| miR-223 | FBXW7 | - | progression | [63] | |
| circ_AHNAK | miR-28 | EIF2B5 | - | progression | [64] |
| circ_CDR1 | miR-135b | - | - | progression | [65] |
| circ_CRIM1 | miR-383 | ZEB2 | progression | - | [66] |
| circ_EPSTI1 | miR-942 | - | progression | - | [67] |
| circ_EXOC6B | miR-421 | RUS1 | - | progression | [68] |
| circ_FAM53B | miR-646 | VAMP2 | progression | - | [69] |
| miR-647 | MDM2 | progression | - | [69] | |
| circ_FBXO7 | miR-96-5p | MTSS1/Wnt | progression | - | [70] |
| circ_ITCH | miR-145 | RASA1 | - | progression | [71] |
| circ_MFN2 | miR-198 | CUL4B | progression, glycolysis | - | [72] |
| circ_MUC16 | miR-1182 | S100B | progression | - | [73] |
| circ_NFIX | miR-647 | JAK1/STAT3 | progression | - | [74] |
| circ_PGAM1 | miR-542 | CDC5L/PEAK1 | progression | - | [75] |
| circ_PHC3 | miR-497 | SOX9 | progression | - | [76] |
| circ_PIP5K1A | miR-661 | IGFBP5 | progression | - | [77] |
| circ-PTK2 | miR-639 | FOXC1 | progression | - | [78] |
| circ_RNF144B | miR-342 | FBXL11 | progression | - | [79] |
| circ_RhoC | miR-302e | VEGFA | progression | - | [80] |
| circ_SETDB1 | miR-129 | MAP3K3 | progression | - | [81] |
| circ_WHSC1 | miR-145 | MUC1 | progression | - | [82] |
| circ_WHSC1 | miR-1182 | hTERT | progression | - | [82] |
| circ_ZNF608 | miR-152 | - | progression | - | [61] |
| circ_UBAP2 | miR-144 | - | - | progression | [83] |
| circ_PLEKHM3 | miR-9 | BRCA1/DNAJB6/KLF4/AKT1 | - | progression | [84] |
| miR-320a | SMG1 | - | progression | [85] | |
| circ_MYLK | miR-652 | - | progression | - | [86] |
| circ_0001741 | miR-188 | FOXN2 | - | proliferation | [87] |
| circ_0004390 | miR-198 | - | proliferation | - | [88] |
| circ_ITCH | miR-10a | - | - | proliferation | [89] |
| circ_PVT1 | miR-149 | - | proliferation | - | [90] |
| circ_UBAP2 | miR-382 | PRPF8 | proliferation | - | [91] |
| circ_ASXL1 | miR-320d | RACGAP1 | proliferation, migration | - | [92] |
| circ_CDK17 | miR-22 | CD147 | proliferation, migration | - | [93] |
| circ_0001535 | miR-593 | PTEN | - | proliferation, migration [94] | |
| circ_0000554 | miR-567 | - | proliferation, invasion | - | [95] |
| circ_0013958 | miR-637 | PLXNB2 | proliferation, invasion | - | [96] |
| circ_ABCB10 | miR-1271 | Capn4/Wnt | proliferation, invasion | - | [97] |
| circ_MTO1 | miR-182 | KLF15 | - | proliferation, invasion | [98] |
| circ_RHOBTB3 | miR-23a | - | - | proliferation, invasion | [99] |
| circ_9119 | miR-21 | PTEN/Akt | - | cell viability | [100] |
| circ_VPS13C | miR-145 | MEK/ERK | cell cycle, motility | - | [101] |
| circ_PLOD2 | miR-378 | - | propagation | - | [102] |
| circ_0051240 | miR-637 | KLK4 | migration, invasion | - | [103] |
| circ_CSPP1 | miR-1236 | - | invasion, migration | - | [104] |
| circ_NFIX | miR-518a | TRIM44 | angiogenesis | - | [105] |
| circ_0026123 | miR-124 | EZH2 | proliferation, metastasis | - | [106] |
| circ_CELSR1 | miR-598 | BRD4 | proliferation, metastasis | - | [107] |
| circ_ATRNL1 | miR-378 | Smad4 | - | angiogenesis, metastasis | [108] |
| circ_ATP2B4 | miR-532 | SREBF1 | metastasis | - | [109] |
| circ_IFNGR2 | miR-378 | ST5 | metastasis | - | [110] |
| circ_0002711 | miR-1244 | ROCK1 | cell growth, glycolysis | - | [111] |
| circ_0005585 | miR-23a/b/ 15a/15b/16 | ESRP1 | colonization | - | [112] |
| circ_0070203 | miR-370 | TGFβR2 | EMT | - | [113] |
| circ_0013561 | miR-23b | ANXA2 | EMT | - | [114] |
| circ_FGFR3 | miR-29a | E2F1 | EMT | - | [115] |
| circ_S-7 | miR-641 | ZEB1, MDM2 | EMT | - | [116] |
| circ_0000745 | miR-3187 | ERBB4/PI3K/AKT | cell stemness | - | [117] |
| circ_0007841 | miR-151 | MEX3C | development | - | [118] |
| circ_002503 | miR-370 | SLC1A5 | development | - | [119] |
| circ_CERS6 | miR-630 | RASSF8 | - | development | [120] |
| circ_0010467 | miR-637 | LIF/STAT3 | platinum resistance | - | [121] |
| circ_0070203 | miR-196b | GAS7 | platinum sensitivity | - | [122] |
| circ_0007841 | miR-532 | NFIB | cisplatin resistance | - | [123] |
| circ_0026123 | miR-543 | RAB1A | cisplatin resistance | - | [124] |
| circ_0063804 | miR-1276 | CLU | cisplatin resistance | - | [125] |
| circ_0067934 | miR-545 | PPA1 | - | cisplatin resistance | [126] |
| circ_Foxp1 | miR-22 | CEBPG | cisplatin resistance | - | [127] |
| miR-150 | FMNL3 | cisplatin resistance | - | [127] | |
| circ_NFIX | miR-942 | NFIB | cisplatin resistance | - | [128] |
| circ_PLPP4 | miR-136 | PIK3R1 | cisplatin resistance | - | [129] |
| circ_0000714 | miR-370 | RAB17, CDK6/RB | paclitaxel resistance | - | [130] |
| circ_0025033 | miR-532 | FOXM1 | paclitaxel resistance | - | [131] |
| circ_0061140 | miR-136 | CBX2 | - | paclitaxel sensitivity | [132] |
| circ_ATL2 | miR-506 | NFIB | paclitaxel resistance | - | [133] |
| circ_CELSR1 | miR-149 | SIK2 | paclitaxel resistance | - | [134] |
| miR-1252 | FOXR2 | paclitaxel resistance | - | [135] | |
| circ_EXOC6B | miR-376c | FOXO3 | - | paclitaxel sensitivity | [136] |
| circ_SETDB1 | miR-508 | ABCC1 | paclitaxel resistance | - | [137] |
| circ_0000735 | miR-526b | p-GP | docetaxel resistance | - | [138] |
| circ_0006404 | miR-346 | p-GP | - | docetaxel | [138] |
| circ_FURIN | miR-423 | MTM1 | testosterone effect | - | [139] |
| circ_MUC16 | miR-199a | ATG13 | autophagy | - | [140] |
| circ_RAB11FIP1 | miR-129 | DSC1 | autophagy | - | [141] |
| circ_zinc finger | miR-212 | superoxide dismutase 2 | glycolysis | - | [142] |
| circRNAs | miRNAs | Targets | Function | Ref. | |
|---|---|---|---|---|---|
| * Increasing | ** Decreasing | ||||
| circ_CDK17 | miR-22 | CD147 | proliferation, migration | - | [93] |
| circ_Foxp1 | CEBPG | cisplatin resistance | - | [127] | |
| circ_0007444 | miR-23a | Dicer1 | - | progression | [49] |
| circ_RHOBTB3 | - | - | proliferation, invasion | [99] | |
| circ_0005585 | miR-23a/b | ESRP1 | colonization | - | [112] |
| circ_0013561 | miR-23b | ANXA2 | EMT | - | [114] |
| circ_RAB11FIP1 | miR-129 | DSC1 | autophagy | - | [141] |
| circ_SETDB1 | MAP3K3 | progression | - | [81] | |
| circ_0000471 | miR-135b | dusp5 | - | progression | [46] |
| circ_CDR1 | - | - | progression | [65] | |
| circ_0061140 | miR-136 | CBX2 | - | paclitaxel sensitivity | [132] |
| circ_PLPP4 | PIK3R1 | cisplatin resistance | - | [129] | |
| circ_0009910 | miR-145 | - | progression | - | [52] |
| circ_0015756 | PSAT1 | progression | - | [53] | |
| circ_ITCH | RASA1 | - | progression | [71] | |
| circ_VPS13C | MEK/ERK | cell cycle, motility | - | [101] | |
| circ_WHSC1 | MUC1 | progression | - | [82] | |
| circ_CELSR1 | miR-149 | SIK2 | paclitaxel resistance | - | [134] |
| circ_PVT1 | - | proliferation | - | [90] | |
| circ_ATRNL1 | miR-152 | - | progression | - | [61] |
| circ_ZNF608 | - | progression | - | [61] | |
| circ_0004390 | miR-198 | - | proliferation | - | [88] |
| circ_MFN2 | CUL4B | progression, glycolysis | - | [72] | |
| circ_BNC2 | miR-223 | LARP4 | - | progression | [62] |
| circ_BNC2 | FBXW7 | - | progression | [63] | |
| circ_0025033 | miR-370 | SLC1A5 | development | - | [119] |
| circ_0070203 | TGFβR2 | EMT | - | [113] | |
| circ_0000714 | RAB17, CDK6/RB | paclitaxel resistance | - | [130] | |
| circ_ATRNL1 | miR-378 | Smad4 | - | angiogenesis, metastasis | [108] |
| circ_IFNGR2 | ST5 | metastasis | - | [110] | |
| circ_PLOD2 | - | propagation | - | [102] | |
| circ_0078607 | miR-518a | Fas | - | progression | [60] |
| circ_NFIX | TRIM44 | angiogenesis | - | [105] | |
| circ_0007841 | miR-532 | NFIB | cisplatin | - | [123] |
| circ_0025033 | FOXM1 | paclitaxel resistance | - | [131] | |
| circ_ATP2B4 | SREBF1 | metastasis | - | [109] | |
| circ_0010467 | miR-637 | LIF/STAT3 | platinum resistance | - | [121] |
| circ_0013958 | PLXNB2 | proliferation, invasion | - | [96] | |
| circ_0051240 | KLK4 | migration, invasion | - | [103] | |
| circ_EPSTI1 | miR-942 | - | progression | - | [67] |
| circ_PIP5K1A | NFIB | cisplatin resistance | - | [128] | |
| circ_MUC16 | miR-1182 | S100B | progression | - | [73] |
| circ_WHSC1 | hTERT | progression | - | [82] | |
| circRNAs | Levels | Function | Associations | OC | Ref. | |
|---|---|---|---|---|---|---|
| Stimulation | Inhibition | Subtype | ||||
| circ_RS-7 | up | - | - | FIGO stage | EOC | [158] |
| lymph node | ||||||
| distant metastasis | ||||||
| circ_HIPK3 | up | proliferation, migration | apoptosis | FIGO stage | EOC | [159] |
| invasion | lymph node | |||||
| circ_EXOC6B | up | - | - | FIGO stage | EOC | [160] |
| circ_N4BP2L2 | up | - | - | FIGO stage | EOC | [161] |
| circ_RNA1656 | down | - | - | FIGO stage | HGSOC | [162] |
| circ_0003972 | down | - | - | - | EOC | [163] |
| circ_0007288 | down | - | - | lymph node | EOC | [163] |
| circ_0078607 | down | apoptosis | proliferation | - | HGSOC | [164] |
| circLARP4 | down | - | - | FIGO, lymph node | EOC | [165] |
| circ-001567 | proliferation | apoptosis | E-/N-cadherin | EOC | [166] | |
| circ-NOLC1 | up | proliferation, migration | - | FIGO stage | EOC | [167] |
| invasion | differentiation | |||||
| circ_0013958 | up | proliferation, migration | - | FIGO stage | EOC | [168] |
| invasion | lymph node | |||||
| circ_BNC2 | down | - | - | FIGO stage | EOC | [169] |
| lymph node | ||||||
| circ_SETDB1 | up | relapse | - | FIGO stage | HGSOC | [170] |
| lymph node | ||||||
| circ-ABCB10 | up | proliferation | apoptosis | FIGO stage | EOC | [171] |
| differentiation | ||||||
| tumor size | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarzenbach, H. Interplay of microRNAs and circRNAs in Epithelial Ovarian Cancer. Non-Coding RNA 2024, 10, 51. https://doi.org/10.3390/ncrna10050051
Schwarzenbach H. Interplay of microRNAs and circRNAs in Epithelial Ovarian Cancer. Non-Coding RNA. 2024; 10(5):51. https://doi.org/10.3390/ncrna10050051
Chicago/Turabian StyleSchwarzenbach, Heidi. 2024. "Interplay of microRNAs and circRNAs in Epithelial Ovarian Cancer" Non-Coding RNA 10, no. 5: 51. https://doi.org/10.3390/ncrna10050051
APA StyleSchwarzenbach, H. (2024). Interplay of microRNAs and circRNAs in Epithelial Ovarian Cancer. Non-Coding RNA, 10(5), 51. https://doi.org/10.3390/ncrna10050051





