The C-Terminal Domain of Liquorilactobacillus nagelii Dextransucrase Mediates the Production of Larger Dextrans Compared to Liquorilactobacillus hordei
Abstract
:1. Introduction
2. Results
2.1. Expression of Dextransucrase Variants
2.2. Characterization of the Native Extracellular and Heterologously Expressed Dextransucrase Variants
2.3. Analysis of Dextrans Formed by the Native Extracellular and Heterologously Expressed Dextransucrase Variants
3. Discussion
4. Materials and Methods
4.1. Strains, Media and Growth Conditions
4.2. Molecular Techniques and Plasmid Construction
4.3. Isolation and Verification of the Dextransucrases
4.4. Activity Measurements of Dextransucrases
4.5. Enzymatic Dextran Formation
4.6. Determination of Molecular and Macromolecular Structures of the Formed Dextrans
4.7. Statistical Analysis and Data Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahmoud, Y.A.G.; El-Naggar, M.E.; Abdel-Megeed, A.; El-Newehy, M. Recent advancements in microbial polysaccharides: Synthesis and applications. Polymers 2021, 13, 4136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, F.; Yuan, R. Applications of natural polymer-based hydrogels in the food industry. In Hydrogels Based on Natural Polymers; Chen, Y., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 357–410. [Google Scholar]
- Lapasin, R.; Pricl, S. Industrial applications of polysaccharides. In Rheology of Industrial Polysaccharides: Theory and Applications; Lapasin, R., Pricl, S., Eds.; Springer: Boston, MA, USA, 1995; pp. 134–161. [Google Scholar]
- Zannini, E.; Waters, D.M.; Coffey, A.; Arendt, E.K. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 2016, 100, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.; Alves, V.D.; Reis, M.A.M. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Mody, K. Microbial exopolysaccharides: Variety and potential applications. In Microbial Production of Biopolymers and Polymer Precursors: Applications and Perspectives; Rehm, B.H.A., Ed.; Caister Academic Press: Palmerston North, New Zealand, 2009; Volume 1, pp. 229–253. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides produced by lactic acid bacteria: From biosynthesis to health-promoting properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Schmid, J. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr. Opin. Biotechnol. 2018, 53, 130–136. [Google Scholar] [CrossRef]
- van Hijum, S.A.F.T.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L.; van Geel-Schutten, I.G.H. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 157–176. [Google Scholar] [CrossRef] [Green Version]
- Nabot, M.; Guérin, M.; Sivakumar, D.; Remize, F.; Garcia, C. Variability of bacterial homopolysaccharide production and properties during food processing. Biology 2022, 11, 171. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Bai, Y.; Pijning, T.; Van Leeuwen, S.S.; Dijkhuizen, L. Structure-function relationships of family GH70 glucansucrase and 4,6-alpha-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell Mol. Life Sci. 2016, 73, 2681–2706. [Google Scholar] [CrossRef] [Green Version]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, alpha-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef] [Green Version]
- Moulis, C.; Joucla, G.; Harrison, D.; Fabre, E.; Potocki-Veronese, G.; Monsan, P.; Remaud-Simeon, M. Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 2006, 281, 31254–31267. [Google Scholar] [CrossRef] [Green Version]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol. Adv. 2018, 36, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Svensson, B.; Russell, R.R.B. Location of repeat elements in glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol. Lett. 2000, 192, 53–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozonnet, S.; Dols-Laffargue, M.; Fabre, E.; Pizzut, S.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.-M. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 2002, 184, 5753–5761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmuth, H.; Wittrock, S.; Kralj, S.; Dijkhuizen, L.; Hofer, B.; Seibel, J. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: Toward tailor-made polymer and oligosaccharide products. Biochemistry 2008, 47, 6678–6684. [Google Scholar] [CrossRef] [Green Version]
- Kralj, S.; van Geel-Schutten, I.G.H.; Faber, E.J.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Rational transformation of Lactobacillus reuteri 121 reuteransucrase into a dextransucrase. Biochemistry 2005, 44, 9206–9216. [Google Scholar] [CrossRef] [Green Version]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; Dijkstra, B.W.; Dijkhuizen, L. Glycosidic bond specificity of glucansucrases: On the role of acceptor substrate binding residues. Biocatal. Biotransformation 2012, 30, 366–376. [Google Scholar] [CrossRef] [Green Version]
- van Leeuwen, S.S.; Kralj, S.; Eeuwema, W.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural characterization of bioengineered α-D-glucans produced by mutant glucansucrase GTF180 enzymes of Lactobacillus reuteri strain 180. Biomacromolecules 2009, 10, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Bodvik, R.; Dedinaite, A.; Karlson, L.; Bergström, M.; Bäverbäck, P.; Pedersen, J.S.; Edwards, K.; Karlsson, G.; Varga, I.; Claesson, P.M. Aggregation and network formation of aqueous methylcellulose and hydroxypropylmethylcellulose solutions. Colloids Surf. A Physicochem. Eng. Asp. 2010, 354, 162–171. [Google Scholar] [CrossRef]
- Zhang, L.; Ruan, D.; Gao, S. Dissolution and regeneration of cellulose in NaOH/thiourea aqueous solution. J. Polym. Sci. Part B: Polym. Phys. 2002, 40, 1521–1529. [Google Scholar] [CrossRef]
- Schmid, J.; Bechtner, J.; Vogel, R.F.; Jakob, F. A systematic approach to study the pH-dependent release, productivity and product specificity of dextransucrases. Microb. Cell Factories 2019, 18, 153. [Google Scholar] [CrossRef]
- Hundschell, C.S.; Braun, A.; Wefers, D.; Vogel, R.F.; Jakob, F. Size-Dependent variability in flow and viscoelastic behavior of levan produced by Gluconobacter albidus TMW 2.1191. Foods 2020, 9, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hundschell, C.S.; Jakob, F.; Wagemans, A.M. Molecular weight dependent structure of the exopolysaccharide levan. Int. J. Biol. Macromol. 2020, 161, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Wilkinson, S.; Daenen, L.; Arendt, E.K. An update on water kefir: Microbiology, composition and production. Int. J. Food Microbiol. 2021, 345, 109128. [Google Scholar] [CrossRef] [PubMed]
- Fels, L.; Jakob, F.; Vogel, R.F.; Wefers, D. Structural characterization of the exopolysaccharides from water kefir. Carbohydr. Polym. 2018, 189, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, M. The microbial flora of sugary kefir grain (the gingerbeer plant): Biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. MIRCEN J. Appl. Microbiol. Biotechnol. 1989, 5, 223–238. [Google Scholar] [CrossRef]
- Eckel, V.; Vogel, R.F.; Jakob, F. In situ production and characterization of cloud forming dextrans in fruit-juices. Int. J. Food Microbiol. 2019, 306, 108261. [Google Scholar] [CrossRef] [PubMed]
- Bechtner, J.; Wefers, D.; Schmid, J.; Vogel, R.F.; Jakob, F. Identification and comparison of two closely related dextransucrases released by water kefir borne Lactobacillus hordei TMW 1.1822 and Lactobacillus nagelii TMW 1.1827. Microbiology 2019, 165, 956–966. [Google Scholar] [CrossRef]
- Bechtner, J.; Hassler, V.; Wefers, D.; Vogel, R.F.; Jakob, F. Insights into extracellular dextran formation by Liquorilactobacillus nagelii TMW 1.1827 using secretomes obtained in the presence or absence of sucrose. Enzym. Microb. Technol. 2021, 143, 109724. [Google Scholar] [CrossRef]
- Bechtner, J.; Ludwig, C.; Kiening, M.; Jakob, F.; Vogel, R.F. Living the sweet life: How Liquorilactobacillus hordei TMW 1.1822 changes its behavior in the presence of sucrose in comparison to glucose. Foods 2020, 9, 1150. [Google Scholar] [CrossRef]
- Vujičić-Žagar, A.; Pijning, T.; Kralj, S.; López, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Fels, L.; Wefers, D.; Behr, J.; Jakob, F.; Vogel, R.F. Lactobacillus hordei dextrans induce Saccharomyces cerevisiae aggregation and network formation on hydrophilic surfaces. Int. J. Biol. Macromol. 2018, 115, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Yeon, M.J.; Choi, N.-S.; Chang, Y.-H.; Jung, M.Y.; Song, J.J.; Kim, J.S. Purification and characterization of a novel glucansucrase from Leuconostoc lactis EG001. Microbiol. Res. 2010, 165, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Rühmkorf, C.; Bork, C.; Mischnick, P.; Rübsam, H.; Becker, T.; Vogel, R.F. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol. 2013, 34, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Côté, G.L.; Skory, C.D. Cloning, expression, and characterization of an insoluble glucan-producing glucansucrase from Leuconostoc mesenteroides NRRL B-1118. Appl. Microbiol. Biotechnol. 2012, 93, 2387–2394. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, G.H.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiology 2004, 150, 2099–2112. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Misaka, T.; Abe, K.; Kobayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186. [Google Scholar] [CrossRef]
- Kristjánsson, M.M.; Kinsella, J.E. Protein and enzyme stability: Structural, thermodynamic, and experimental aspects. In Advances in Food and Nutrition Research; Kinsella, J.E., Ed.; Academic Press: Cambridge, MA, USA, 1991; Volume 35, pp. 237–316. [Google Scholar]
- Ratkowsky, D.A.; Olley, J.; Ross, T. Unifying temperature effects on the growth rate of bacteria and the stability of globular proteins. J. Theor. Biol. 2005, 233, 351–362. [Google Scholar] [CrossRef]
- Brison, Y.; Pijning, T.; Malbert, Y.; Fabre, É.; Mourey, L.; Morel, S.; Potocki-Véronèse, G.; Monsan, P.; Tranier, S.; Remaud-Siméon, M.; et al. Functional and structural characterization of α-(1->2) branching sucrase derived from DSR-E glucansucrase. J. Biol. Chem. 2012, 287, 7915–7924. [Google Scholar] [CrossRef] [Green Version]
- Waldherr, F.W.; Doll, V.M.; Meissner, D.; Vogel, R.F. Identification and characterization of a glucan-producing enzyme from Lactobacillus hilgardii TMW 1.828 involved in granule formation of water kefir. Food Microbiol. 2010, 27, 672–678. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Vuillemin, M.; Bondy, P.; Remaud-Simeon, M.; Moulis, C. Processivity of dextransucrases synthesizing very-high-molar-mass dextran is mediated by sugar-binding pockets in domain V. J. Biol. Chem. 2020, 295, 5602–5613. [Google Scholar] [CrossRef] [Green Version]
- Funane, K.; Ishii, T.; Ono, H.; Kobayashi, M. Changes in linkage pattern of glucan products induced by substitution of Lys residues in the dextransucrase. FEBS Lett. 2005, 579, 4739–4745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irague, R.; Massou, S.; Moulis, C.; Saurel, O.; Milon, A.; Monsan, P.; Remaud-Siméon, M.; Portais, J.-C.; Potocki-Véronèse, G. NMR-based structural glycomics for high-throughput screening of carbohydrate-active enzyme specificity. Anal. Chem. 2011, 83, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Kirby, A.J.; Hollfelder, F. Enzymes under the nanoscope. Nature 2008, 456, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold spring harbor, NY, USA, 1989. [Google Scholar]
- Kushner, S.R. An improved method for transformation of Escherichia coli with ColE1-derived plasmids. Genet. Eng. Elsevier 1978, 173, 17–23. [Google Scholar]
- Münkel, F.; Bechtner, J.; Eckel, V.; Fischer, A.; Herbi, F.; Jakob, F.; Wefers, D. Detailed structural characterization of glucans produced by glucansucrases from Leuconostoc citreum TMW 2.1194. J. Agric. Food Chem. 2019, 67, 6856–6866. [Google Scholar] [CrossRef]
- Yuryev, V.; Tomasik, P.; Bertoft, E. Starch: Achievements in Understanding of Structure and Functionality; Nova Science Publishers: New York, NY, USA, 2007; pp. 1–315. [Google Scholar]
- Ua-Arak, T.; Jakob, F.; Vogel, R.F. Influence of levan-producing acetic acid bacteria on buckwheat-sourdough breads. Food Microbiol. 2017, 65, 95–104. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
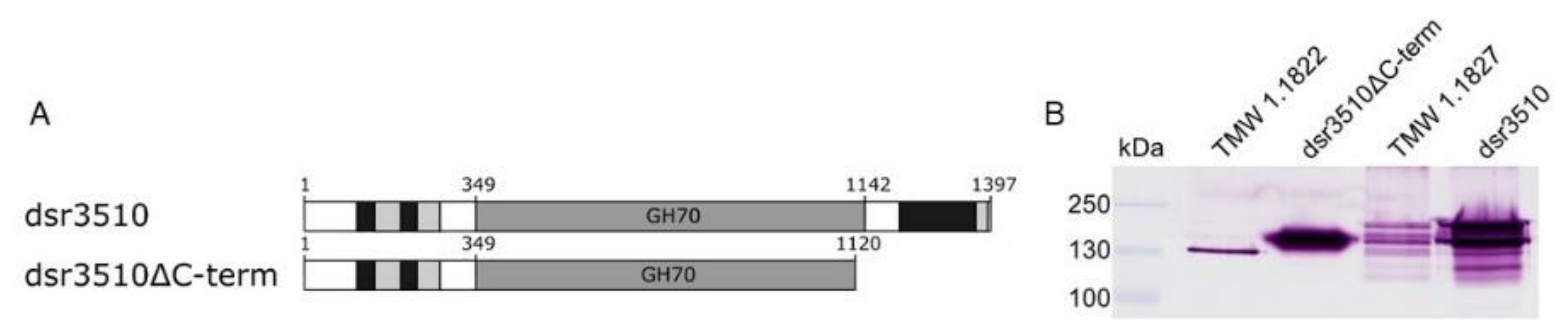

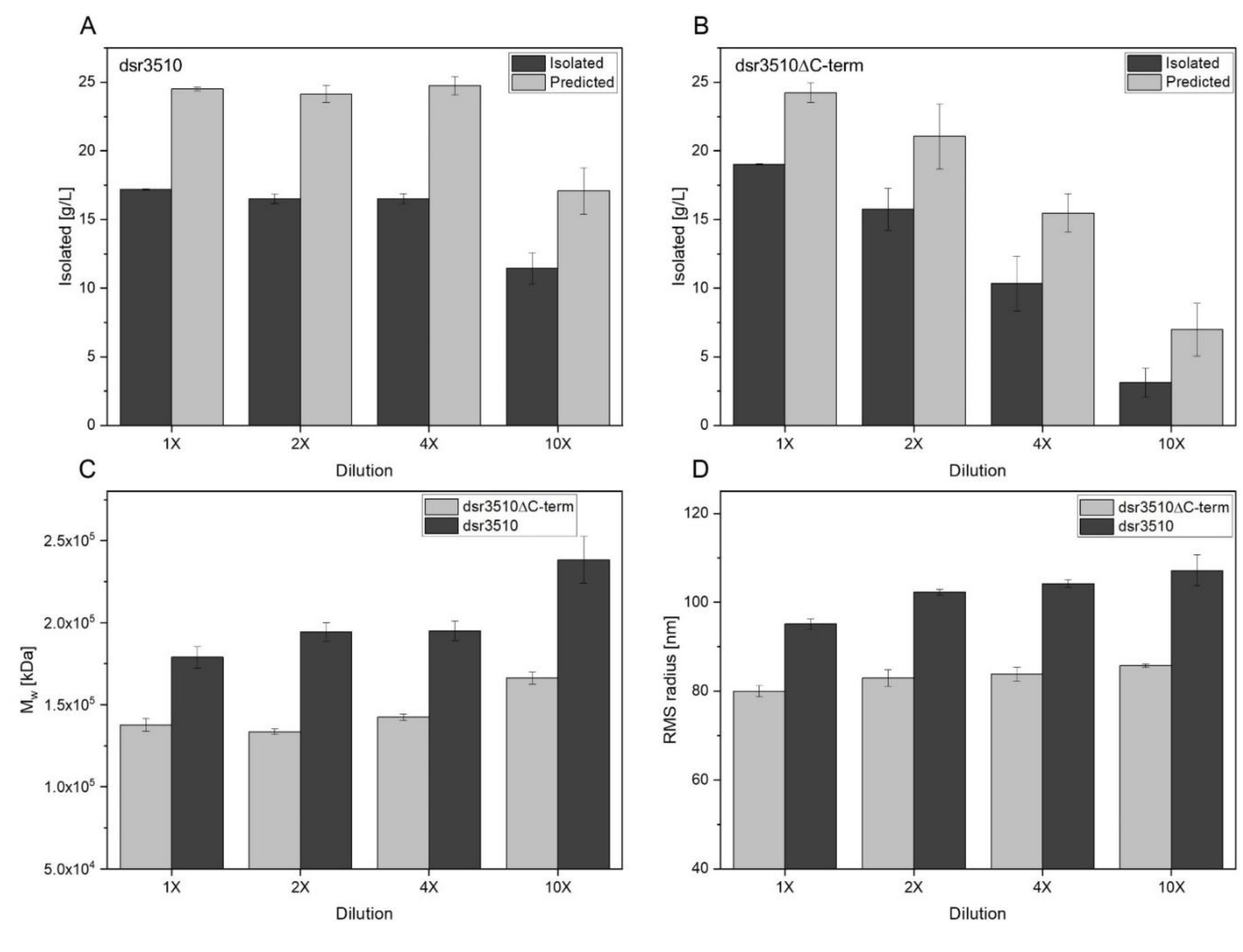
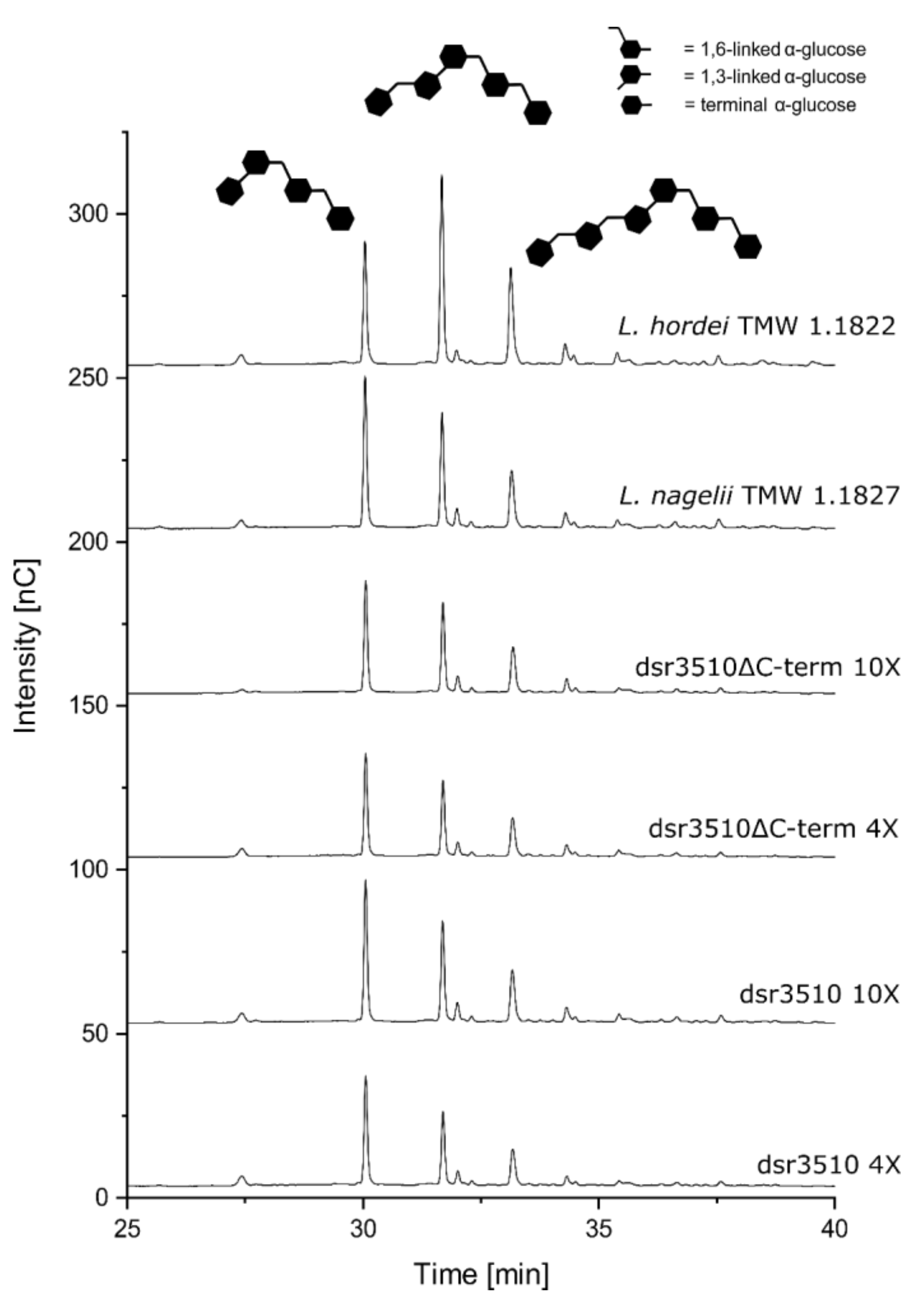
| Dextransucrase Variant | KM [mM] | vmax [mmol/min × L−1] |
|---|---|---|
| L. nagelii TMW 1.1827 | 12.99 ± 3.74 * | 1.03 ± 0.07 * |
| dsr3510 | 10.97 ± 1.56 | 0.200 ± 0.006 |
| dsr3510ΔC-term | 12.57 ± 0.73 | 0.089 ± 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bechtner, J.; Hassler, V.; Wefers, D.; Ehrmann, M.; Jakob, F. The C-Terminal Domain of Liquorilactobacillus nagelii Dextransucrase Mediates the Production of Larger Dextrans Compared to Liquorilactobacillus hordei. Gels 2022, 8, 171. https://doi.org/10.3390/gels8030171
Bechtner J, Hassler V, Wefers D, Ehrmann M, Jakob F. The C-Terminal Domain of Liquorilactobacillus nagelii Dextransucrase Mediates the Production of Larger Dextrans Compared to Liquorilactobacillus hordei. Gels. 2022; 8(3):171. https://doi.org/10.3390/gels8030171
Chicago/Turabian StyleBechtner, Julia, Verena Hassler, Daniel Wefers, Matthias Ehrmann, and Frank Jakob. 2022. "The C-Terminal Domain of Liquorilactobacillus nagelii Dextransucrase Mediates the Production of Larger Dextrans Compared to Liquorilactobacillus hordei" Gels 8, no. 3: 171. https://doi.org/10.3390/gels8030171
APA StyleBechtner, J., Hassler, V., Wefers, D., Ehrmann, M., & Jakob, F. (2022). The C-Terminal Domain of Liquorilactobacillus nagelii Dextransucrase Mediates the Production of Larger Dextrans Compared to Liquorilactobacillus hordei. Gels, 8(3), 171. https://doi.org/10.3390/gels8030171






