Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly(L-lysine)-Based Amphiphilic Copolypeptides
Abstract
:1. Introduction
2. Results and Discussion
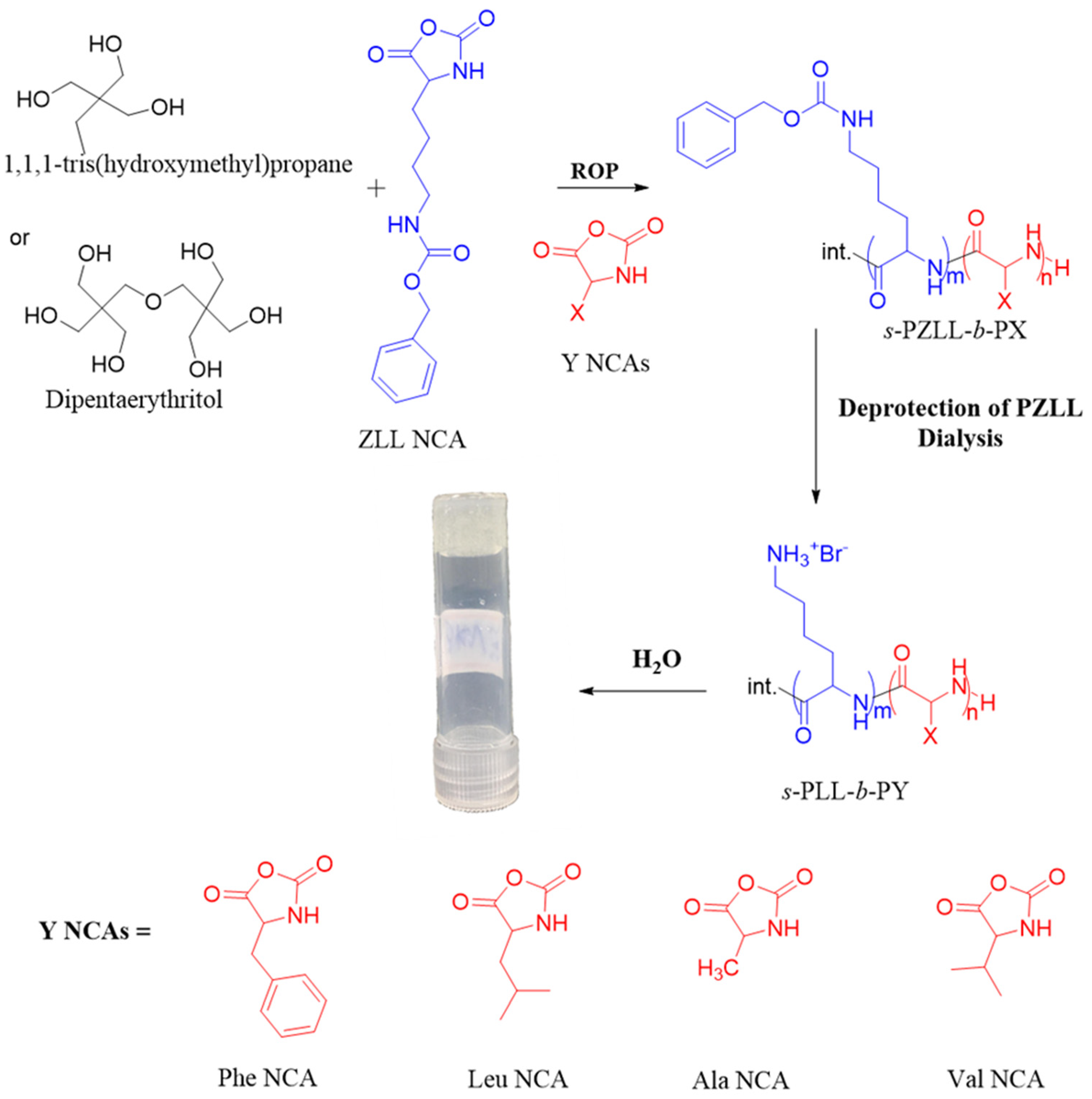
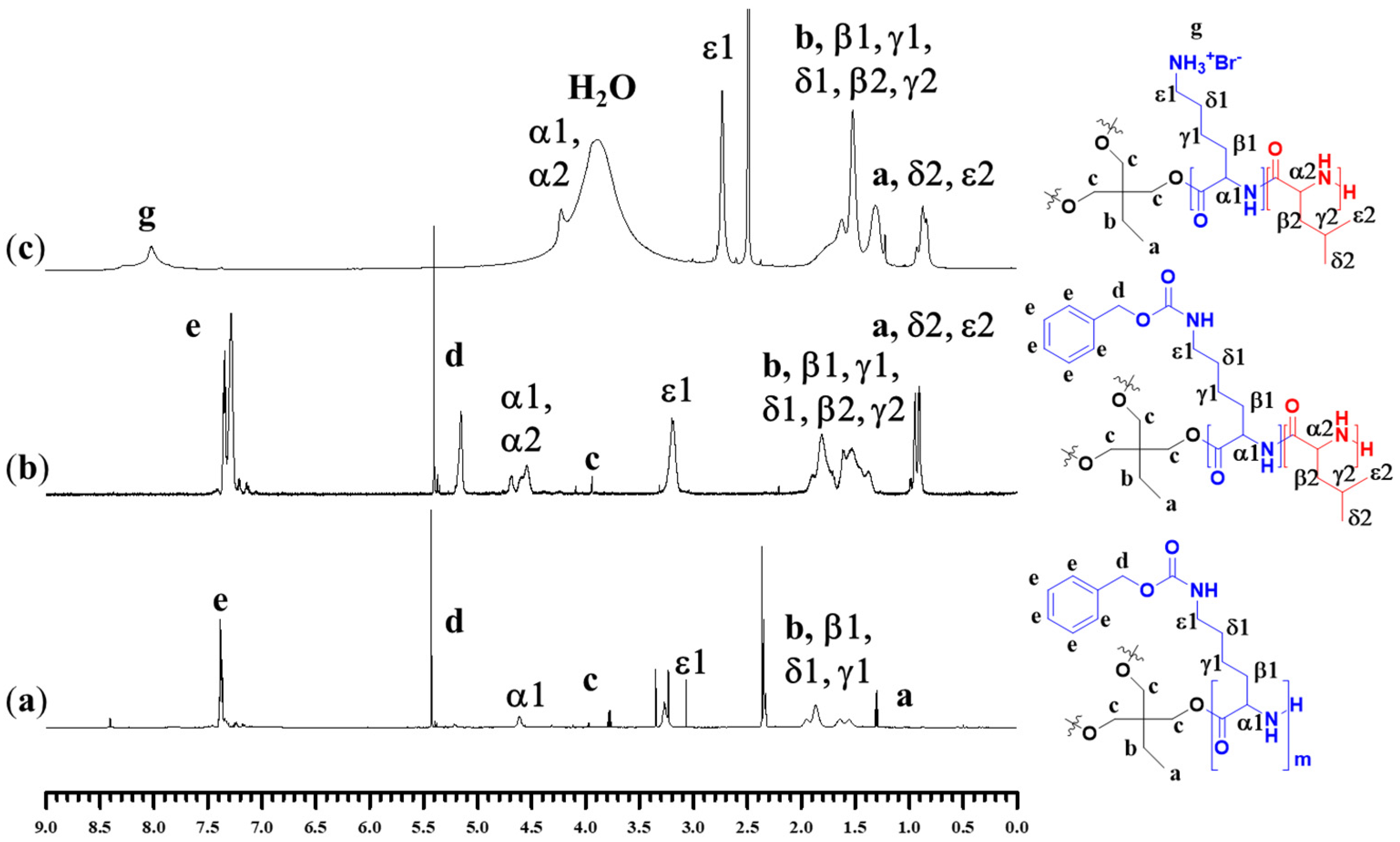
2.1. Synthesis and Characterization of Polypeptides
2.2. Hydrogelation of Block Polypeptides
2.3. Molecular Structure of Polypeptide Hydrogels
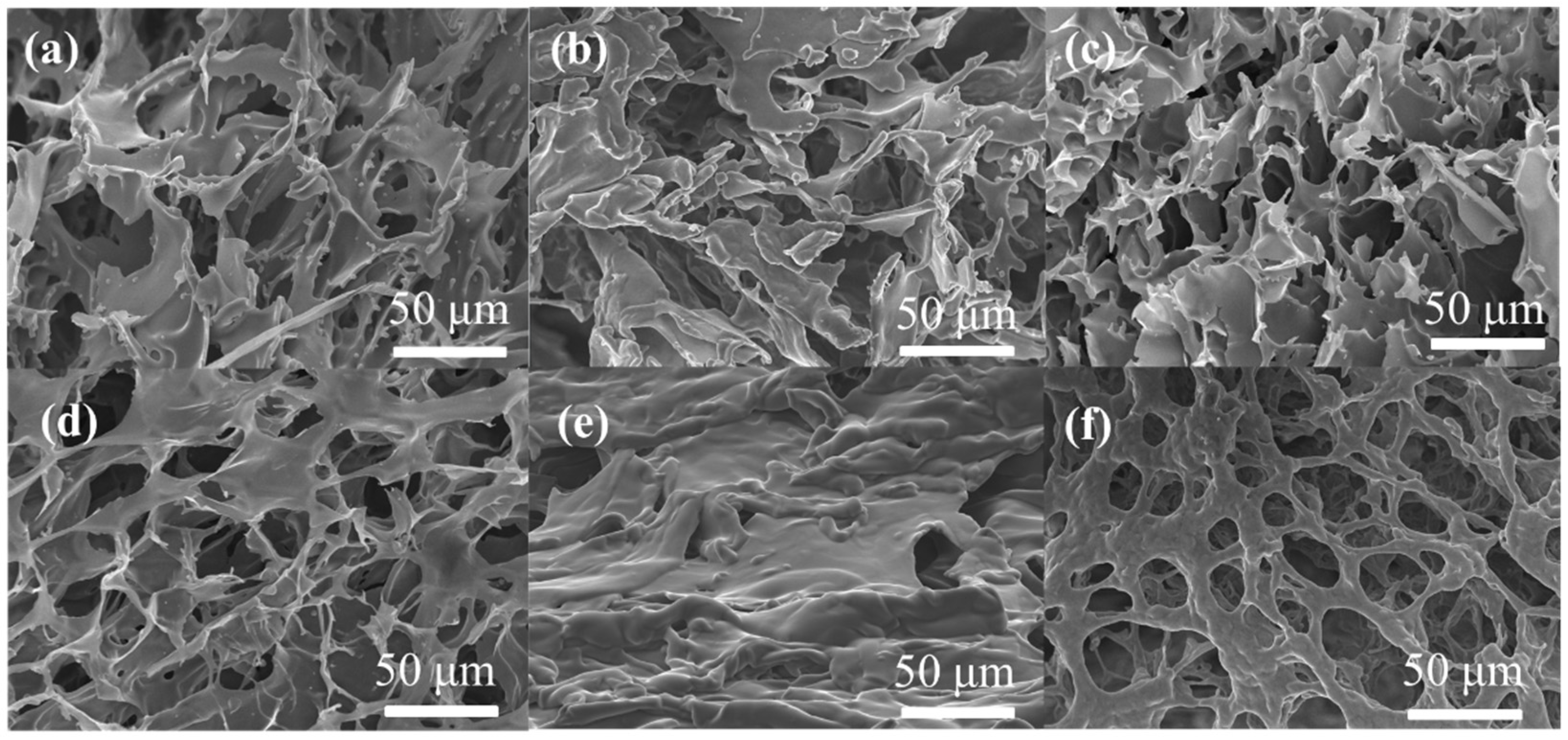
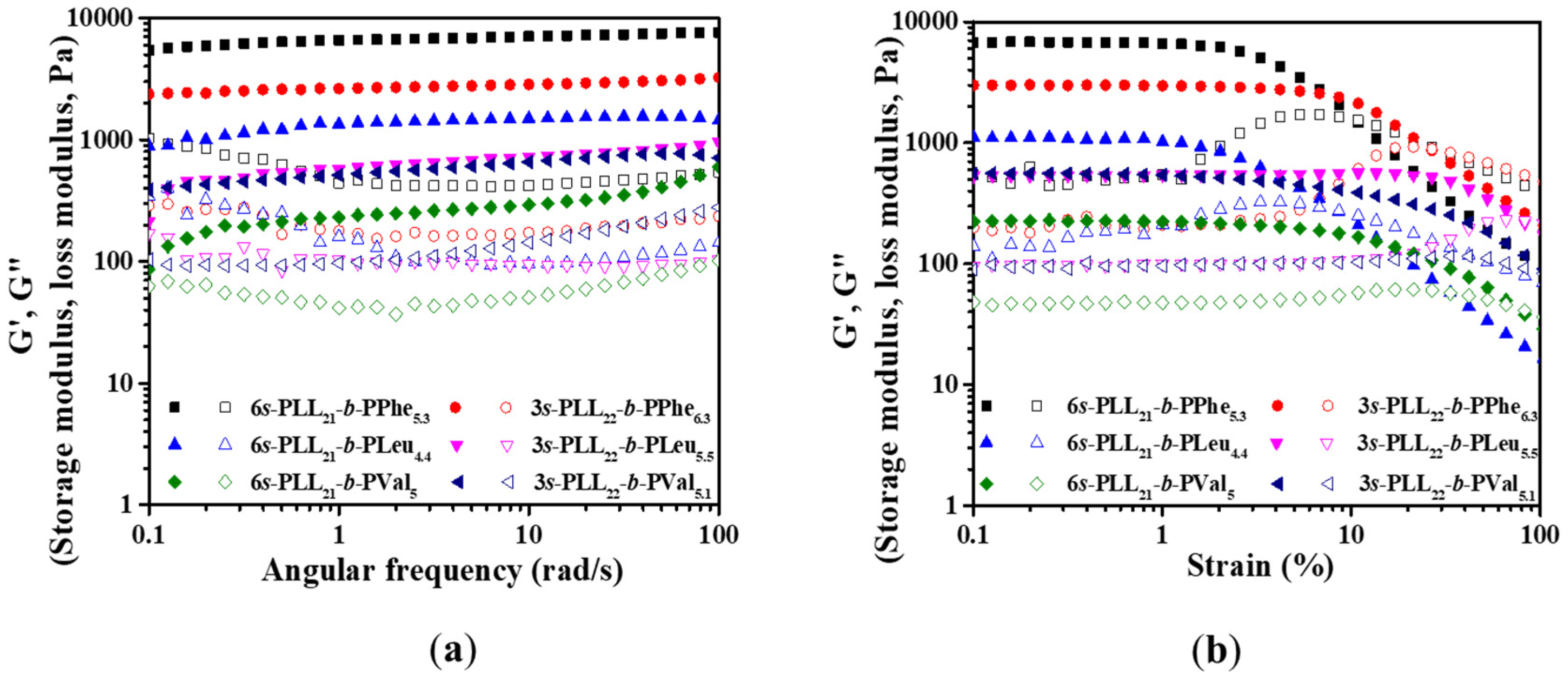
2.4. Morphology and Mechanical Properties of Polypeptide Hydrogels
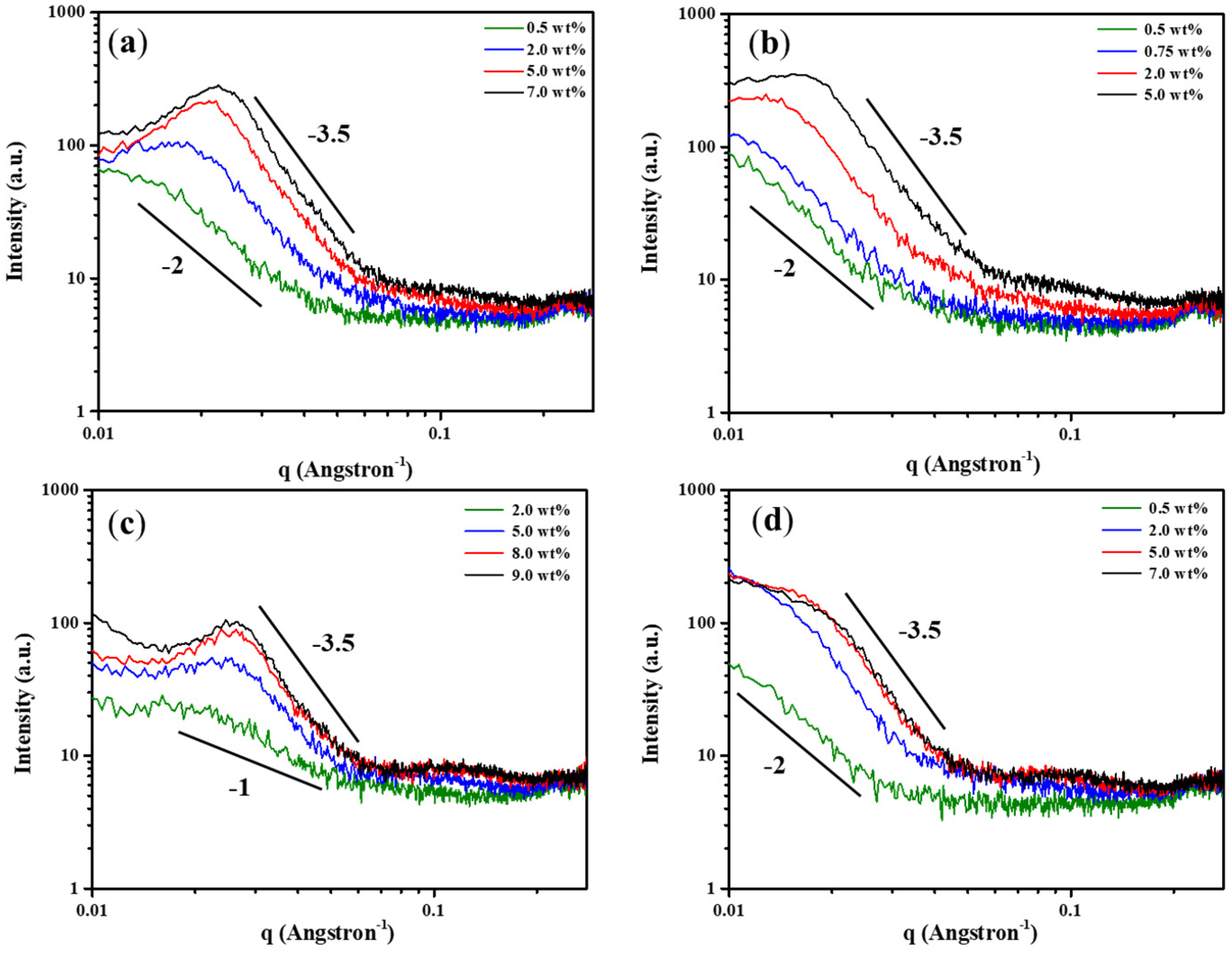
2.5. Molecular Assembly of Polypeptide Hydrogels
2.6. Gelation Mechanism of Polypeptide Hydrogels
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Star-Shaped Poly(L-lysine)-Based Block Copolypeptides
4.3. Characterization of Block Polypeptides
4.4. Preparation Polypeptide Hydrogels and Determination of Critical Gelation Concentration (CGC)
4.5. Characterization of Polypeptide Secondary Conformation
4.6. Characterization of Polypeptide Hydrogels
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Appel, E.A.; del Barrio, J.; Loh, X.J.; Scherman, O.A. Supramolecular polymeric hydrogels. Chem. Soc. Rev. 2012, 41, 6195–6214. [Google Scholar] [CrossRef]
- Anderson, A.J.; Culver, H.R.; Bryant, S.J.; Bowman, C.N. Viscoelastic and thermoreversible networks crosslinked by non-covalent interactions between “clickable” nucleic acid oligomers and DNA. Polym. Chem. 2020, 11, 2959–2968. [Google Scholar] [CrossRef]
- Strandman, S.; Zhu, X. Self-healing supramolecular hydrogels based on reversible physical interactions. Gels 2016, 2, 16. [Google Scholar] [CrossRef] [Green Version]
- Barrett-Catton, E.; Ross, M.L.; Asuri, P. Multifunctional Hydrogel Nanocomposites for Biomedical Applications. Polymers 2021, 13, 856. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.-N.; Su, C.-F.; Huang, C.-C.; Jan, J.-S. Biomimetic hydrogels based on L-Dopa conjugated gelatin as pH-responsive drug carriers and antimicrobial agents. Colloids Surf. B Biointerfaces 2020, 196, 111316. [Google Scholar] [CrossRef] [PubMed]
- Hogan, K.J.; Mikos, A.G. Biodegradable thermoresponsive polymers: Applications in drug delivery and tissue engineering. Polymer 2020, 211, 123063. [Google Scholar] [CrossRef]
- Gao, Q.; Zhang, C.; Wang, M.; Wu, Y.; Gao, C.; Zhu, P. Injectable pH-responsive poly (γ-glutamic acid)-silica hybrid hydrogels with high mechanical strength, conductivity and cytocompatibility for biomedical applications. Polymer 2020, 197, 122489. [Google Scholar] [CrossRef]
- Ravichandran, R.; Sundarrajan, S.; Venugopal, J.R.; Mukherjee, S.; Ramakrishna, S. Advances in polymeric systems for tissue engineering and biomedical applications. Macromol. Biosci. 2012, 12, 286–311. [Google Scholar] [CrossRef]
- Li, Y.; Rodrigues, J.; Tomás, H. Injectable and biodegradable hydrogels: Gelation, biodegradation and biomedical applications. Chem. Soc. Rev. 2012, 41, 2193–2221. [Google Scholar] [CrossRef]
- Pham, T.-N.; Jiang, Y.-S.; Su, C.-F.; Jan, J.-S. In situ formation of silver nanoparticles-contained gelatin-PEG-dopamine hydrogels via enzymatic cross-linking reaction for improved antibacterial activities. Int. J. Biol. Macromol. 2020, 146, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-F.; Chen, G.-Y.; Chang, C.-H.; Su, Y.-C.; Chen, Y.-C.; Jiang, Y.-s.; Jan, J.-S. TRAIL encapsulated to polypeptide-crosslinked nanogel exhibits increased anti-inflammatory activities in Klebsiella pneumoniae-induced sepsis treatment. Mater. Sci. Eng. C. 2019, 102, 85–95. [Google Scholar] [CrossRef]
- Hsu, F.-M.; Hu, M.-H.; Jiang, Y.-S.; Lin, B.-Y.; Hu, J.-J.; Jan, J.-S. Antibacterial polypeptide/heparin composite hydrogels carrying growth factor for wound healing. Mater. Sci. Eng. C 2020, 112, 110923. [Google Scholar] [CrossRef]
- Sun, Y.; Wollenberg, A.L.; O’Shea, T.M.; Cui, Y.; Zhou, Z.H.; Sofroniew, M.V.; Deming, T.J. Conformation-directed formation of self-healing diblock copolypeptide hydrogels via polyion complexation. J. Am. Chem. Soc. 2017, 139, 15114–15121. [Google Scholar] [CrossRef]
- Huang, J.; Hastings, C.L.; Duffy, G.P.; Kelly, H.M.; Raeburn, J.; Adams, D.J.; Heise, A. Supramolecular hydrogels with reverse thermal gelation properties from (oligo) tyrosine containing block copolymers. Biomacromolecules 2013, 14, 200–206. [Google Scholar] [CrossRef]
- O’Brien, S.; Brannigan, R.P.; Ibanez, R.; Wu, B.; O’Dwyer, J.; O’Brien, F.J.; Cryan, S.-A.; Heise, A. Biocompatible polypeptide-based interpenetrating network (IPN) hydrogels with enhanced mechanical properties. J. Mater. Chem. B 2020, 8, 7785–7791. [Google Scholar] [CrossRef]
- Hou, S.-S.; Fan, N.-S.; Tseng, Y.-C.; Jan, J.-S. Self-Assembly and Hydrogelation of Coil–Sheet Poly (l-lysine)-block-poly (l-threonine) Block Copolypeptides. Macromolecules 2018, 51, 8054–8063. [Google Scholar] [CrossRef]
- Gaspard, J.; Silas, J.A.; Shantz, D.F.; Jan, J.-S. Supramolecular assembly of lysine-b-glycine block copolypeptides at different solution conditions. Supramol. Chem. 2010, 22, 178–185. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Arham, M.; Jan, J.-S. Alkyl chain grafted poly (l-lysine): Self-assembly and biomedical application as carriers. Soft Matter 2011, 7, 3975–3983. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Chen, Y.-F.; Shen, X.-Y.; Hu, J.-J.; Jan, J.-S. Reduction-and pH-Sensitive lipoic acid-modified Poly (l-lysine) and polypeptide/silica hybrid hydrogels/nanogels. Polymer 2016, 86, 32–41. [Google Scholar] [CrossRef]
- Cheng, J.; Deming, T.J. Synthesis of polypeptides by ring-opening polymerization of α-amino acid N-carboxyanhydrides. In Peptide-Based Materials; Deming, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 310, p. 126. [Google Scholar]
- Aoi, K.; Tsutsumiuchi, K.; Okada, M. Glycopeptide Synthesis by an. alpha.-Amino Acid N-Carboxyanhydride (NCA) Method: Ring-Opening Polymerization of a Sugar-Substituted NCA. Macromolecules 1994, 27, 875–877. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, X.; Fan, Z.; Du, J. Ring-Opening Polymerization of N-Carboxyanhydride-Induced Self-Assembly for Fabricating Biodegradable Polymer Vesicles. ACS Macro Lett. 2019, 8, 1216–1221. [Google Scholar] [CrossRef]
- Zhou, P.; Dai, X.-G.; Kong, J.; Ling, J. Synthesis of Well-defined Poly (tetrahydrofuran)-b-Poly (a-amino acid) s via Cationic Ring-opening Polymerization (ROP) of Tetrahydrofuran and Nucleophilic ROP of N-thiocarboxyanhydrides. Chin. J. Polym. Sci. 2021, 39, 702–708. [Google Scholar] [CrossRef]
- Marzano, M.; Falanga, A.P.; Marasco, D.; Borbone, N.; D’Errico, S.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Evaluation of an analogue of the marine ε-PLL peptide as a ligand of G-quadruplex DNA structures. Mar. Drugs 2020, 18, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deming, T.J. Synthesis of side-chain modified polypeptides. Chem. Rev. 2016, 116, 786–808. [Google Scholar] [CrossRef] [Green Version]
- Hou, S.-S.; Hsu, Y.-Y.; Lin, J.-H.; Jan, J.-S. Alkyl-poly (L-threonine)/cyclodextrin supramolecular hydrogels with different molecular assemblies and gel properties. ACS Macro Lett. 2016, 5, 1201–1205. [Google Scholar] [CrossRef]
- Hsiao, L.-W.; Lai, Y.-D.; Lai, J.-T.; Hsu, C.-C.; Wang, N.-Y.; Steven, S.-S.W.; Jan, J.-S. Cross-linked polypeptide-based gel particles by emulsion for efficient protein encapsulation. Polymer 2017, 115, 261–272. [Google Scholar] [CrossRef]
- Shen, X.-Y.; Tang, C.-C.; Jan, J.-S. Synthesis and hydrogelation of star-shaped poly (L-lysine) polypeptides modified with different functional groups. Polymer 2018, 151, 108–116. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, S.; Wan, Y.; Fu, W.; Li, Z. Hydrogels assembled from star-shaped polypeptides with a dendrimer as the core. Soft Matter 2015, 11, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.; Borase, T.; Payne, C.; O’Dwyer, J.; Cryan, S.-A.; Heise, A. Hydrogels from amphiphilic star block copolypeptides. RSC Adv. 2016, 6, 23370–23376. [Google Scholar] [CrossRef]
- Tang, C.-C.; Zhang, S.-H.; My Phan, T.H.; Tseng, Y.-C.; Jan, J.-S. Block length and topology affect self-assembly and gelation of poly(L-lysine)-block-poly(S-benzyl-l-cysteine) block copolypeptides. Polymer 2021, 228, 123891. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Lai, Y.-D.; Chang, C.-H.; Tsai, Y.-C.; Tang, C.-C.; Jan, J.-S. Star-shaped polypeptides exhibit potent antibacterial activities. Nanoscale 2019, 11, 11696–11708. [Google Scholar] [CrossRef]
- Breedveld, V.; Nowak, A.P.; Sato, J.; Deming, T.J.; Pine, D.J. Rheology of block copolypeptide solutions: Hydrogels with tunable properties. Macromolecules 2004, 37, 3943–3953. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Qi, D.; Wang, J.; Yu, S.; He, C.; Deng, M. Effects of ethyl-L-glutamated and phenylalanine ratio/sequence on the secondary structure and gelation properties of their PEGylated copolymers. Polymer 2020, 191, 122276. [Google Scholar] [CrossRef]
- Thornton, P.D.; Billah, S.M.R.; Cameron, N.R. Enzyme-Degradable Self-Assembled Hydrogels From Polyalanine-Modified Poly (ethylene glycol) Star Polymers. Macromol. Rapid Commun. 2013, 34, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Dayananda, K.; Choi, E.K.; Park, H.J.; Kim, J.S.; Lee, D.S. Synthesis and characterization of poly (l-glutamic acid)-block-poly (l-phenylalanine). Polymer 2009, 50, 2252–2257. [Google Scholar] [CrossRef]
- Kousar, A.; Liu, J.; Mehwish, N.; Wang, F.; Dang-i, A.; Feng, C. pH-Regulated supramolecular chirality of phenylalanine-based hydrogels. Mater. Today Chem. 2019, 11, 217–224. [Google Scholar] [CrossRef]
- Triftaridou, A.I.; Chécot, F.; Iliopoulos, I. Poly (N, N-dimethylacrylamide)-block-Poly (L-lysine) Hybrid Block Copolymers: Synthesis and Aqueous Solution Characterization. Macromol. Chem. Phys. 2010, 211, 768–777. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Xiao, C.; Ding, J.; Zhuang, X.; Huang, Y.; Chen, X. Decisive role of hydrophobic side groups of polypeptides in thermosensitive gelation. Biomacromolecules 2012, 13, 2053–2059. [Google Scholar] [CrossRef]
- Zhang, S.; Fu, W.; Li, Z. Supramolecular hydrogels assembled from nonionic poly (ethylene glycol)-b-polypeptide diblocks containing OEGylated poly-L-glutamate. Polym. Chem. 2014, 5, 3346–3351. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Huang, Y.-C.; Jan, J.-S. Molecular assembly of alkyl chain-grafted poly (L-lysine) tuned by backbone chain length and grafted alkyl chain. RSC Adv. 2015, 5, 22783–22791. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Huang, Y.-F.; Huang, Y.-C.; Wen, T.-C.; Jan, J.-S. Alkyl chain-grafted poly (L-lysine) vesicles with tunable molecular assembly and membrane permeability. ACS Macro Lett. 2014, 3, 220–223. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Shiau, A.-L.; Chang, S.-J.; Fan, N.-S.; Wang, C.-T.; Wu, C.-L.; Jan, J.-S. One-dimensional poly (L-lysine)-block-poly (L-threonine) assemblies exhibit potent anticancer activity by enhancing membranolysis. Acta Biomater. 2017, 55, 283–295. [Google Scholar] [CrossRef]
- Holowka, E.P.; Pochan, D.J.; Deming, T.J. Charged polypeptide vesicles with controllable diameter. J. Am. Chem. Soc. 2005, 127, 12423–12428. [Google Scholar] [CrossRef]
- Nowak, A.P.; Breedveld, V.; Pakstis, L.; Ozbas, B.; Pine, D.J.; Pochan, D.; Deming, T.J. Rapidly recovering hydrogel scaffolds from self-assembling diblock copolypeptide amphiphiles. Nature 2002, 417, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.L. Explanations for the cause of shear thickening in concentrated colloidal suspensions. J. Rheol. 1998, 42, 111–123. [Google Scholar] [CrossRef]
- Chu, B. Laser Light Scattering: Basic Principles and Practice; Courier Corporation: North Chelmsford, MA, USA, 2007. [Google Scholar]
- Tsai, Y.-L.; Tseng, Y.-C.; Chen, Y.-M.; Wen, T.-C.; Jan, J.-S. Zwitterionic polypeptides bearing carboxybetaine and sulfobetaine: Synthesis, self-assembly, and their interactions with proteins. Polym. Chem. 2018, 9, 1178–1189. [Google Scholar] [CrossRef]
- Chan, B.A.; Xuan, S.; Horton, M.; Zhang, D. 1, 1, 3, 3-Tetramethylguanidine-promoted ring-opening polymerization of N-butyl N-carboxyanhydride using alcohol initiators. Macromolecules 2016, 49, 2002–2012. [Google Scholar] [CrossRef]
- Jommanee, N.; Chanthad, C.; Manokruang, K. Preparation of injectable hydrogels from temperature and pH responsive grafted chitosan with tuned gelation temperature suitable for tumor acidic environment. Carbohydr. Polym. 2018, 198, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Maeda, T.; Hotta, A. PEG-based nanocomposite hydrogel: Thermo-responsive sol-gel transition and degradation behavior controlled by the LA/GA ratio of PLGA-PEG-PLGA. Polym. Degrad. Stab. 2018, 147, 222–228. [Google Scholar] [CrossRef]


| Polypeptides | CGC (wt%) | Secondary Structure | ||
|---|---|---|---|---|
| Random Coil (%) | α-Helix (%) | β-Sheet/β-Turn (%) | ||
| 3s-PLL22-b-PPhe6.3 | 4.5 | 48.3 | 0 | 37.6/14.2 |
| 6s-PLL21-b-PPhe5.3 | 1.0 | 48.6 | 0 | 35.8/15.6 |
| 3s-PLL22-b-PLeu5.5 | 7.0 | 53.7 | 1.8 | 26.0/18.6 |
| 6s-PLL21-b-PLeu4.4 | 4.0 | 51.2 | 8.5 | 20.6/19.7 |
| 3s-PLL22-b-PVal5.1 | 5.5 | 47.2 | 0 | 38.9/13.9 |
| 6s-PLL21-b-PVal5 | 4.0 | 46.1 | 0 | 39.9/14.0 |
| Samples | Concentration (wt%) | Rg (Å) |
|---|---|---|
| 6s-PLL20-b-PPhe5 | 5.0 | 58.5 |
| 6s-PLL20-b-PLeu4.2 | 5.0 | 97.5 |
| 6s-PLL20-b-PVal4.8 | 5.0 | 47.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, T.H.M.; Huang, C.-C.; Tsai, Y.-J.; Hu, J.-J.; Jan, J.-S. Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly(L-lysine)-Based Amphiphilic Copolypeptides. Gels 2021, 7, 131. https://doi.org/10.3390/gels7030131
Phan THM, Huang C-C, Tsai Y-J, Hu J-J, Jan J-S. Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly(L-lysine)-Based Amphiphilic Copolypeptides. Gels. 2021; 7(3):131. https://doi.org/10.3390/gels7030131
Chicago/Turabian StylePhan, Thi Ha My, Ching-Chia Huang, Yi-Jen Tsai, Jin-Jia Hu, and Jeng-Shiung Jan. 2021. "Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly(L-lysine)-Based Amphiphilic Copolypeptides" Gels 7, no. 3: 131. https://doi.org/10.3390/gels7030131
APA StylePhan, T. H. M., Huang, C.-C., Tsai, Y.-J., Hu, J.-J., & Jan, J.-S. (2021). Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly(L-lysine)-Based Amphiphilic Copolypeptides. Gels, 7(3), 131. https://doi.org/10.3390/gels7030131








