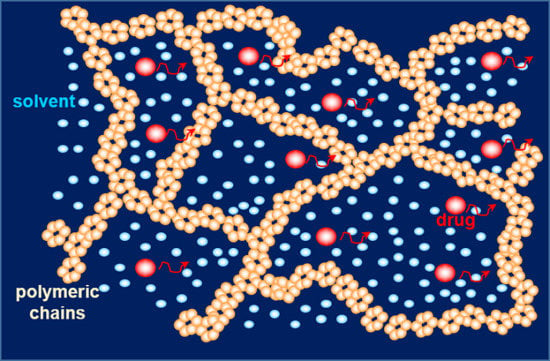
Theoretical Importance of PVP-Alginate Hydrogels Structure on Drug Release Kinetics
Abstract
:1. Introduction
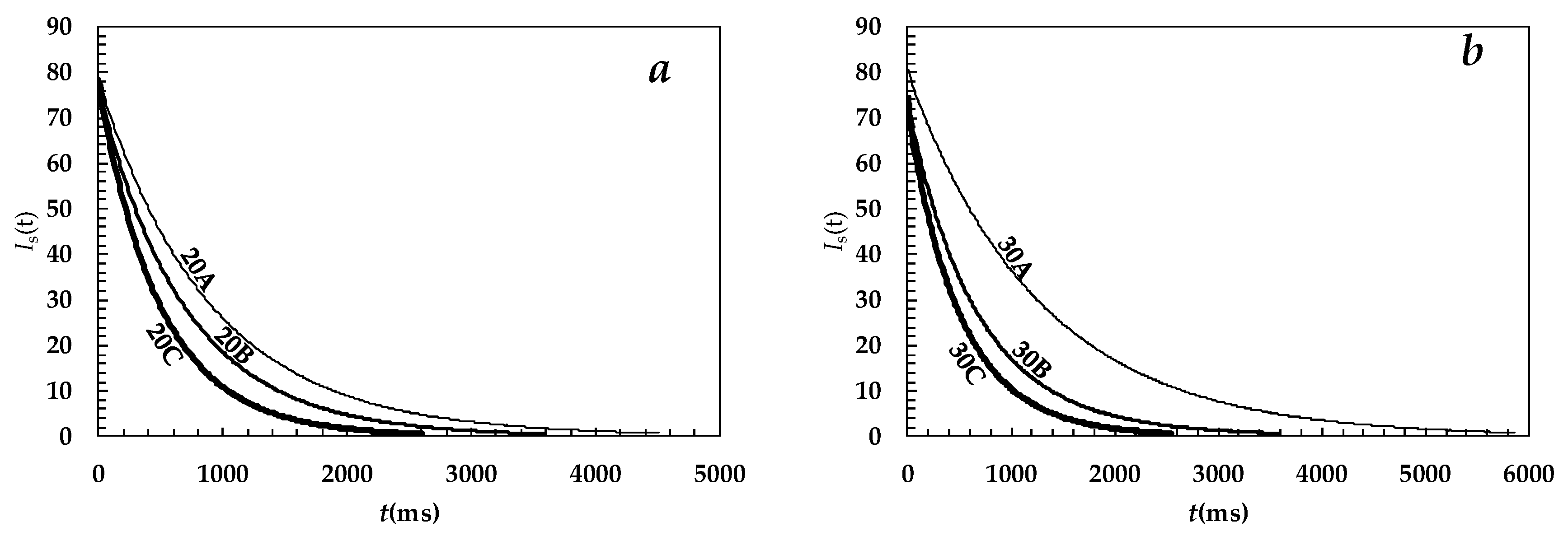
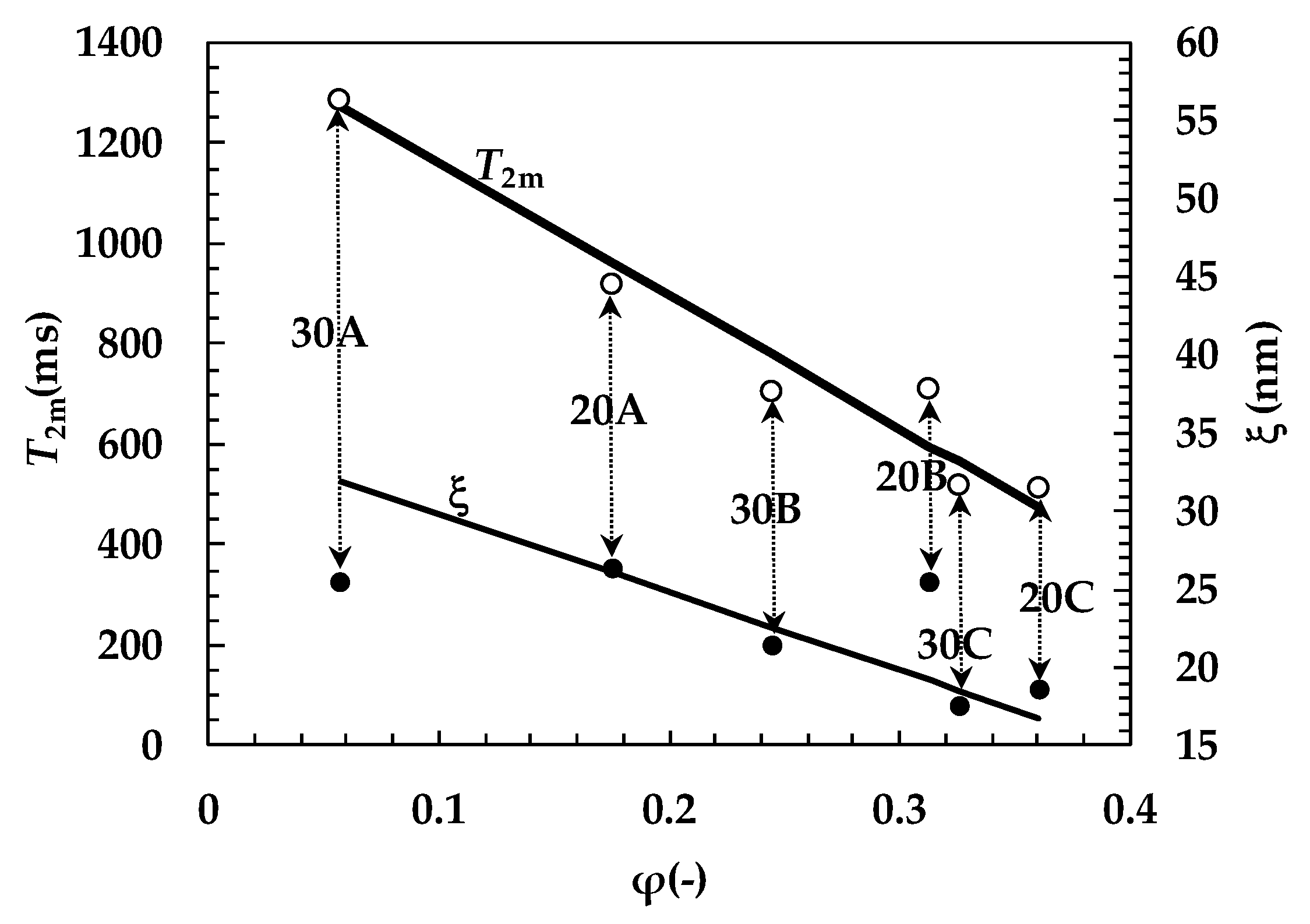
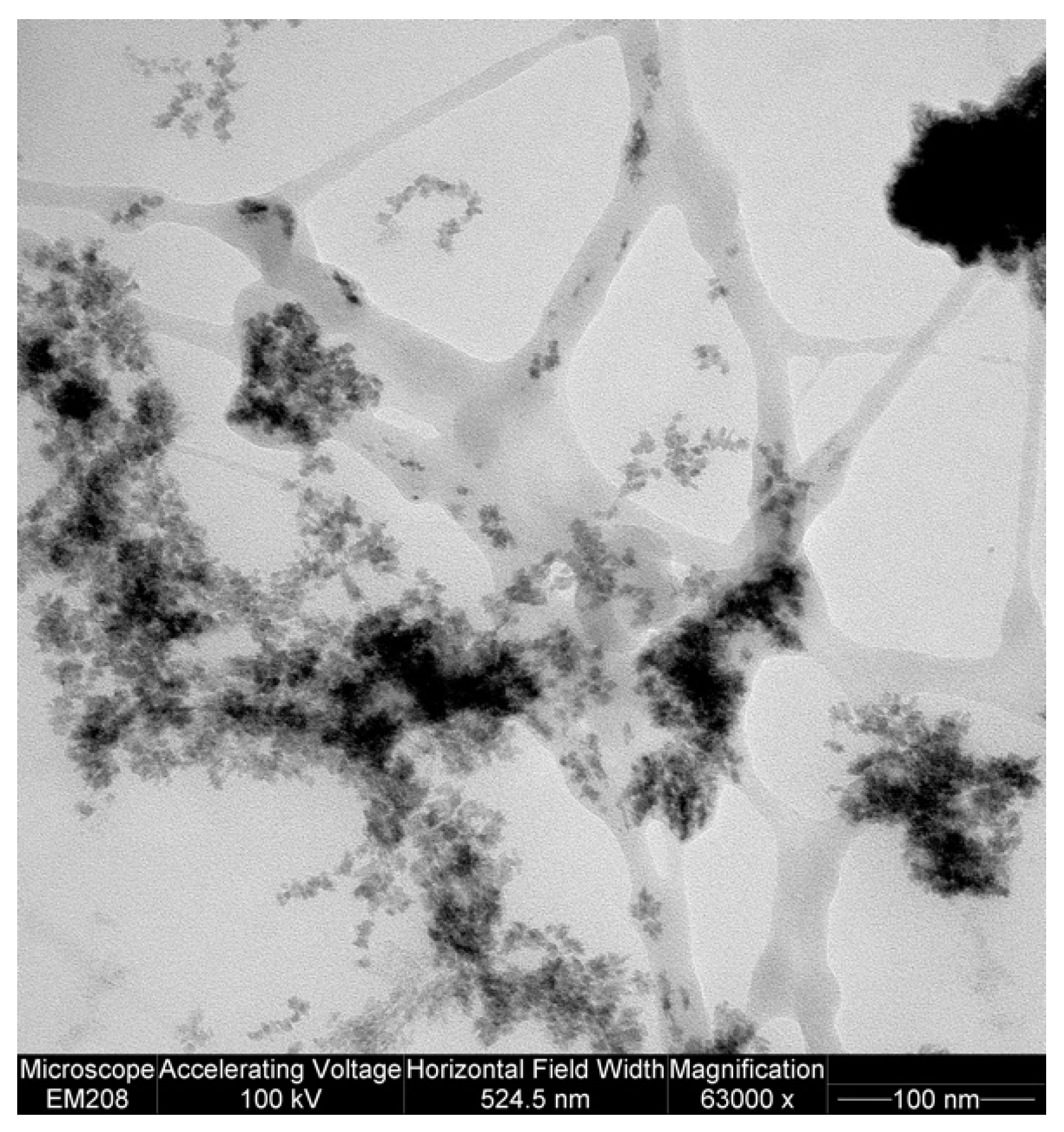
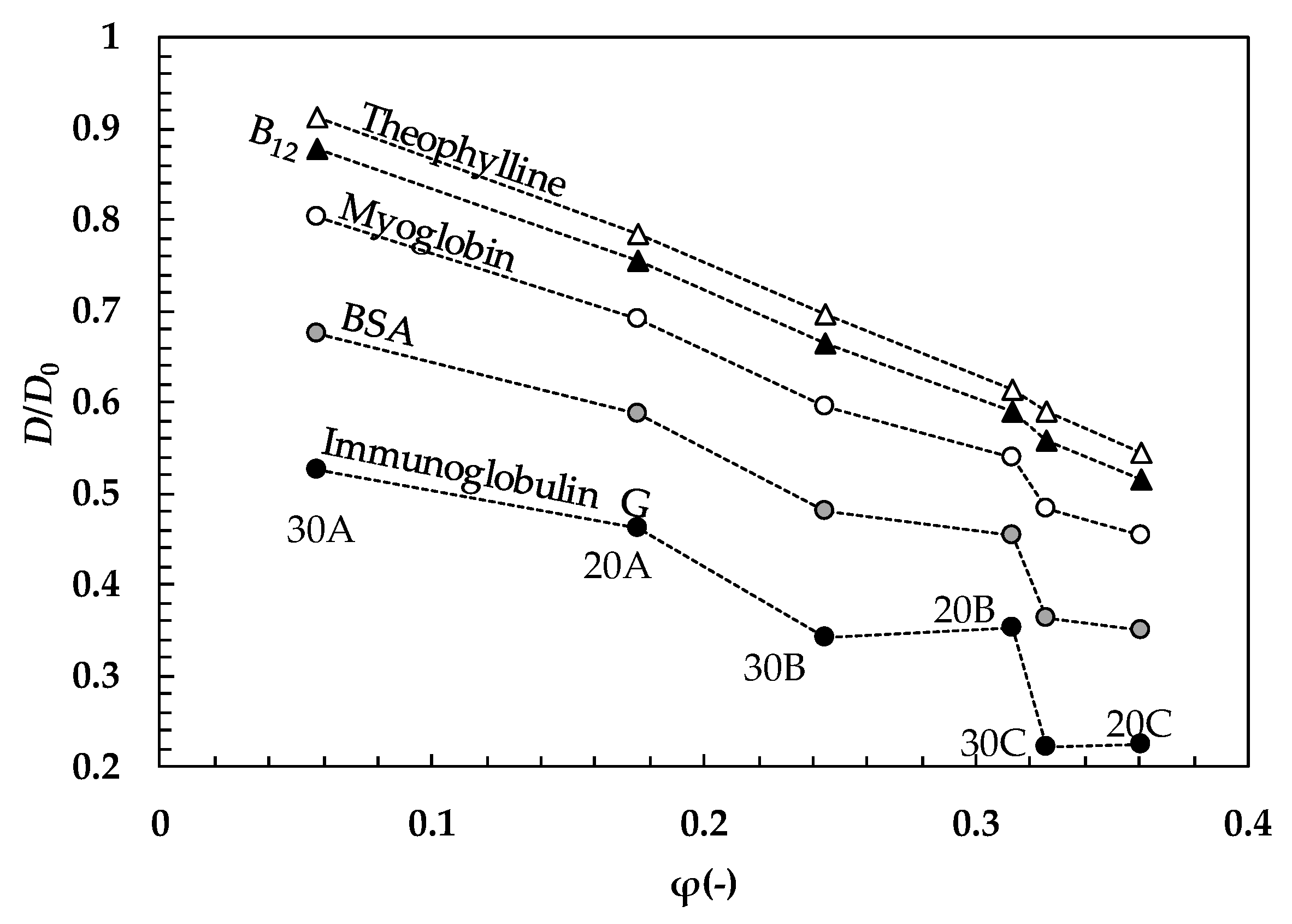
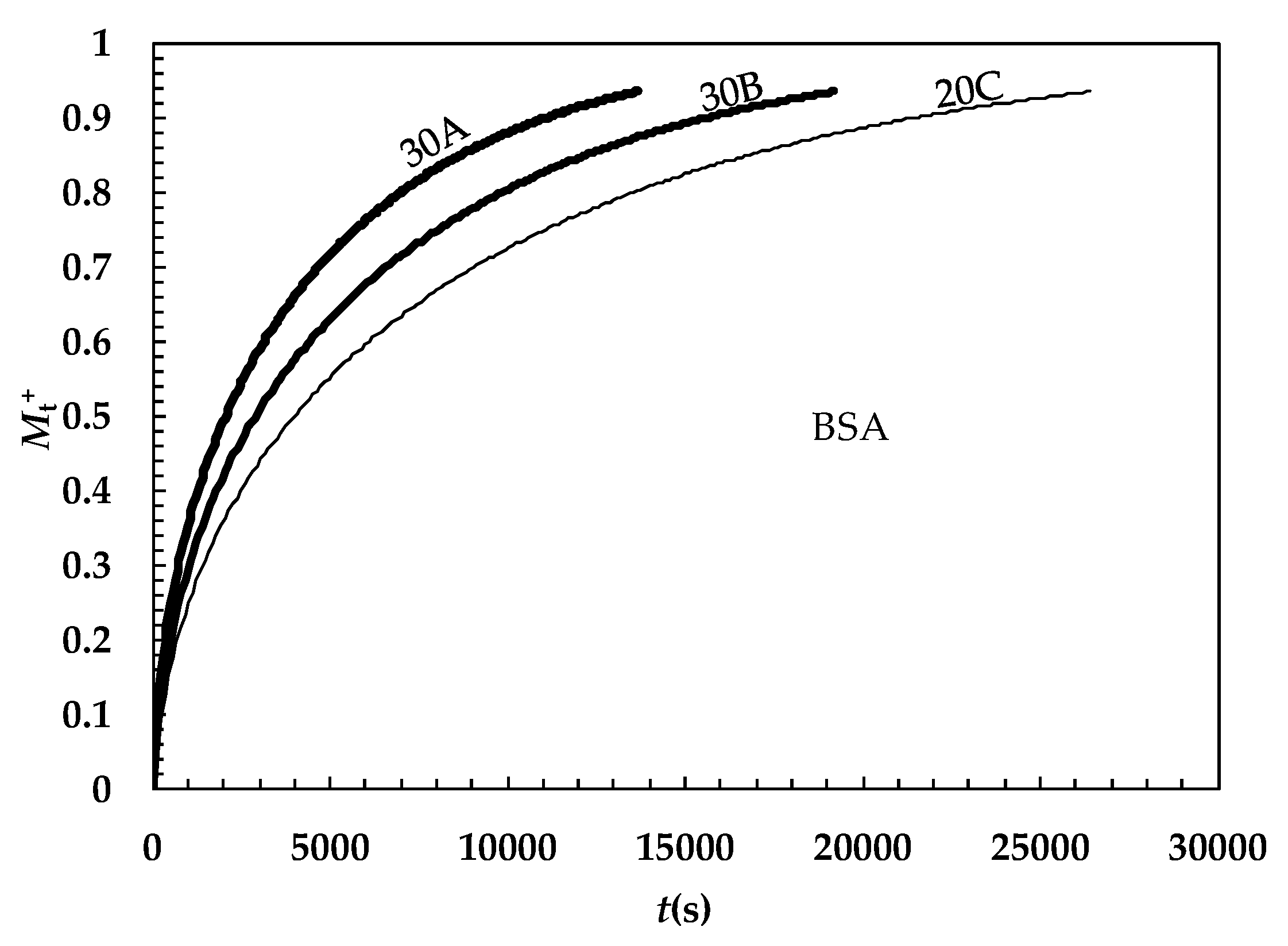
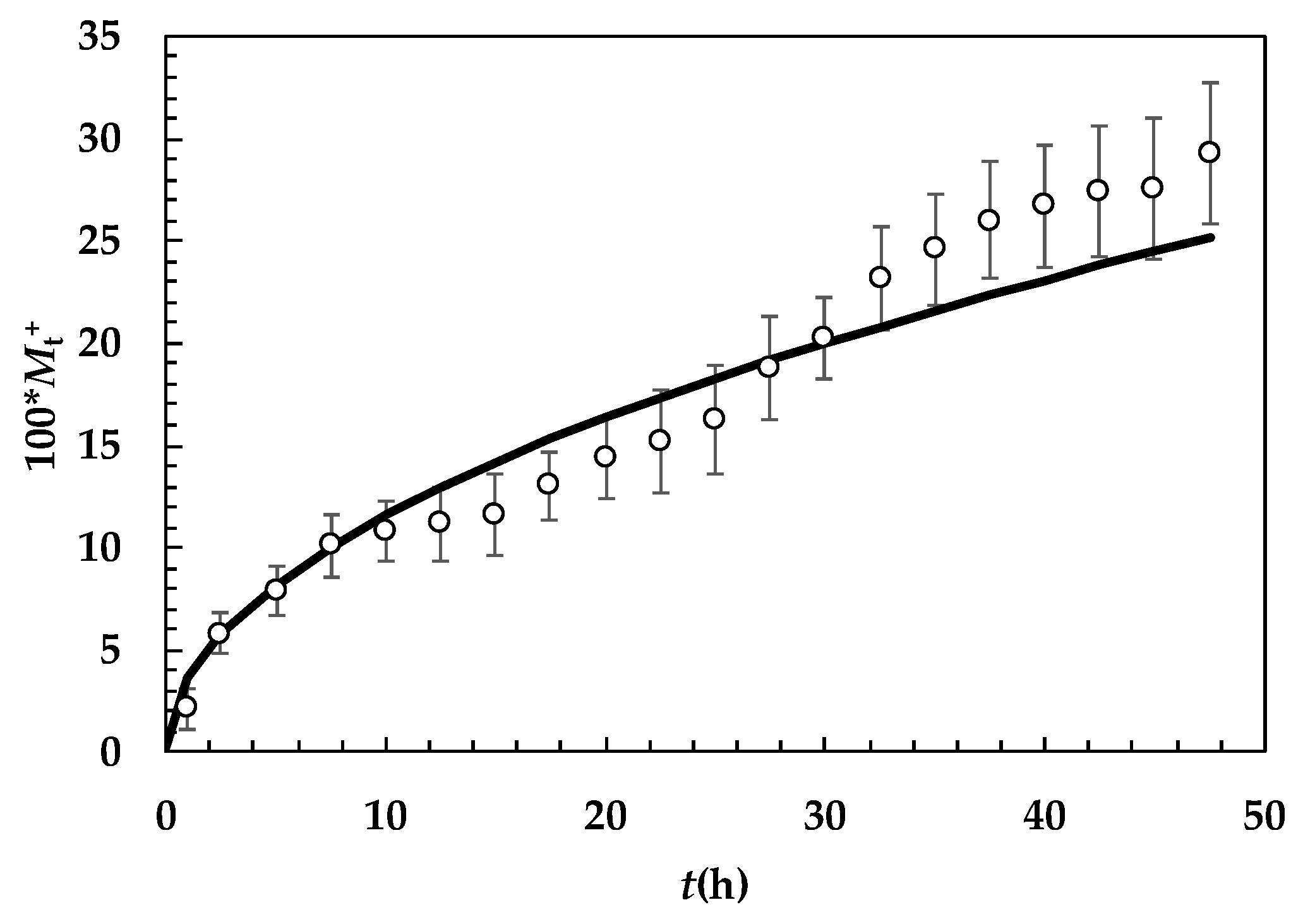
2. Results and Discussion
3. Conclusions
4. Materials and Methods
4.1. Interpenetrating Polymeric Network (IPN) Preparation
4.2. Rheology
4.3. LF-NMR
4.4. TEM
4.5. Release Test
Author Contributions
Funding
Conflicts of Interest
References and Note
- Lapasin, R.; Pricl, S. Rheology of Industrial Polysaccharides: Theory and Applications; Blackie Academic & Professional: London, UK, 1995; Chapter 1. [Google Scholar]
- Lin, C.-C.; Metters, A.T. Hydrogels in controlled release formulations: Network design and mathematical modeling. Adv. Drug Deliv. Rev. 2006, 58, 1379–1408. [Google Scholar] [CrossRef]
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef]
- Posocco, B.; Dreussi, E.; de Santa, J.; Toffoli, G.; Abrami, M.; Musiani, F.; Grassi, M.; Farra, R.; Tonon, F.; Grassi, G.; et al. Polysaccharides for the delivery of antitumor drugs. Materials 2015, 8, 2569–2615. [Google Scholar] [CrossRef]
- Matricardi, P.; Alhaique, F.; Coviello, T. Introduction. In Polysaccharide Hydrogels: Characterization and Biomedical Applications; Matricardi, P., Alhaique, F., Coviello, T., Eds.; Pan Standford Publishing: Singapore, 2015; Chapter 1. [Google Scholar]
- Matricardi, M.; di Meo, C.; Coviello, C.; Hennink, W.E.; Alhaique, F. Interpenetrating polymer networks polysaccharide hydrogels for drug delivery and tissue engineering. Adv. Drug Deliv. Rev. 2013, 65, 1172–1187. [Google Scholar] [CrossRef]
- Grassi, M.; Grassi, G. Application of mathematical modelling in sustained release delivery systems. Expert Opin. Drug Deliv. 2014, 11, 1299–1321. [Google Scholar] [CrossRef]
- Grassi, M.; Lapasin, R.; Pricl, S. Modeling of drug release from a swellable matrix. Chem. Eng. Commun. 1998, 169, 79–109. [Google Scholar] [CrossRef]
- Miller-Chou, A.A.; Koening, J.K. A review of polymer dissolution. Prog. Polym. Sci. 2003, 28, 1223–1270. [Google Scholar] [CrossRef]
- Hasa, D.; Voinovich, D.; Perissutti, B.; Grassi, M.; Bonifacio, A.; Sergo, V.; Cepek, C.; Chierotti, M.R.; Gobetto, R.; Dall’Acqua, S.; et al. Enhanced oral bioavailability of vinpocetine through mechanochemical salt formation: Physico-chemical characterization and in vivo studies. Pharm. Res. 2011, 28, 1870–1880. [Google Scholar] [CrossRef]
- Grassi, M.; Colombo, I.; Lapasin, R. Drug release from an ensemble of swellable crosslinked polymer particles. J. Control. Release 2000, 68, 97–113. [Google Scholar] [CrossRef]
- Coceani, N.; Magarotto, L.; Ceschia, D.; Colombo, I.; Grassi, M. Theoretical and experimental analysis of drug release from an ensemble of polymeric particles containing amorphous and nano-crystalline drug. Chem. Eng. Sci. 2012, 71, 345–355. [Google Scholar] [CrossRef]
- Grassi, G.; Hasa, H.; Voinovich, D.; Perissutti, B.; Dapas, B.; Farra, R.; Franceschinis, E.; Grassi, M. Simultaneous release and ADME processes of poorly water-soluble drugs: Mathematical modeling. Mol. Pharm. 2010, 7, 1488–1497. [Google Scholar] [CrossRef]
- Singh, M.; Lumpkin, J.; Rosenblat, J. Mathematical modeling of drug release from hydrogel matrices via a diffusion coupled with desorption mechanism. J. Control. Release 1994, 32, 17–25. [Google Scholar] [CrossRef]
- Singh, M.; Lumpkin, J.; Rosenblat, J. Effect of electrostatic interactions on polylysine release rates from collagen matrices and comparison with model predictions. J. Control. Release 1995, 35, 165–179. [Google Scholar] [CrossRef]
- Huang, J.; Wigent, R.J.; Schwartz, J.B. Drug-polymer interaction and its significance on the physical stability of nifedipine amorphous dispersion in microparticles of an ammonio methacrylate copolymer and ethylcellulose binary blend. J. Pharm. Sci. 2008, 97, 251–262. [Google Scholar] [CrossRef]
- Paulsson, M.; Edsman, K. Controlled drug release from gels using lipophilic interactions of charged substances with surfactants and polymers. J. Colloid Interface Sci. 2002, 248, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E.; Park, K.; Peppas, N.A. Molecular imprinting within hydrogels. Adv. Drug Deliv. Rev. 2002, 54, 149–161. [Google Scholar] [CrossRef]
- Milcovich, G.; Lettieri, S.; Antunes, F.E.; Medronho, B.; Fonseca, A.C.; Coelho, J.F.G.; Marizza, P.; Perrone, F.; Farra, R.; Dapas, B.; et al. Recent advances in smart biotechnology: Hydrogels and nanocarriers for tailored bioactive molecules depot. Adv. Colloid Interface Sci. 2017, 249, 163–180. [Google Scholar] [CrossRef]
- Fanesi, G.; Abrami, M.; Zecchin, F.; Giassi, I.; Dal Ferro, E.; Boisen, A.; Grassi, G.; Bertoncin, P.; Grassi, M.; Marizza, P. Combined Used of Rheology and LF-NMR for the Characterization of PVP-Alginates Gels Containing Liposomes. Pharm. Res. 2018, 35, 171–182. [Google Scholar] [CrossRef]
- Draper, N.R.; Smith, H. Applied Regression Analysis; John Wiley & Sons Inc.: New York, NY, USA, 1966. [Google Scholar]
- Marizza, P.; Abrami, M.; Keller, S.S.; Posocco, P.; Laurini, E.; Goswami, K.; Skov, A.L.; Boisen, A.; Larobina, D.; Grassi, G.; et al. Synthesis and characterization of UV photocrosslinkable hydrogels with poly(N-vinyl-2-pyrrolidone): Determination of the network mesh size distribution. J. Polym. Mater. Polym. Biomater. 2016, 65, 516–525. [Google Scholar] [CrossRef]
- Flory, P.J. Principles of Polymer Chemistry; Cornell University Press: Ithaca, NY, USA, 1953. [Google Scholar]
- Schurz, J. Rheology of polymer solutions of the network type. Prog. Polym. Sci. 1991, 16, 1–53. [Google Scholar] [CrossRef]
- Chui, M.; Phillips, R. Measurement of the porous microstructure of hydrogels by nuclear magnetic resonance. J. Colloid Interface Sci. 1995, 174, 336–344. [Google Scholar] [CrossRef]
- Scherer, G.W. Hydraulic radius and mesh size of gels. J. Sol Gel Sci. Technol. 1994, 1, 285–291. [Google Scholar] [CrossRef]
- Abrami, M.; Chiarappa, G.; Farra, R.; Grassi, G.; Marizza, P.; Grassi, M. Use of low field NMR for the characterization of gels and biological tissues. Admet Dmpk 2018, 6, 34–46. [Google Scholar] [CrossRef]
- Coviello, T.; Matricardi, P.; Alhaique, F.; Farra, R.; Tesei, G.; Fiorentino, S.; Asaro, F.; Milcovich, G.; Grassi, M. Guar gum/borax hydrogel: Rheological, low field NMR and release characterizations. Express Polym. Lett. 2013, 7, 733–746. [Google Scholar] [CrossRef]
- Amsden, B. Solute Diffusion within Hydrogels. Mechanisms and Models. Macromolecules 1998, 31, 8382–8395. [Google Scholar] [CrossRef]
- Lustig, S.R.; Peppas, N.A. Solute diffusion in swollen membranes. IX. Scaling laws for solute diffusion in gels. J. Appl. Polym. Sci. 1988, 36, 735–747. [Google Scholar] [CrossRef]
- Dalmoro, A.; Barba, A.A.; Lamberti, G.; Grassi, M.; d’Amore, M. Pharmaceutical Applications of Biocompatible Polymer Blends Containing Sodium Alginate. Adv. Polym. Technol. 2012, 31, 219–230. [Google Scholar] [CrossRef]
- Grassi, M.; Grassi, G.; Lapasin, R.; Colombo, I. Understanding Drug Release and Absorption Mechanisms a Physical and Mathematical Approach; CRC Press Imprint of Taylor & Francis: Boca Raton, FL, USA, 2007; Chapter 4. [Google Scholar]
- Luo, Z.; Zhang, G. Scaling for sedimentation and diffusion of poly(ethylene glycol) in water. J. Phys. Chem. B 2009, 113, 12462–12465. [Google Scholar] [CrossRef]
- Hong, P.; Koza, S.; Bouvier, E.S.P. Size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J. Liq. Chromatogr. Release Technol. 2012, 35, 2923–2950. [Google Scholar]
- Stellwagen, N.C.; Magnusdottir, S.; Gelfi, C.; Righetti, P.G. Measuring the translational diffusion coefficients of small DNA molecules by capillary electrophoresis. Biopolymers 2001, 58, 390–397. [Google Scholar] [CrossRef]
- Patil, S.M.; Keire, D.A.; Chen, K. Comparison of NMR and dynamic light scattering for measuring diffusion coefficients of formulated insulin: Implications for particle size distribution measurements in drug products. AAPS J. 2017, 19, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef]
- Macchioni, A.; Ciancaleoni, G.; Zuccaccia, C.; Zuccaccia, D. Determining accurate molecular sizes in solution through NMR diffusion spectroscopy. Chem. Soc. Rev. 2008, 37, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hou, T. Application of molecular dynamics simulations in molecular property prediction ii: Diffusion coefficient. J. Comput. Chem. 2011, 32, 3505–3519. [Google Scholar] [CrossRef]
- Weiss, L.B.; Dahirel, V.; Marry, V.; Jardat, M. Computation of the hydrodynamic radius of charged nanoparticles from nonequilibrium molecular dynamics. J. Phys. Chem. B 2018, 122, 5940–5950. [Google Scholar] [CrossRef] [PubMed]
- Durchschlag, H.; Zipper, P. Modeling the hydration of proteins: Prediction of structural and hydrodynamic parameters from X-ray diffraction and scattering data. Eur. Biophys. J. 2003, 32, 487–502. [Google Scholar] [CrossRef]
- De la Torre, J.G. The HYDRO software suite for the prediction of solution properties of rigid and flexible macromolecules and nanoparticles. In Analytical Ultracentrifugation: Instrumentation, Software, and Applications; Uchiyama, S., Arisaka, F., Stafford, W.F., Laue, T., Eds.; Springer: Tokyo, Japan, 2016; pp. 195–217. [Google Scholar]
- Atomic coordinates: myoglobin, PDB ID: 1VXB; BSA, PDB ID: 3V03. Vitamin B12 and theophylline molecular models were built and optimized according to the computational procedure described in Reference [44].
- Marson, D.; Laurini, E.; Posocco, P.; Fermeglia, M.; Pricl, S. Cationic carbosilane dendrimers and oligonucleotide binding: An energetic affair. Nanoscale 2015, 7, 3876–3887. [Google Scholar] [CrossRef]
- Kuijpers, A.J.; Engbers, G.H.M.; Feijen, J.; de Smed, S.C.; Meyvis, T.K.L.; Demeester, J.; Krijgsveld, J.; Zaat, S.A.J.; Dankert, J. Characterization of the network structure of carbodiimide cross-linked gelatin gels. Macromolecules 1999, 32, 3325–3334. [Google Scholar] [CrossRef]
- Meiboom, S.; Gill, D. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar] [CrossRef]
- Desmon, K. Techniques for Electron Microscopy; Blackwell Scientific Publication: Oxford, UK; Edinburg, TX, USA, 1965. [Google Scholar]
- Sabatini, D.D.; Bensch, K.; Barnett, R.J. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J. Cell. Biol. 1963, 17, 19–58. [Google Scholar] [CrossRef]

| System | λ1 (s) | g1 (Pa) | g2 (Pa) | g3 (Pa) | g4 (Pa) | ge (Pa) |
|---|---|---|---|---|---|---|
| 20A | (9.2 ± 2) × 10−2 | 8.2 ± 1.2 | 7.7 ± 1.4 | 15.1 ± 1.7 | - | 364 ± 18 |
| 20B | (5.9 ± 0.9) × 10−2 | 10.4 ± 0.8 | 11.1 ± 0.7 | 4.9 ± 0.9 | 17.3 ± 3.0 | 279 ± 9 |
| 20C | (10.0 ± 1) × 10−2 | 39.3 ± 1.3 | 25.5±1.5 | 19.5 ± 4.0 | 178 ± 23.0 | 854 ± 25 |
| 30A | (6.7 ± 3) × 10−2 | 13.0 ± 2 | 10.6 ± 2.0 | 9.1 ± 3.0 | 8.2 ± 8.0 | 337 ± 21 |
| 30B | (2.9 ± 2.6) × 10−2 | 200.6 ± 79 | 48.6 ± 26 | 16.0 ± 7.0 | 19.4 ± 7.6 | 453 ± 30 |
| 30C | (9.1 ± 1.1) × 10−2 | 136.6 ± 6 | 30.0 ± 5.0 | 33.0 ± 7.0 | 95.5 ± 25 | 1041 ± 32 |
| System | G (Pa) | ρx (mol/cm3) | ξ (nm) |
|---|---|---|---|
| 20A | 395 ± 28 | (1.7 ± 0.10) × 10−7 | 26.0 ± 0.6 |
| 20B | 434 ± 8 | (1.9 ± 0.03) × 10−7 | 25.0 ± 0.1 |
| 20C | 1117 ± 35 | (4.9 ± 0.15) × 10−7 | 18.6 ± 0.2 |
| 30A | 432 ± 23 | (1.9 ± 0.10) × 10−7 | 25.5 ± 0.5 |
| 30B | 737 ± 89 | (3.2 ± 0.40) × 10−7 | 21.4 ± 0.9 |
| 30C | 1335 ± 42 | (5.8 ± 0.20) × 10−7 | 17.5 ± 0.2 |
| System | A1% | T21 (ms) | A2% | T22 (ms) | T2m (ms) | (1/T2)m (ms−1) |
|---|---|---|---|---|---|---|
| 20A | 36 ± 1.5 | 1162 ± 14 | 64 ± 1.5 | 782 ± 5 | 920 ± 22 | (11.3 ± 0.2) × 10−4 |
| 20B | 80 ± 1.1 | 771 ± 3 | 20 ± 1.1 | 449 ± 10 | 708 ± 11 | (14.8 ± 0.3) × 10−4 |
| 20C | 58 ± 1.1 | 595 ± 2 | 42 ± 1.1 | 393 ± 7 | 509 ± 8 | (20.0 ± 0.4) × 10−4 |
| 30A | 68 ± 17.0 | 1349 ± 36 | 32 ± 17 | 1141 ± 79 | 1283 ± 310 | (7.8 ± 2.0) × 10−4 |
| 30B | 26 ± 1.2 | 1029 ± 36 | 74 ± 1.2 | 587 ± 79 | 703 ± 16 | (15.1 ± 0.3) × 10−4 |
| 30C | 79 ± 2.0 | 571 ± 5 | 21 ± 2 | 321 ± 159 | 518 ± 15 | (20.4 ± 0.9) × 10−4 |
| System | A1% | ξ1 (nm) | A2% | ξ2 (nm) | ξ (nm) | Rf (nm) |
|---|---|---|---|---|---|---|
| 20A | 36 ± 1.5 | 39.6 | 64 ± 1.5 | 22.1 | 26.0 ± 0.6 | 3.8 |
| 20B | 80 ± 1.1 | 30.3 | 20 ± 1.1 | 15.4 | 25.0 ± 0.1 | 5.1 |
| 20C | 58 ± 1.1 | 23.7 | 42 ± 1.1 | 14.4 | 18.6 ± 0.2 | 4.1 |
| 30A | 68 ± 17.0 | 28.2 | 32 ± 17 | 21.2 | 25.5 ± 0.5 | 2.0 |
| 30B | 26 ± 1.2 | 39.4 | 74 ± 1.2 | 18.4 | 21.4 ± 0.9 | 3.7 |
| 30C | 79 ± 2.0 | 21.1 | 21 ± 2 | 10.8 | 17.5 ± 0.2 | 3.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abrami, M.; Marizza, P.; Zecchin, F.; Bertoncin, P.; Marson, D.; Lapasin, R.; de Riso, F.; Posocco, P.; Grassi, G.; Grassi, M. Theoretical Importance of PVP-Alginate Hydrogels Structure on Drug Release Kinetics. Gels 2019, 5, 22. https://doi.org/10.3390/gels5020022
Abrami M, Marizza P, Zecchin F, Bertoncin P, Marson D, Lapasin R, de Riso F, Posocco P, Grassi G, Grassi M. Theoretical Importance of PVP-Alginate Hydrogels Structure on Drug Release Kinetics. Gels. 2019; 5(2):22. https://doi.org/10.3390/gels5020022
Chicago/Turabian StyleAbrami, Michela, Paolo Marizza, Francesca Zecchin, Paolo Bertoncin, Domenico Marson, Romano Lapasin, Filomena de Riso, Paola Posocco, Gabriele Grassi, and Mario Grassi. 2019. "Theoretical Importance of PVP-Alginate Hydrogels Structure on Drug Release Kinetics" Gels 5, no. 2: 22. https://doi.org/10.3390/gels5020022
APA StyleAbrami, M., Marizza, P., Zecchin, F., Bertoncin, P., Marson, D., Lapasin, R., de Riso, F., Posocco, P., Grassi, G., & Grassi, M. (2019). Theoretical Importance of PVP-Alginate Hydrogels Structure on Drug Release Kinetics. Gels, 5(2), 22. https://doi.org/10.3390/gels5020022