Multi-Omics Analyses Reveal the Antifungal Mechanism of Phenazine-1-Carboxylic Acid Against Pseudogymnoascus destructans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Reagents
2.2. Revival of P. destructans and In Vitro Antifungal Assays
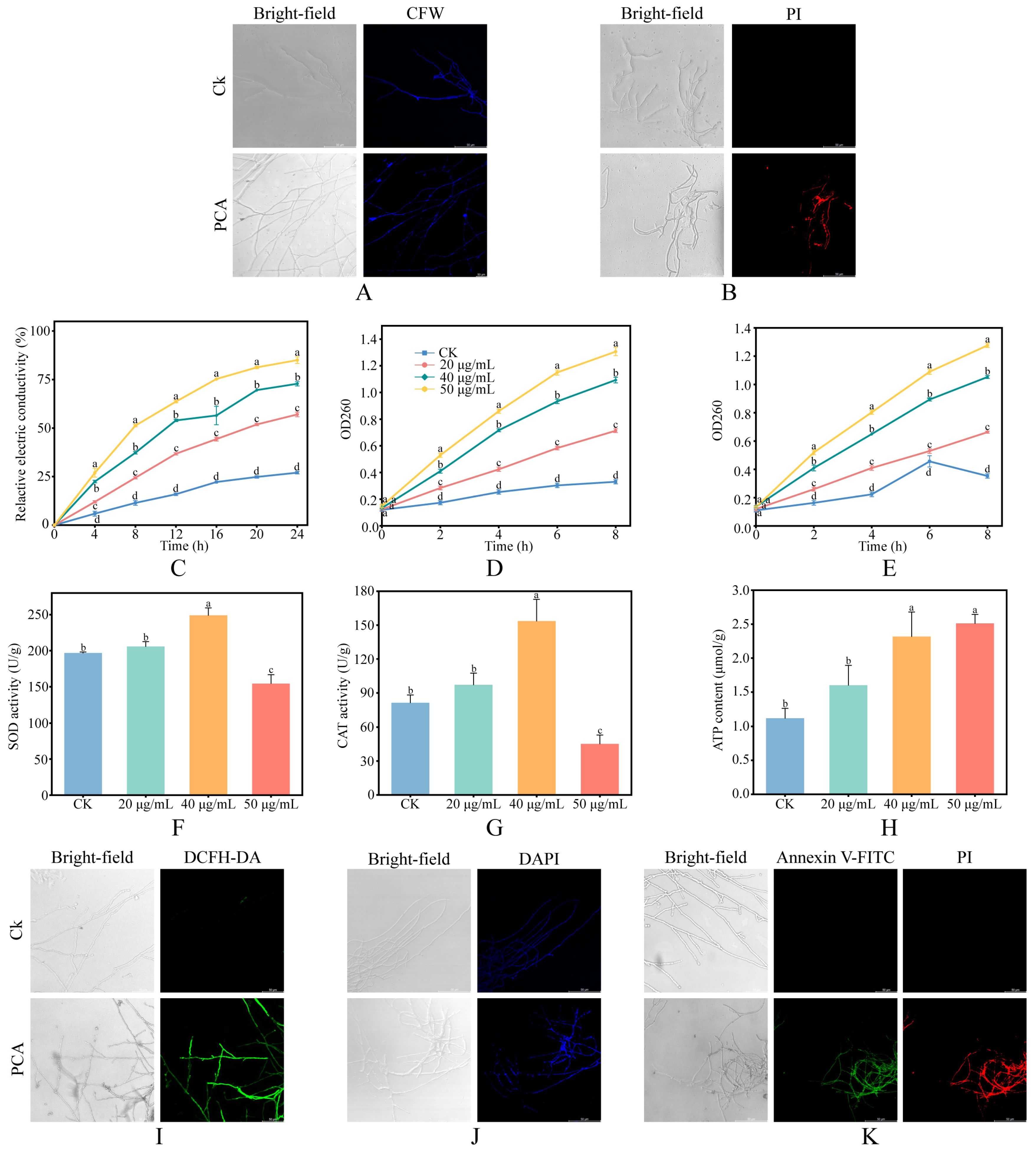
2.3. Assessment of Cell Wall and Membrane Integrity
2.4. Detection of Oxidative Stress and Energy Metabolism
2.5. Detection of DNA Damage and Apoptosis
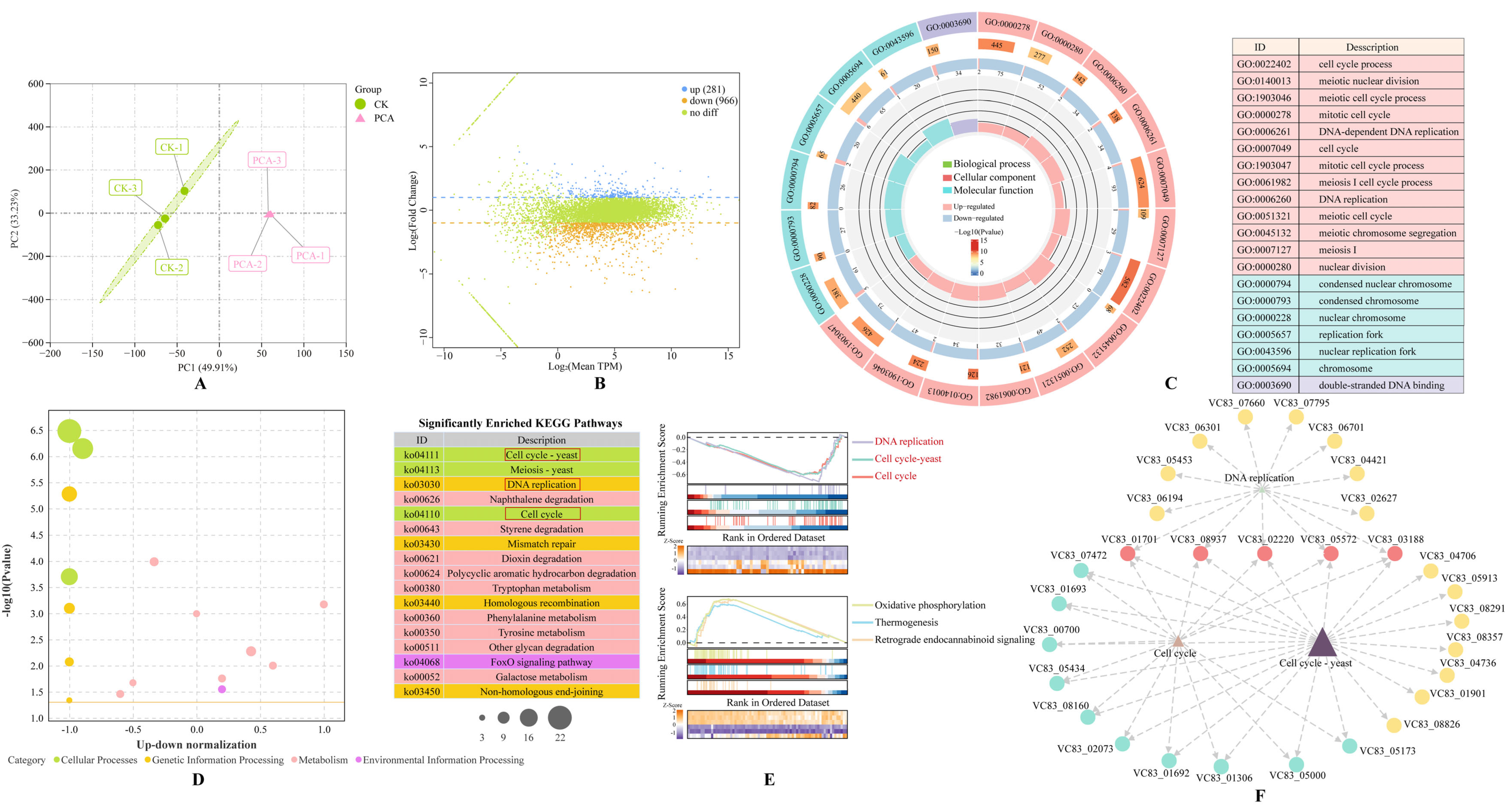
2.6. Transcriptomic Analysis
2.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
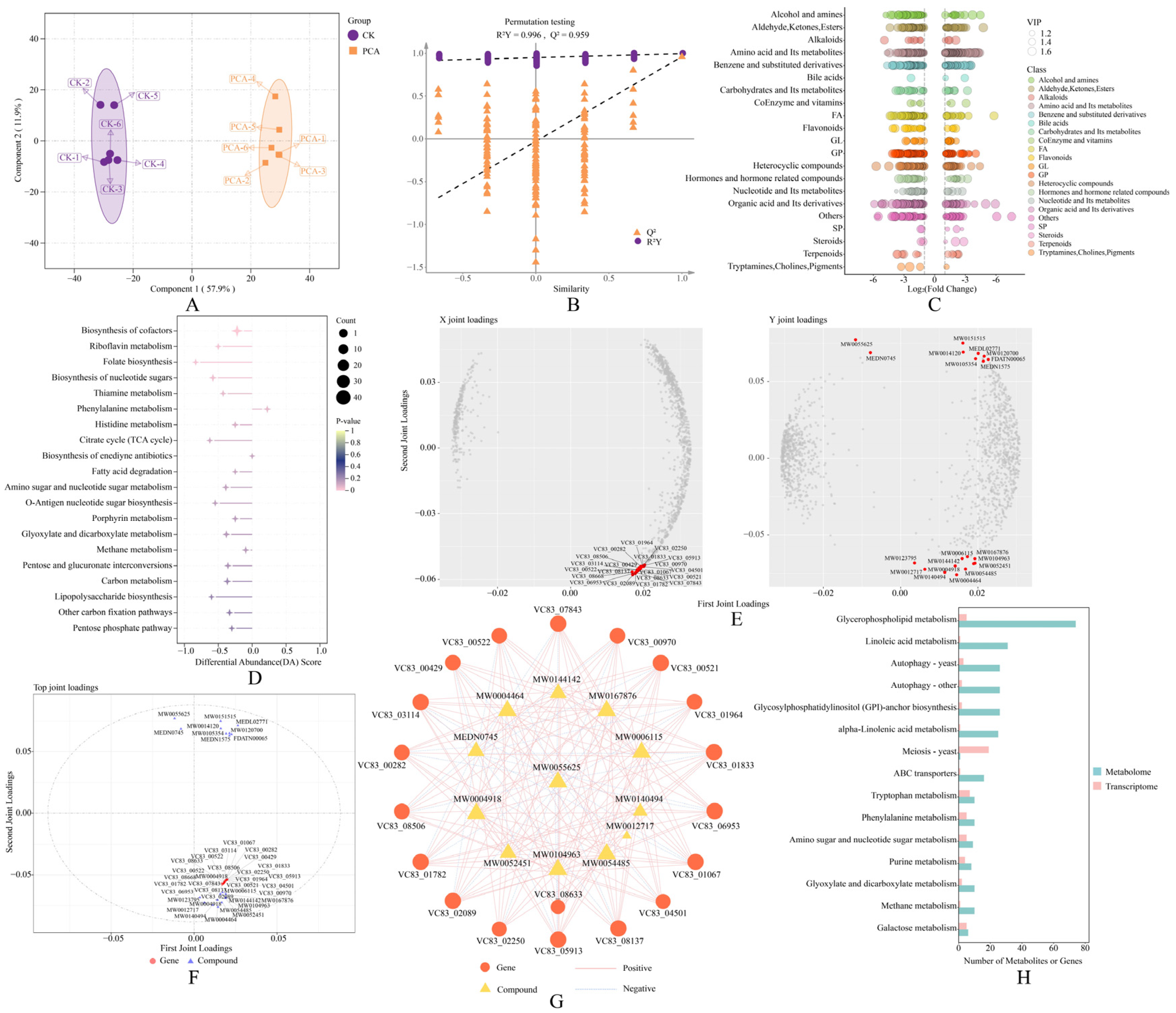
2.8. Metabolomics Analysis
2.9. Integrative Analysis of Transcriptomics and Metabolomics
2.10. Statistical Analysis
3. Result
3.1. Antifungal Activity of PCA Against P. destructans
3.2. Effects of PCA on Cell Wall and Membrane Integrity of P. destructans
3.3. Effects of PCA on Oxidative Stress, Energy Metabolism, and Apoptosis of P. destructans
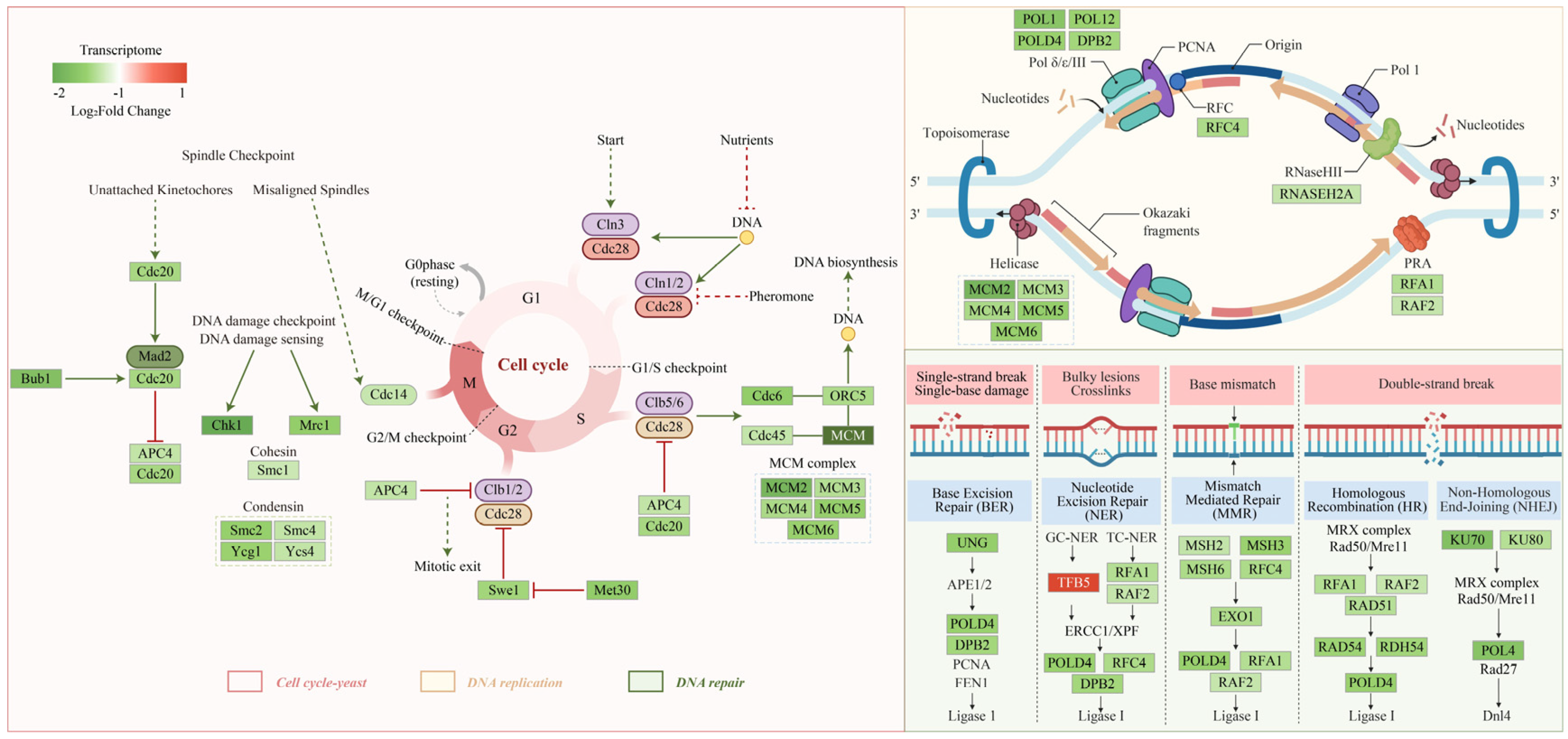
3.4. Transcriptomic Analysis of P. destructans in Response to PCA Treatment
3.5. RT-qPCR Verification of RNA-Seq Data
3.6. Metabolomic Analysis of P. destructans in Response to PCA Treatment
3.7. Integration of Transcriptomic and Metabolomic Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Blehert, D.S.; Hicks, A.C.; Behr, M.; Meteyer, C.U.; Berlowski-Zier, B.M.; Buckles, E.L.; Coleman, J.T.; Darling, S.R.; Gargas, A.; Niver, R.; et al. Bat white-nose syndrome: An emerging fungal pathogen? Science 2009, 323, 227. [Google Scholar] [CrossRef] [PubMed]
- Vanderwolf, K.J.; McAlpine, D.F.; Ryan, C.C.; Broders, H.G. Pseudogymnoascus destructans environmental reservoir decreases 11 years after an outbreak of white-nose syndrome. Ecosphere 2025, 16, e70149. [Google Scholar] [CrossRef]
- Hoyt, J.R.; Sun, K.; Parise, K.L.; Lu, G.; Langwig, K.E.; Jiang, T.; Yang, S.; Frick, W.F.; Kilpatrick, A.M.; Foster, J.T.; et al. Widespread bat white-nose syndrome fungus, northeastern China. Emerg. Infect. Dis. 2016, 22, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, J.R.; Langwig, K.E.; Sun, K.; Parise, K.L.; Li, A.; Wang, Y.; Huang, X.; Worledge, L.; Miller, H.; White, J.P.; et al. Environmental reservoir dynamics predict global infection patterns and population impacts for the fungal disease white-nose syndrome. Proc. Natl. Acad. Sci. USA 2020, 117, 7255–7262. [Google Scholar] [CrossRef]
- Li, A.; Leng, H.; Li, Z.; Jin, L.; Sun, K.; Feng, J. Temporal dynamics of the bat wing transcriptome: Insight into gene-expression changes that enable protection against pathogen. Virulence 2023, 14, 2156185. [Google Scholar] [CrossRef]
- Li, Z.; Li, A.; Hoyt, J.R.; Dai, W.; Leng, H.; Li, Y.; Li, W.; Liu, S.; Jin, L.; Sun, K.; et al. Activity of bacteria isolated from bats against Pseudogymnoascus destructans in China. Microb. Biotechnol. 2022, 15, 469–481. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, Y.; Huang, L.; Pu, Y.; Sun, X.; Feng, J.; Sun, K. Antifungal mechanism of ketone volatile organic compounds against Pseudogymnoascus destructans. Virulence 2025, 16, 2569627. [Google Scholar] [CrossRef]
- Xun, W.; Gong, B.; Liu, X.; Yang, X.; Zhou, X.; Jin, L. Antifungal mechanism of phenazine-1-carboxylic acid against Pestalotiopsis kenyana. Int. J. Mol. Sci. 2023, 24, 11274. [Google Scholar] [CrossRef]
- Xu, Y. Green microorganism based antimicrobial: Shenqinmycin (M18). Fine Spec. Chem. 2004, 20, 8–9. [Google Scholar]
- Cimmino, A.; Bahmani, Z.; Castaldi, S.; Masi, M.; Isticato, R.; Abdollahzadeh, J.; Amini, J.; Evidente, A. Phenazine-1-carboxylic acid (PCA), produced for the first time as an antifungal metabolite by Truncatella angustata, a causal agent of grapevine trunk diseases (GTDs) in Iran. J. Agric. Food. Chem. 2021, 69, 12143–12147. [Google Scholar] [CrossRef] [PubMed]
- Simionato, A.S.; Navarro, M.O.; de Jesus, M.L.; Barazetti, A.R.; da Silva, C.S.; Simões, G.C.; Balbi-Peña, M.I.; de Mello, J.C.; Panagio, L.A.; de Almeida, R.S. The effect of phenazine-1-carboxylic acid on mycelial growth of Botrytis cinerea produced by Pseudomonas aeruginosa LV strain. Front. Microbiol. 2017, 8, 1102. [Google Scholar] [CrossRef]
- Xu, S.; Pan, X.; Luo, J.; Wu, J.; Zhou, Z.; Liang, X.; He, Y.; Zhou, M. Effects of phenazine-1-carboxylic acid on the biology of the plant-pathogenic bacterium Xanthomonas oryzae pv. oryzae. Pestic. Biochem. Physiol. 2015, 117, 39–46. [Google Scholar] [CrossRef]
- Huang, J.; Chen, S.; He, W.; Xiao, Y.; Wang, N.; Huang, L. Phenazine-1-carboxylic acid has a broad-spectrum antifungal effect by targeting isocitrate lyase. J. Agric. Food Chem. 2025, 73, 5007–5019. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shan, M.; Li, A.; Wang, K.; Wei, Z.; Shen, M.; Lu, J.; Sun, K.; Li, Z.; Feng, J. Mechanisms of volatile organic compounds from bat cave environments against Pseudogymnoascus destructans in vitro. Appl. Environ. Microbiol. 2025, 91, e0118725. [Google Scholar] [CrossRef] [PubMed]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Micalizzi, E.W.; Mack, J.N.; White, G.P.; Avis, T.J.; Smith, M.L. Microbial inhibitors of the fungus Pseudogymnoascus destructans, the causal agent of white-nose syndrome in bats. PLoS ONE 2017, 12, e0179770. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Schipor, S.; Vladoiu, S.; Baciu, A.E.; Niculescu, A.M.; Caragheorgheopol, A.; Iancu, I.; Plesa, A.; Popescu, A.; Manda, D.; Scientific, A. A comparative analysis of three methods used for RNA quantitation. Rom. Rep. Phys. 2016, 68, 1178–1188. [Google Scholar]
- Chen, S. fastp 1.0: An ultra-fast all-round tool for FASTQ data quality control and preprocessing. iMeta 2025, 4, e70078. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, G.; Li, H.; Huang, L.; Cheng, Y.; Liu, J.; Song, R.; Wang, X. Integrated multi-omics analysis reveals that Gongying San ameliorates subclinical mastitis by modulating intestinal microbiota and metabolites in dairy cows. Front. Vet. Sci. 2025, 12, 1589900. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2013, 2, 13. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddani, S.E.; Uh, H.W.; Jongbloed, G.; Hayward, C.; Klaric, L.; Kielbasa, S.M.; Houwing-Duistermaat, J. Integrating omics datasets with the OmicsPLS package. BMC Bioinformatics 2018, 19, 371. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.M.; Dumville, I.; Nabholz, B.; Zhelyazkova, V.; Stecker, R.M.; Blomberg, A.S.; Dool, S.E.; Fritze, M.; Tilak, M.K.; Bashta, A.T.; et al. Two distinct host-specialized fungal species cause white-nose disease in bats. Nature 2025, 642, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, H.; Luo, Y.; Ouyang, H.; Hu, H.; Jin, C. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 2007, 64, 1014–1027. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, S.; Xu, Y.; Zhang, J.; Wu, P.; Luo, L.; Jiang, L. Efficacy of pterostilbene inhibition of postharvest anthracnose on papaya fruit and antifungal mechanisms against Colletotrichum gloeosporioides. Postharvest. Biol. Tec. 2025, 221, 113304. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, B. Role of cell membrane homeostasis in the pathogenicity of pathogenic filamentous fungi. Virulence 2024, 15, 2299183. [Google Scholar] [CrossRef]
- Yin, Q.; Yang, R.; Ren, Y.; Yang, Z.; Li, T.; Huang, H.; Tang, Q.; Li, D.; Jiang, S.; Wu, X.; et al. Transcriptomic, biochemical, and morphological study reveals the mechanism of inhibition of Pseudopestalotiopsis camelliae-sinensis by phenazine-1-carboxylic acid. Front. Microbiol. 2021, 12, 618476. [Google Scholar] [CrossRef]
- Arroyo, J.; Farkas, V.; Sanz, A.B.; Cabib, E. Strengthening the fungal cell wall through chitin-glucan cross-links: Effects on morphogenesis and cell integrity. Cell. Microbiol. 2016, 18, 1239–1250. [Google Scholar] [CrossRef]
- Lin, S.; Wang, Y.; Lu, Q.; Zhang, B.; Wu, X. Combined transcriptome and metabolome analyses reveal the potential mechanism for the inhibition of Penicillium digitatum by X33 antimicrobial oligopeptide. Bioresour. Bioprocess 2021, 8, 120. [Google Scholar] [CrossRef]
- Chen, C.; Cai, N.; Chen, J.; Wan, C. UHPLC-Q-TOF/MS-based metabolomics approach reveals the antifungal potential of pinocembroside against citrus green mold phytopathogen. Plants 2019, 9, 17. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Risch, T.S.; Savary, B.J. Isolation and identification of an extracellular subtilisin-like serine protease secreted by the bat pathogen Pseudogymnoascus destructans. PLoS ONE 2015, 10, e0120508. [Google Scholar] [CrossRef] [PubMed]
- Reeder, S.M.; Palmer, J.M.; Prokkola, J.M.; Lilley, T.M.; Reeder, D.M.; Field, K.A. Pseudogymnoascus destructans transcriptome changes during white-nose syndrome infections. Virulence 2017, 8, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A.J.; He, X.; Qiu, Y.; Davis, M.J.; Vedula, P.; Lyons, D.M.; Park, Y.D.; Hardison, S.E.; Malachowski, A.N.; Osterholzer, J.J.; et al. Cryptococcal heat shock protein 70 homolog Ssa1 contributes to pulmonary expansion of Cryptococcus neoformans during the afferent phase of the immune response by promoting macrophage M2 polarization. J. Immunol. 2015, 194, 5999–6010. [Google Scholar] [CrossRef]
- Neves-da-Rocha, J.; Santos-Saboya, M.J.; Lopes, M.E.R.; Rossi, A.; Martinez-Rossi, N.M. Insights and perspectives on the role of proteostasis and heat shock proteins in fungal infections. Microorganisms 2023, 11, 1878. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Li, Q.; Zhang, S.; Lv, Y.; Zhai, H.; Wei, S.; Ma, P.; Hu, Y. Transcriptomic and biochemical analyses revealed antifungal mechanism of trans-anethole on Aspergillus flavus growth. Appl. Microbiol. Biotechnol. 2023, 107, 7213–7230. [Google Scholar] [CrossRef]
- Flieger, M.; Bandouchova, H.; Cerny, J.; Chudickova, M.; Kolarik, M.; Kovacova, V.; Martinkova, N.; Novak, P.; Sebesta, O.; Stodulkova, E.; et al. Vitamin B2 as a virulence factor in Pseudogymnoascus destructans skin infection. Sci. Rep. 2016, 6, 33200. [Google Scholar] [CrossRef]
- Fan, W.; Li, B.; Du, N.; Hui, T.; Cao, Y.; Li, X.; Ren, H. Energy metabolism as the target of 3-phenyllactic acid against Rhizopus oryzae. Int. J. Food. Microbiol. 2022, 369, 109606. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, C.; Xiao, J.; Sun, S.; Gao, T.; Zhu, B.; Zhang, D. Metabolomic and transcriptome analysis of the inhibitory effects of Bacillus subtilis strain Z-14 against Fusarium oxysporum causing vascular wilt diseases in cucumber. J. Agri. Food Chem. 2023, 71, 2644–2657. [Google Scholar] [CrossRef]
- Larcombe, D.E.; Bohovych, I.M.; Pradhan, A.; Ma, Q.; Hickey, E.; Leaves, I.; Cameron, G.; Avelar, G.M.; de Assis, L.J.; Childers, D.S.; et al. Glucose-enhanced oxidative stress resistance—A protective anticipatory response that enhances the fitness of Candida albicans during systemic infection. PLoS Pathogens 2023, 19, e1011505. [Google Scholar] [CrossRef]
- Chang, W.; Wu, X.; Cheng, A.; Zhang, L.; Ji, M.; Lou, H. Retigeric acid B exerts antifungal effect through enhanced reactive oxygen species and decreased cAMP. Biochim. Biophys Acta 2011, 1810, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Zou, X.; Wei, Y.; Jiang, S.; Xu, F.; Wang, H.; Zhan, P.; Shao, X. ROS stress and cell membrane disruption are the main antifungal mechanisms of 2-phenylethanol against Botrytis cinerea. J. Agri. Food Chem. 2022, 70, 14468–14479. [Google Scholar] [CrossRef] [PubMed]
- Traynor, A.M.; Sheridan, K.J.; Jones, G.W.; Calera, J.A.; Doyle, S. Involvement of sulfur in the biosynthesis of essential metabolites in pathogenic fungi of animals, particularly Aspergillus spp.: Molecular and therapeutic implications. Front. Microbiol. 2019, 10, 2859. [Google Scholar] [CrossRef]
- Perez-Martin, J.; Bardetti, P.; Castanheira, S.; de la Torre, A.; Tenorio-Gomez, M. Virulence-specific cell cycle and morphogenesis connections in pathogenic fungi. Semin. Cell. Dev. Biol. 2016, 57, 93–99. [Google Scholar] [CrossRef]
- Kelliher, C.M.; Leman, A.R.; Sierra, C.S.; Haase, S.B. Investigating conservation of the cell-cycle-regulated transcriptional program in the fungal pathogen, Cryptococcus neoformans. PLoS Genet. 2016, 12, e1006453. [Google Scholar] [CrossRef]
- Stoeber, K.; Tlsty, T.D.; Happerfield, L.; Thomas, G.A.; Romanov, S.; Bobrow, L.; Williams, E.D.; Williams, G.H. DNA replication licensing and human cell proliferation. J. Cell. Sci. 2001, 114, 2027–2041. [Google Scholar] [CrossRef]
- Tian, Y.; Zhou, Y.; Chen, F.; Qian, S.; Hu, X.; Zhang, B.; Liu, Q. Research progress in MCM family: Focus on the tumor treatment resistance. Biomed. Pharmacother. 2024, 173, 116408. [Google Scholar] [CrossRef]
- Trouiller, B.; Schaefer, D.G.; Charlot, F.; Nogue, F. MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res. 2006, 34, 232–242. [Google Scholar] [CrossRef]
- Fan, L.; Wei, Y.; Chen, Y.; Ouaziz, M.; Jiang, S.; Xu, F.; Wang, H.; Shao, X. Transcriptome analysis reveals the mechanism of antifungal peptide epinecidin-1 against Botrytis cinerea by mitochondrial dysfunction and oxidative stress. Pestic. Biochem. Physiol. 2024, 202, 105932. [Google Scholar] [CrossRef]
- Li, D.; Xu, Y.; Zhang, D.; Quan, H.; Mylonakis, E.; Hu, D.; Li, M.; Zhao, L.; Zhu, L.; Wang, Y.; et al. Fluconazole assists berberine to kill fluconazole-resistant Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 6016–6027. [Google Scholar] [CrossRef]
- Huang, H.; Sun, L.; Bi, K.; Zhong, G.; Hu, M. The effect of phenazine-1-carboxylic acid on the morphological, physiological, and molecular characteristics of Phellinus noxius. Molecules 2016, 21, 613. [Google Scholar] [CrossRef]
- Sobel, J.D. Resistance to fluconazole of Candida albicans in vaginal isolates: A 10-year study in a clinical referral center. Antimicrob. Agents Chemother. 2023, 67, e0018123. [Google Scholar] [CrossRef]
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An overlooked pesticide class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Huang, Z.; Sun, S.; Jin, Z.; Ji, Y.; Lu, J.; Xu, T.; Sun, K.; Li, Z.; Feng, J. Multi-Omics Analyses Reveal the Antifungal Mechanism of Phenazine-1-Carboxylic Acid Against Pseudogymnoascus destructans. J. Fungi 2026, 12, 16. https://doi.org/10.3390/jof12010016
Huang Z, Sun S, Jin Z, Ji Y, Lu J, Xu T, Sun K, Li Z, Feng J. Multi-Omics Analyses Reveal the Antifungal Mechanism of Phenazine-1-Carboxylic Acid Against Pseudogymnoascus destructans. Journal of Fungi. 2026; 12(1):16. https://doi.org/10.3390/jof12010016
Chicago/Turabian StyleHuang, Zihao, Shaopeng Sun, Zhouyu Jin, Yantong Ji, Jiaqi Lu, Ting Xu, Keping Sun, Zhongle Li, and Jiang Feng. 2026. "Multi-Omics Analyses Reveal the Antifungal Mechanism of Phenazine-1-Carboxylic Acid Against Pseudogymnoascus destructans" Journal of Fungi 12, no. 1: 16. https://doi.org/10.3390/jof12010016
APA StyleHuang, Z., Sun, S., Jin, Z., Ji, Y., Lu, J., Xu, T., Sun, K., Li, Z., & Feng, J. (2026). Multi-Omics Analyses Reveal the Antifungal Mechanism of Phenazine-1-Carboxylic Acid Against Pseudogymnoascus destructans. Journal of Fungi, 12(1), 16. https://doi.org/10.3390/jof12010016




