New Records of Canker-Causing Pathogens of Acacia spp. and Pithecellobium dulce in Southern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Surveys and Fungal Isolation
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analysis
2.4. Pathogenicity Test
3. Results
3.1. Surveys and Fungal Isolation
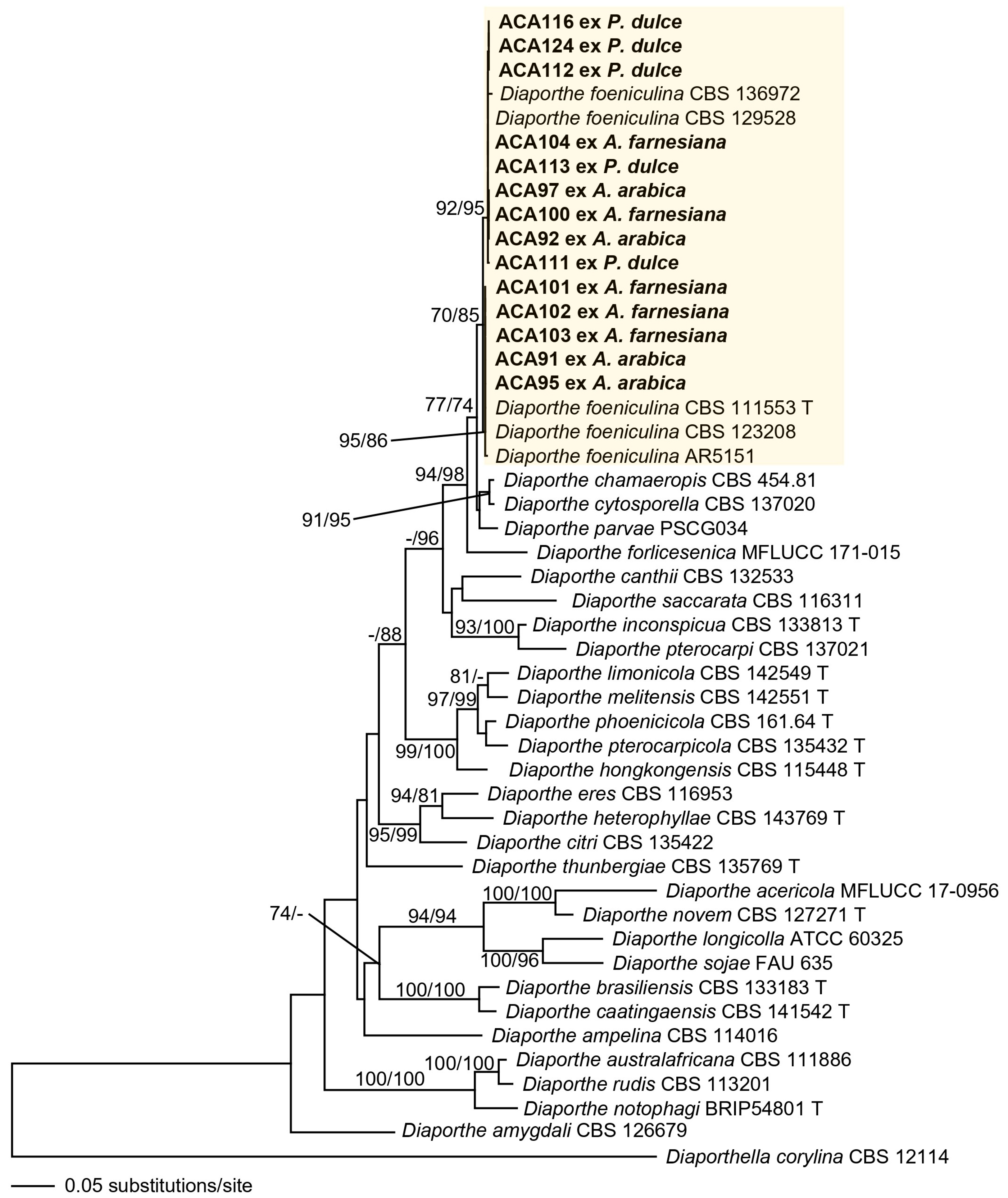
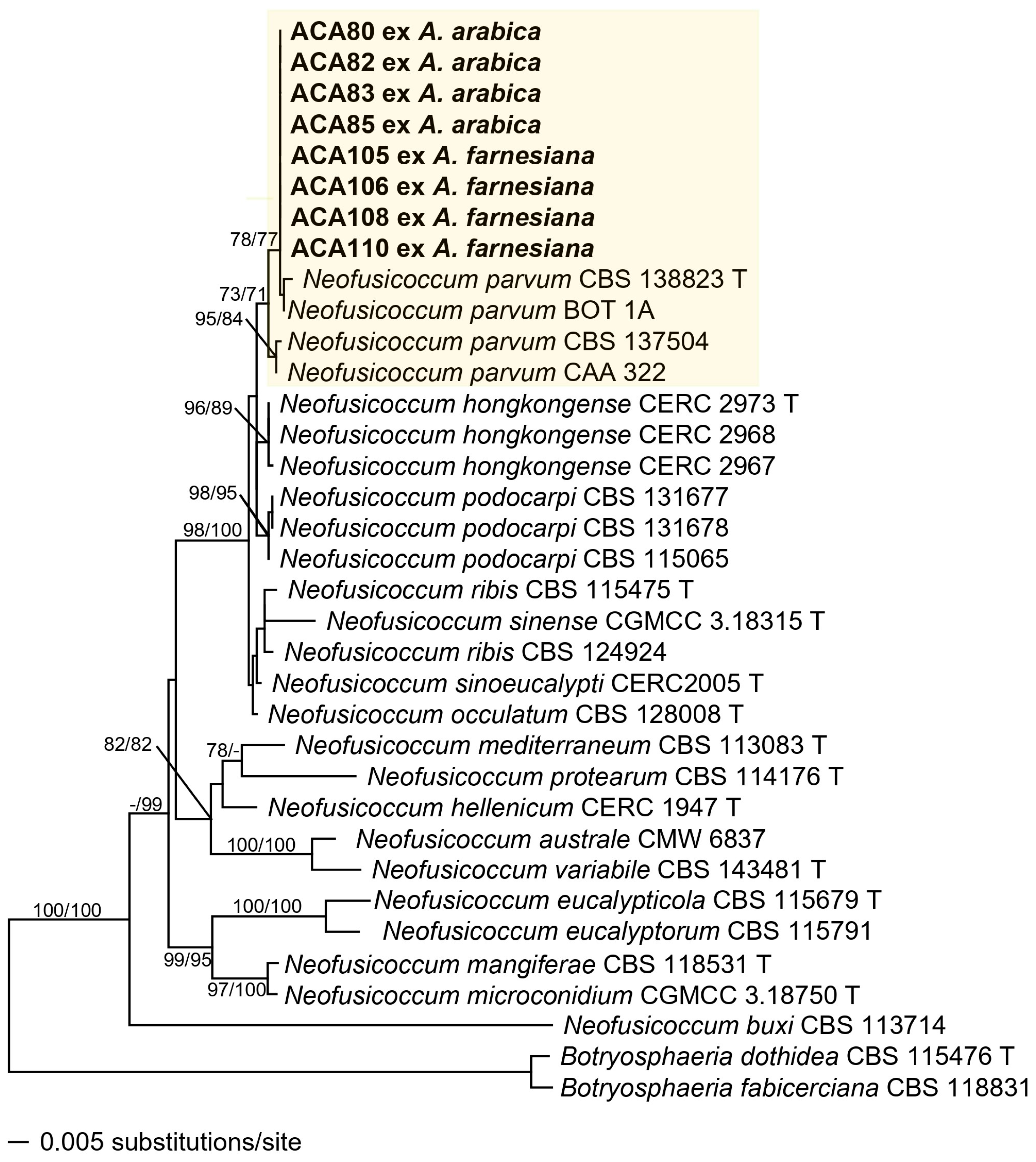
3.2. Phylogenetic Analysis
3.3. Pathogenicity Test
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yahara, T.; Javadi, F.; Onoda, Y.; de Queiroz, L.P.; Faith, D.P.; Prado, D.E.; Akasaka, M.; Kadoya, T.; Ishihama, F.; Davies, S.; et al. Global Legume Diversity Assessment: Concepts, Key Indicators, and Strategies. Taxon 2013, 62, 249–266. [Google Scholar] [CrossRef]
- Lewis, G.P.; Schrire, B.D.; Mackinder, B.A.; Rico, L.; Clark, R. A 2013 Linear Sequence of Legume Genera Set in a Phylogenetic Context—A Tool for Collections Management and Taxon Sampling. S. Afr. J. Bot. 2013, 89, 76–84. [Google Scholar] [CrossRef]
- Giuliani, C.; Giovanetti, M.; Foggi, B.; Mariotti Lippi, M. Two Alien Invasive Acacias in Italy: Differences and Similarities in Their Flowering and Insect Visitors. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2016, 150, 285–294. [Google Scholar] [CrossRef]
- Murugesan, S.; Lakshmanan, D.K.; Arumugam, V.; Alexander, R.A. Nutritional and Therapeutic Benefits of Medicinal Plant Pithecellobium Dulce (Fabaceae): A Review. J. Appl. Pharm. Sci. 2019, 9, 130–139. [Google Scholar] [CrossRef]
- Boa, E.; Lenn’e, J.M. Diseases of Nitrogen Fixing Trees in Developing Countries: An Annotated List; Natural Resources Institute, Overseas Development Administration: Chatham, UK, 1994; ISBN 0859543757. [Google Scholar]
- Old, K.M.; See, L.S.; Sharma, J.K.; Yuan, Z.Q. A Manual of Diseases of Tropical Acacias in Australia, South-East Asia, and India; Center for International Forestry Research: Jakarta, Indonesia, 2000; ISBN 9798764447. [Google Scholar]
- Luo, Y.; Niederholzer, F.; Camiletti, B.X.; Michailides, T.J. Survey on Latent Infection of Canker-Causing Pathogens in Budwood and Young Trees from Almond and Prune Nurseries in California. Plant Dis. 2024, 108, 550–557. [Google Scholar] [CrossRef]
- Tjahjono, B. Minimising Disease Incidence in Acacia in the Nursery. In Heart Rot and Root Rot in Tropical Plantations; Potter, K., Ed.; Australian Centre for International Agricultural Research: Canberra, Australia, 2006. [Google Scholar]
- Nasution, A.; Glen, M.; Beadle, C.; Mohammed, C. Ceratocystis Wilt and Canker—A Disease That Compromises the Growing of Commercial Acacia-Based Plantations in the Tropics. Aust. For. 2019, 82, 80–93. [Google Scholar] [CrossRef]
- Tarigan, M.; Roux, J.; Van Wyk, M.; Tjahjono, B.; Wingfield, M.J. A New Wilt and Dieback Disease of Acacia mangium Associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. S. Afr. J. Bot. 2011, 77, 292–304. [Google Scholar] [CrossRef]
- Wingfield, M.; Wingfield, B.; Warburton, P.; Japarudin, Y.; Lapammu, M.; Abdul Rauf, M.; Boden, D.; Barnes, I. Ceratocystis wilt of Acacia mangium in Sabah: Understanding the disease and reducing its impact. J. Trop. For. Sci. 2023, 35, 51–66. [Google Scholar] [CrossRef]
- Roux, J.; Wingfield, M. Ceratocystis Species: Emerging Pathogens of Non-Native Plantation Eucalyptus and Acacia Species. South. For. A J. For. Sci. 2009, 71, 115–120. [Google Scholar] [CrossRef]
- Syazwan, S.A.; Mohd-Farid, A.; Wan-Muhd-Azrul, W.-A.; Syahmi, H.M.; Zaki, A.M.; Ong, S.P.; Mohamed, R. Survey, Identification, and Pathogenicity of Ceratocystis fimbriata Complex Associated with Wilt Disease on Acacia mangium in Malaysia. Forests 2021, 12, 1782. [Google Scholar] [CrossRef]
- Polizzi, G.; Catara, V. First Report of Leaf Spot Caused by Cylindrocladium pauciramosum on Acacia retinodes, Arbutus unedo, Feijoa sellowiana, and Dodonaea viscosa in Southern Italy. Plant Dis. 2001, 85, 803. [Google Scholar] [CrossRef]
- Costanzo, M.B.; Gusella, G.; Fiorenza, A.; Leonardi, G.R.; Aiello, D.; Polizzi, G. Lasiodiplodia citricola, a New Causal Agent of Acacia Spp. Dieback. New Dis. Rep. 2022, 45, e12094. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A Genus of Endophytic, Saprobic and Plant Pathogenic Fungi. Persoonia—Mol. Phylogeny Evol. Fungi 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as Endophytes and Latent Pathogens of Woody Plants: Diversity, Ecology and Impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Michael, A., Innis, D.H., Gelfand, J.J., Sninsky, T.J., Eds.; White Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Alves, A.; Crous, P.W.; Correia, A.; Phillips, A.J.L.; Alves, A. Morphological and Molecular Data Reveal Cryptic Speciation in Lasiodiplodia theobromae. Fungal Divers. 2008, 28, 1–13. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Groenewald, J.Z.; Woodhall, J.; Armengol, J.; Cinelli, T.; Eichmeier, A.; Ezra, D.; Fontaine, F.; Gramaje, D.; Gutierrez-Aguirregabiria, A.; et al. Diaporthe Diversity and Pathogenicity Revealed from a Broad Survey of Grapevine Diseases in Europe. Persoonia—Mol. Phylogeny Evol. Fungi 2018, 40, 135–153. [Google Scholar] [CrossRef]
- Zhang, W.; Groenewald, J.Z.; Lombard, L.; Schumacher, R.K.; Phillips, A.J.L.; Crous, P.W. Evaluating Species in Botryosphaeriales. Persoonia—Mol. Phylogeny Evol. Fungi 2021, 46, 63–115. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Smith, S.A.; Dunn, C.V. Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 2008, 24, 715–716. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. RaxmlGUI: A Graphical Front-End for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0a165; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Izzi, C.F.; Acosta, A.; Carranza, M.L.; Ciaschetti, G.; Conti, F.; Martino, L.D.; D’orazio, G.; Frattaroli, A.; Pirone, G.; Stanisci, A. Il Censimento Della Flora Vascolare Degli Ambienti Dunali Costieri Dell’Italia Centrale. Fitosociologia 2007, 44, 129–137. [Google Scholar]
- Sidoti, A. Canker and Decline Caused by Neofusiccocum parvum on Acacia melanoxylon in Italy. For.—Riv. Di Selvic. Ed Ecol. For. 2016, 13, 41–46. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Kraus, C.; Markakis, E.; Alves, A.; Armengol, J.; Eichmeier, A.; Compant, S.; Gramaje, D. Fungal Trunk Diseases of Fruit Trees in Europe: Pathogens, Spread and Future Directions. Phytopathol. Mediterr. 2023, 61, 563–599. [Google Scholar] [CrossRef]
- Zlatković, M.; Keča, N.; Wingfield, M.J.; Jami, F.; Slippers, B. Botryosphaeriaceae Associated with the DieBack of Ornamental Trees in the Western Balkans. Antonie Van Leeuwenhoek 2016, 109, 543–564. [Google Scholar] [CrossRef]
- Heath, R.N.; Roux, J.; Slippers, B.; Drenth, A.; Pennycook, S.R.; Wingfield, B.D.; Wingfield, M.J. Occurrence and Pathogenicity of Neofusicoccum parvum and N. mangiferae on Ornamental Tibouchina Species. For. Pathol. 2011, 41, 48–51. [Google Scholar] [CrossRef]
- Sakalidis, M.L.; Slippers, B.; Wingfield, B.D.; Hardy, G.S.J.; Burgess, T.I. The Challenge of Understanding the Origin, Pathways and Extent of Fungal Invasions: Global Populations of the Neofusicoccum parvum–N. ribis Species Complex. Divers. Distrib. 2013, 19, 873–883. [Google Scholar] [CrossRef]
- Gusella, G.; Costanzo, M.B.; Aiello, D.; Polizzi, G. Characterization of Neofusicoccum parvum Causing Canker and Dieback on Brachychiton Species. Eur. J. Plant Pathol. 2021, 161, 999–1005. [Google Scholar] [CrossRef]
- Gusella, G.; Aiello, D.; Polizzi, G. First Report of Leaf and Twig Blight of Indian Hawthorn (Rhaphiolepis indica) Caused by Neofusicoccum parvum in Italy. J. Plant Pathol. 2020, 102, 275. [Google Scholar] [CrossRef]
- Gusella, G.; Di Pietro, C.; Leonardi, G.R.; Aiello, D.; Polizzi, G. Canker and Dieback of Camphor Tree (Cinnamomum camphora) Caused by Botryosphaeriaceae in Italy. J. Plant Pathol. 2023, 105, 1675–1681. [Google Scholar] [CrossRef]
- Aiello, D.; Gusella, G.; Fiorenza, A.; Guarnaccia, V.; Polizzi, G. Identification of Neofusicoccum parvum Causing Canker and Twig Blight on Ficus carica in Italy. Phytopathol. Mediterr. 2020, 59, 213–218. [Google Scholar] [CrossRef]
- Fiorenza, A.; Gusella, G.; Vecchio, L.; Aiello, D.; Polizzi, G. Diversity of Botryosphaeriaceae Species Associated with Canker and Dieback of Avocado (Persea americana) in Italy. Phytopathol. Mediterr. 2023, 62, 47–63. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Vitale, A.; Cirvilleri, G.; Aiello, D.; Susca, A.; Epifani, F.; Perrone, G.; Polizzi, G. Characterisation and Pathogenicity of Fungal Species Associated with Branch Cankers and Stem-End Rot of Avocado in Italy. Eur. J. Plant Pathol. 2016, 146, 963–976. [Google Scholar] [CrossRef]
- Gusella, G.; Leonardi, G.R.; La Quatra, G.; Aiello, D.; Voglmayr, H.; Polizzi, G. Re-Evaluating the Etiology of Citrus “Dothiorella Gummosis” in Italy. Plant Dis. 2025. [Google Scholar] [CrossRef] [PubMed]
- Fungal Database. Available online: https://Fungi.Ars.Usda.Gov (accessed on 10 November 2025).
- Annesi, T.; Luongo, L.; Vitale, S.; Galli, M.; Belisario, A. Characterization and Pathogenicity of Phomopsis theicola Anamorph of Diaporthe foeniculina Causing Stem and Shoot Cankers on Sweet Chestnut in Italy. J. Phytopathol. 2016, 164, 412–416. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Tziros, G.T.; Kavroulakis, N. First Report of Diaporthe foeniculina Associated with Branch Canker of Avocado in Greece. Plant Dis. 2020, 104, 3057. [Google Scholar] [CrossRef]
- Gusella, G.; Gugliuzzo, A.; Guarnaccia, V.; Martino, I.; Aiello, D.; Costanzo, M.B.; Russo, A.; Groenewald, J.Z.; Crous, P.W.; Polizzi, G. Fungal Species Causing Canker and Wilt of Ficus carica and Evidence of Their Association by Bark Beetles in Italy. Plant Dis. 2024, 108, 2136–2147. [Google Scholar] [CrossRef]
- León, M.; Berbegal, M.; Rodríguez-Reina, J.M.; Elena, G.; Abad-Campos, P.; Ramón-Albalat, A.; Olmo, D.; Vicent, A.; Luque, J.; Miarnau, X.; et al. Identification and Characterization of Diaporthe Spp. Associated with Twig Cankers and Shoot Blight of Almonds in Spain. Agronomy 2020, 10, 1062. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Emerging Citrus Diseases in Europe Caused by Species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef]
- Luo, Y.; Gu, S.; Felts, D.; Puckett, R.D.; Morgan, D.P.; Michailides, T.J. Development of QPCR Systems to Quantify Shoot Infections by Canker-Causing Pathogens in Stone Fruits and Nut Crops. J. Appl. Microbiol. 2017, 122, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Jaime, A.; Aroca, A.; Raposo, R.; García-Jiménez, J.; Armengol, J. Occurrence of Fungal Pathogens Associated with Grapevine Nurseries and the Decline of Young Vines in Spain. J. Phytopathol. 2006, 154, 598–602. [Google Scholar] [CrossRef]
- McCracken, A.R.; Berrie, A.; Barbara, D.J.; Locke, T.; Cooke, L.R.; Phelps, K.; Swinburne, T.R.; Brown, A.E.; Ellerker, B.; Langrell, S.R.H. Relative Significance of Nursery Infections and Orchard Inoculum in the Development and Spread of Apple Canker (Nectria galligena) in Young Orchards. Plant Pathol. 2003, 52, 553–566. [Google Scholar] [CrossRef]


| Species | Strain a | Host | Country | GenBank Accession Number b | ||
|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | ||||
| Diaporthe foeniculina | ACA 91 | Acacia arabica | Italy, Giarre | PX649194 | PX662033 | PX662010 |
| Diaporthe foeniculina | ACA 92 | Acacia arabica | Italy, Giarre | PX644738 | PX662034 | PX662011 |
| Diaporthe foeniculina | ACA 95 | Acacia arabica | Italy, Giarre | PX649195 | PX662035 | PX662012 |
| Diaporthe foeniculina | ACA 96 | Acacia arabica | Italy, Giarre | PX644739 | PX662036 | PX662013 |
| Diaporthe foeniculina | ACA 97 | Acacia arabica | Italy, Giarre | PX644740 | PX662037 | PX662014 |
| Diaporthe foeniculina | ACA 98 | Acacia arabica | Italy, Giarre | PX644741 | PX662038 | PX662015 |
| Diaporthe foeniculina | ACA 99 | Acacia farnesiana | Italy, Giarre | PX644742 | PX662039 | PX662016 |
| Diaporthe foeniculina | ACA 100 | Acacia farnesiana | Italy, Giarre | PX644743 | PX662040 | PX662017 |
| Diaporthe foeniculina | ACA 101 | Acacia farnesiana | Italy, Giarre | PX649196 | PX662041 | PX662018 |
| Diaporthe foeniculina | ACA 102 | Acacia farnesiana | Italy, Giarre | PX649197 | PX662042 | PX662019 |
| Diaporthe foeniculina | ACA 103 | Acacia farnesiana | Italy, Giarre | PX649198 | PX662043 | PX662020 |
| Diaporthe foeniculina | ACA 104 | Acacia farnesiana | Italy, Giarre | PX644744 | PX662044 | PX662021 |
| Diaporthe foeniculina | ACA 111 | Pithecellobium dulce | Italy, Giarre | PX644745 | PX662045 | PX662022 |
| Diaporthe foeniculina | ACA 112 | Pithecellobium dulce | Italy, Giarre | PX644746 | PX662046 | PX662023 |
| Diaporthe foeniculina | ACA 113 | Pithecellobium dulce | Italy, Giarre | PX644747 | PX662047 | PX662024 |
| Diaporthe foeniculina | ACA 116 | Pithecellobium dulce | Italy, Giarre | PX644748 | PX662048 | PX662025 |
| Diaporthe foeniculina | ACA 117 | Pithecellobium dulce | Italy, Giarre | PX649199 | PX662049 | PX662026 |
| Diaporthe foeniculina | ACA 118 | Pithecellobium dulce | Italy, Giarre | PX649200 | PX662050 | PX662027 |
| Diaporthe foeniculina | ACA 119 | Pithecellobium dulce | Italy, Giarre | PX644749 | PX662051 | PX662028 |
| Diaporthe foeniculina | ACA 122 | Pithecellobium dulce | Italy, Giarre | PX644750 | PX662052 | PX662029 |
| Diaporthe foeniculina | ACA 123 | Pithecellobium dulce | Italy, Giarre | PX649201 | PX662053 | PX662030 |
| Diaporthe foeniculina | ACA 124 | Pithecellobium dulce | Italy, Giarre | PX644751 | PX662054 | PX662031 |
| Diaporthe foeniculina | ACA 126 | Pithecellobium dulce | Italy, Giarre | PX644752 | PX662055 | PX662032 |
| Neofusicoccum parvum | ACA 80 | Acacia arabica | Italy, Giarre | PX651383 | PX662056 | PX662064 |
| Neofusicoccum parvum | ACA 82 | Acacia arabica | Italy, Giarre | PX651384 | PX662057 | PX662065 |
| Neofusicoccum parvum | ACA 83 | Acacia arabica | Italy, Giarre | PX651385 | PX662058 | PX662066 |
| Neofusicoccum parvum | ACA 85 | Acacia farnesiana | Italy, Giarre | PX651386 | PX662059 | PX662067 |
| Neofusicoccum parvum | ACA 105 | Pithecellobium dulce | Italy, Giarre | PX651387 | PX662060 | PX662068 |
| Neofusicoccum parvum | ACA 106 | Pithecellobium dulce | Italy, Giarre | PX651388 | PX662061 | PX662069 |
| Neofusicoccum parvum | ACA 108 | Pithecellobium dulce | Italy, Giarre | PX651389 | PX662062 | PX662070 |
| Neofusicoccum parvum | ACA 110 | Pithecellobium dulce | Italy, Giarre | PX651390 | PX662063 | PX662071 |
| Species | Strain a | Host | Country | GenBank Accession Number b | ||
|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | ||||
| Botryosphaeria dothidea | CBS 115476 T | Prunus sp. | Switzerland | AY236949 | AY236898 | AY236927 |
| Botryosphaeria fabicerciana | CBS 118831 | Syzygium cordatum | South Africa | DQ316084 | MT592032 | MT592468 |
| Diaporthe acericola | MFLUCC 17-0956 | Acer negundo | Italy | KY964224 | KY964180 | KY964074 |
| Diaporthe ampelina | CBS 114016 | Vitis vinifera | France | AF230751 | GQ250351 | JX275452 |
| Diaporthe amygdali | CBS 126679 | Prunus dulcis | Portugal | KC343022 | KC343748 | KC343990 |
| Diaporthe australafricana | CBS 111886 | Vitis vinifera | Australia | KC343038 | KC343764 | KC344006 |
| Diaporthe brasiliensis | CBS 133183 T | Aspidosperma tomentosus | Brazil | KC343042 | KC343768 | KC344010 |
| Diaporthe caatingaensis | CBS 141542 T | Tacinga inamoena | Brazil | KY085926 | KY115603 | KY115600 |
| Diaporthe chamaeropis | CBS 454.81 | Chamaerops humilis | Greece | KC343048 | KC343774 | KC344016 |
| Diaporthe canthii | CBS 132533 | Canthium inerme | South Africa | JX069864 | KC843120 | KC843230 |
| Diaporthe citri | CBS 135422 | Citrus sp. | USA | KC843311 | KC843071 | KC843187 |
| Diaporthe cytosporella | CBS 137020 | Citrus limon | Spain | KC843307 | KC843116 | KC843221 |
| Diaporthe eres | CBS 116953 | Pyrus pyrifolia | New Zealand | KC343147 | KC343873 | KC344115 |
| Diaporthe foeniculina | CBS 111553 T | Foeniculum vulgare | Portugal | KC343101 | KC343827 | KC344069 |
| Diaporthe foeniculina | CBS 129528 | Rhus pendulina | South Africa | JF951146 | KC843100 | KC843205 |
| Diaporthe foeniculina | CBS 123208 | Foeniculum vulgare | Portugal | EU814480 | GQ250315 | JX275464 |
| Diaporthe foeniculina | CBS 136972 | Vaccinium corymbosum | Italy | KJ160565 | KJ160597 | MF418509 |
| Diaporthe foeniculina | AR5151 | Citrus latifolia | USA | KC843303 | KC843112 | KC843217 |
| Diaporthe forlicesenica | MFLUCC 17-1015 | Dorycnium hirsutum | Italy | KY964215 | KY964171 | KY964099 |
| Diaporthe hongkongensis | CBS 115448 T | Dichroa febrifuga | China | KC343119 | KC343845 | KC344087 |
| Diaporthe heterophyllae | CBS 143769 T | Acacia heterophylla | France | MG600222 | MG600224 | MG600226 |
| Diaporthe inconspicua | CBS 133813 T | Maytenus ilicifolia | Brazil | KC343123 | KC343849 | KC344091 |
| Diaporthe limonicola | CBS 142549 T | Citrus limon | Malta | MF418422 | MF418501 | MF418582 |
| Diaporthe longicolla | ATCC 60325 | Glycine max | USA | KJ590728 | KJ590767 | KJ610883 |
| Diaporthe melitensis | CBS 142551 T | Citrus limon | Malta | MF418424 | MF418503 | MF418584 |
| Diaporthe notophagi | BRIP54801 T | Notophagus cunninghamii | Australia | JX862530 | JX862536 | KF170922 |
| Diaporthe novem | CBS 127271 T | Glycine max | Croatia | KC343156 | KC343882 | KC344124 |
| Diaporthe parvae | PSCG034 | Pyrus bretschneideri | China | MK626919 | MK654858 | MK691248 |
| Diaporthe phoenicicola | CBS 161.64 T | Areca catechu | India | KC343032 | KC343758 | KC344000 |
| Diaporthe pterocarpi | CBS 137021 | Pterocarpus indicus | Thailand | JQ619901 | JX275418 | JX275462 |
| Diaporthe pterocarpicola | CBS 135432 T | Pterocarpus indicus | Thailand | JQ619887 | JX275403 | JX275441 |
| Diaporthe rudis | CBS 113201 | Vits vinifera | Portugal | KC343234 | KC343960 | KC344202 |
| Diaporthe sojae | FAU 635 | Glycine max | USA | KJ590719 | KJ590762 | KJ610875 |
| Diaporthe saccarata | CBS 116311 | Protea repens | South Africa | KC343190 | KC343916 | KC344158 |
| Diaporthe thunbergiae | CBS 135769 T | Thunbergia laurifolia | Thailand | JQ619893 | JX275409 | JX275449 |
| Diaporthella corylina | CBS 12114 T | Corylus sp. | China | KC343004 | KC343730 | KC343972 |
| Neofusicoccum buxi | CBS 113714 | Buxus sempervirens | Sweden | KX464164 | KX464677 | KX464954 |
| Neofusicoccum australe | CMW 6837 | Acacia sp. | Australia | AY339262 | AY339270 | AY339254 |
| Neofusicoccum eucalypticola | CBS 115679 T | Eucalyptus grandis | Australia | AY615141 | AY615133 | AY615125 |
| Neofusicoccum eucalyptorum | CBS 115791 | Eucalyptus grandis | South Africa | AF283686 | AY236891 | AY236920 |
| Neofusicoccum hellenicum | CERC 1947 T | Pistacia vera | Greece | KP217053 | KP217061 | KP217069 |
| Neofusicoccum hongkongense | CERC 2973 T | Araucaria cunninghamii | China | KX278052 | KX278157 | KX278261 |
| Neofusicoccum hongkongense | CERC 2967 | Araucaria cunninghamii | China | KX278050 | KX278155 | KX278259 |
| Neofusicoccum hongkongense | CERC 2968 | Araucaria cunninghamii | China | KX278051 | KX278156 | KX278260 |
| Neofusicoccum mangiferae | CBS 118531 T | Mangifera indica | Australia | AY615185 | DQ093221 | AY615172 |
| Neofusicoccum mediterraneum | CBS 113083 T | Pistacia vera | U.S.A. | KX464186 | KX464712 | KX464998 |
| Neofusicoccum microconidium | CGMCC 3.18750 T | Eucalyptus urophylla × E. grandis | China | KX278053 | KX278158 | KX278262 |
| Neofusicoccum occulatum | CBS 128008 T | Eucalyptus grandis | Australia | EU301030 | EU339509 | EU339472 |
| Neofusicoccum parvum | CBS 138823 T | Populus nigra | New Zealand | AY236943 | AY236888 | AY236917 |
| Neofusicoccum parvum | CBS 137504 | Vitis vinifera | Algeria | KJ657702 | KJ657715 | KX505915 |
| Neofusicoccum parvum | CBS 140887 | Vitis vinifera cv. Alfrocheiro | Portugal | MT587448 | MT592158 | MT592648 |
| Neofusicoccum parvum | CBS 139672 | Bruguiera gymnorhiza | South Africa | MT587446 | MT592156 | MT592646 |
| Neofusicoccum parvum | BOT 1A | Citrus limon | Italy | MW727244 | MW789904 | MW789889 |
| Neofusicoccum parvum | CAA 322 | Malus domestica | Portugal | KX505906 | KX505894 | KX505916 |
| Neofusicoccum parvum | CPC 32275 | Pomaderris aspera | Australia | MT587451 | MT592160 | MT592651 |
| Neofusicoccum parvum | CPC 35861 | Aloe sp. | South Africa | MT587449 | MT592159 | MT592649 |
| Neofusicoccum parvum | CBS 139671 | Bruguiera gymnorhiza | South Africa | MT587445 | MT592155 | MT592645 |
| Neofusicoccum podocarpi | CBS 131677 | Podocarpus henkelii | South Africa | MT587508 | MT592223 | MT592715 |
| Neofusicoccum podocarpi | CBS 115065 | Wollemia nobilis | Australia | MT587507 | MT592222 | MT592714 |
| Neofusicoccum podocarpi | CBS 131678 | Podocarpus henkelii | South Africa | MT587509 | MT592224 | MT592716 |
| Neofusicoccum protearum | CBS 114176 T | Leucadendron salignum | South Africa | AF452539 | KX464720 | KX465006 |
| Neofusicoccum ribis | CBS 115475 T | Ribes sp. | USA | AY236935 | AY236877 | AY236906 |
| Neofusicoccum ribis | CBS 124924 | Terminalia catappa | Cameroon | FJ900607 | FJ900653 | FJ900634 |
| Neofusicoccum sinense | CGMCC 3.18315 T | Unknown woody plant | China | KY350148 | KY817755 | KY350154 |
| Neofusicoccum sinoeucalypti | CERC2005 T | Eucalyptus urophylla × E. grandis | China | KX278061 | KX278166 | KX278270 |
| Neofusicoccum variabile | CBS 143481 T | Mimusops caffra | South Africa | MH558610 | MH576586 | MH569155 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonardi, G.R.; Vecchio, L.; Gusella, G.; Aiello, D.; Voglmayr, H.; Polizzi, G. New Records of Canker-Causing Pathogens of Acacia spp. and Pithecellobium dulce in Southern Italy. J. Fungi 2025, 11, 874. https://doi.org/10.3390/jof11120874
Leonardi GR, Vecchio L, Gusella G, Aiello D, Voglmayr H, Polizzi G. New Records of Canker-Causing Pathogens of Acacia spp. and Pithecellobium dulce in Southern Italy. Journal of Fungi. 2025; 11(12):874. https://doi.org/10.3390/jof11120874
Chicago/Turabian StyleLeonardi, Giuseppa Rosaria, Laura Vecchio, Giorgio Gusella, Dalia Aiello, Hermann Voglmayr, and Giancarlo Polizzi. 2025. "New Records of Canker-Causing Pathogens of Acacia spp. and Pithecellobium dulce in Southern Italy" Journal of Fungi 11, no. 12: 874. https://doi.org/10.3390/jof11120874
APA StyleLeonardi, G. R., Vecchio, L., Gusella, G., Aiello, D., Voglmayr, H., & Polizzi, G. (2025). New Records of Canker-Causing Pathogens of Acacia spp. and Pithecellobium dulce in Southern Italy. Journal of Fungi, 11(12), 874. https://doi.org/10.3390/jof11120874






