Abstract
Four unprecedented polyketides named isoprenylisobenzofuran B (2), isoprenylisobenzofuran C1/C2 (3), diaporisoindole F1/F2 (4), and isochromophilonol A1/A2 (7) were isolated from ethyl acetate extracts of the newly described endophytic fungus Diaporthe africana. Additionally, the previously reported cyclic depsipeptide eucalactam B (1) was also identified, along with the known compounds diaporisoindole A/B (5), tenellone B (6) and beauvericin (8). The taxonomic identification of the fungus was accomplished using a polyphasic approach combining multi-gene phylogenetic analysis and microscopic morphological characters. The structures 1–8 were determined by a detailed analysis of their spectral data, namely high-resolution electrospray ionization mass spectrometry (HR-ESIMS), 1D/2D nuclear magnetic resonance (NMR) spectroscopy, as well as electronic circular dichroism (ECD) spectra. In addition, chemical methods such as Marfey’s analysis were also employed to determine the stereochemistry in compound 1. All the compounds obtained were evaluated for antimicrobial and in vitro cytotoxic properties. Compounds 3–8 were active against certain fungi and Gram-positive bacteria with MIC values of 8.3 to 66.6 µg/mL. In addition, 3–5 displayed cytotoxic effects (22.0 ≤ IC50 ≤ 59.2 µM) against KB3.1 and L929 cell lines, whereas compounds 6–8 inhibited the growth of seven mammalian cancer cell lines with IC50 ranging from 17.7 to 49.5 µM (6), 0.9 to 12.9 µM (7) and 1.9 to 4.3 µM (8).
1. Introduction
Fungi belonging to the genus Diaporthe (including its anamorph Phomopsis) have proven to be prolific sources of bioactive secondary metabolites with potential applications in pharmaceutical and agricultural fields [1,2]. In recent decades, more than 335 bioactive natural products have been discovered from Diaporthe spp. [1]. Polyketides plausibly constitute the largest and most structurally diverse class of secondary metabolites reported in this genus and account for ~64% of the hitherto isolated compounds in this fungal group [1]. These polyketides mainly include compounds belonging to the family of macrolides, azaphilones, benzofuranones, quinones, chromones, chromanones, xanthones, pyrones, phenols, and oblongolides, as classified by Xu et al. and Chepkirui et al. [1,2]. These substances have diverse biological properties, and the main bioactivities reported involve cytotoxic, antibacterial, and antifungal effects [1]. Given the emergence of multidrug resistance in many pathogens, drug-resistant cancer cells, and the emergence of life-threatening viral diseases, there is an urgent demand for the discovery of potential new pharmacological drugs [3]. Attributed to the widespread chemical and biological diversity of its secondary metabolites, the extensive exploitation of this fungal genus remains opportune for discovering new lead compounds that could possibly be developed as novel pharmaceutical drugs and/or biocontrol agents [4,5]. For instance, the compound emodin, when isolated from the endophytic fungus Diaporthe lithocarpi, has demonstrated remarkable cytotoxic activity against P-388 murine leukemia cells [6]. Additionally, isochromophilones A, G, and 5-chloroisorotiorin, derived from Diaporthe perseae, have shown potent antibacterial effects against various human pathogens, including the Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa [7]. Noteworthy are pestalotiopsones B and F produced by Diaporthe sp. SCSIO 41011, which have displayed significant antiviral activity against three subtypes of the influenza A virus [8]. These striking examples not only emphasize the potential of utilizing these compounds as valuable precursors for developing new derivatives with enhanced potency but also shed light on the vast potential of Diaporthe-derived secondary metabolites in the realm of drug discovery.
As part of our search for new bioactive secondary metabolites from selected endophytic fungal strains inhabiting Cameroonian medicinal plants, numerous metabolites have been isolated from the culture of Diaporthe breyniae [9], some of which displayed antimicrobial and cytotoxic effects. In our continuous endeavors, a new species of Diaporthe was recently discovered, and the investigation of its ethyl acetate (EtOAc) extracts showed moderate antimicrobial activities as well as interesting HPLC-DAD/MS (high-performance liquid chromatography coupled with diode array detection and mass spectrometry) chromatograms, possibly containing unknown compounds. These results prompted an in-depth investigation of this fungus, and we herein report upon the taxonomic identification as well as the isolation, structural elucidation, and biological properties of its secondary metabolites.
2. Materials and Methods
2.1. General Experimental Procedures
Optical rotations were measured in methanol (Uvasol, Merck, Darmstadt, Germany) using an Anton Paar MCP-150 polarimeter (sodium D line, a Nickel alloy sample cell 100 mm/3 mm, 0.7 mL volume, Seelze, Germany) at 20 °C. UV-Vis spectra were acquired using methanol (Uvasol, Merck, Darmstadt, Germany) with a Shimadzu UV-Vis 2450 spectrophotometer (Kyoto, Japan). ECD spectra were recorded on a J-815 spectropolarimeter (JASCO, Pfungstadt, Germany) using a 0.5 mm quartz cuvette and MeOH as the solvent. Nuclear magnetic resonance (NMR) spectra were recorded at a temperature of 298 K with an Avance III 500 spectrometer (Bruker, Billerica, MA, USA, 1H-NMR: 500 MHz and 13C-NMR: 125 MHz) and an Ascend 700 spectrometer equipped with 5 mm TCI cryoprobe (Bruker, Billerica, MA, USA, 1H-NMR: 700 MHz and 13C-NMR: 175 MHz). Chemical shifts were given in parts per million (ppm) and coupling constants in hertz (Hz). NMR spectra were referenced to residual solvent signals with resonances at δH/C 2.50/39.5 for DMSO and at δH/C 7.26/77.2 for CDCl3. Electrospray ionization mass spectra (ESI-MS) were acquired with an UltiMate 3000 Series uHPLC (Thermo Fischer Scientific, Waltman, MA, USA) equipped with a C18 Acquity UPLC BEH column (2.1 × 50 mm, 1.7 µm; Waters, Milford, CT, USA) connected to an amaZon speed ESI-Iontrap-MS (Bruker, Billerica, MA, USA). The HPLC parameters were set as follows: solvent A: MilliQ H2O + 0.1% formic acid, solvent B: acetonitrile (ACN) + 0.1% formic acid, gradient: 5% B for 0.5 min increasing to 100% B in 19.5 min, then an isocratic condition at 100% B for 5 min with a flow rate of 0.6 mL/min, and Diode-Array Detection (DAD) at 210 nm and 190–600 nm. High-resolution electrospray ionization mass spectra (HR-ESIMS) were recorded with an Agilent 1200 Infinity Series HPLC–UV system (Agilent Technologies, Santa Clara, CA, USA) connected to a MaXis ESI-TOF mass spectrometer (scan range 100–2500 m/z, capillary voltage 4500 V, dry temperature 200 °C). The column and HPLC parameters remained consistent with those used for ESI-MS, and the DAD was set in the range of 200–640 nm.
2.2. Fungal Isolation and Identification
Two strains of endophytic fungi were isolated from the bark of the terrestrial plants Asystasia macrophylla and Polyscias fulva, which were collected in Kala Mountain (Center region in Cameroon) and Tonga (West Cameroon), respectively, following the protocol previously described [9].
Hyphal material (1 mm diam) was scratched of actively growing cultures on a YM 6.3 agar (malt extract 10 g/L, yeast extract 4 g/L, D-glucose 4 g/L, agar 20 g/L, pH 6.3 before autoclaving) and was transferred onto a 9-cm-diam Petri dished containing 2% tap water agar supplemented with sterile pine needles (PNA) [10], potato dextrose agar (PDA), oatmeal agar (OA) and malt extract agar (MEA) [11]. The plates were incubated at 21 °C in darkness. Pigment production, colony diameters, and colony characters on PDA, OA, and MEA were noted after 15 d. Colony colors were described using the color chart of the Royal Horticultural Society London (1966) [12]. An examination of morphological characters was conducted by mounting fungal structures in clear lactic acid, and 30 measurements of each structure were conducted. Photomicrographs were taken using an eclipse Ni compound microscope, using a DS-Fi3 (Nikon, Tokyo, Japan) and NIS-Elements imaging software v. 5.20, and a Keyence VHX-970F microscope (Neu-Isenburg, Germany).
The sequences of five loci in total (ITS, cal, his3, tef1, tub2; see [9] for details) were generated and subsequently checked for their phylogenetic placement first in a broad phylogenetic study following the maximum Likelihood criterion implemented in IQTree V2.1.3 [13], assessing statistical support by bootstrapping [14]. In total, 370 taxa were used (retrieved from GenBank, Supplementary Material Table S5) for the gblocks [15] curated MAFFT [16] (as implemented in Geneious 7.1.9) alignment using distinct nucleotide substitution models [17,18]. After the evaluation of the phylogenetic relationship of strain CBS 150080, the data matrix was restricted to a well-supported clade, including 66 Diaporthe spp. (Table 1). The original full-length sequences were aligned using MAFFT, and misalignments were corrected manually. Trees were inferred from both the Maximum-Likelihood criterion using IQTree and Bayesian methodology (parallelized version of MrBayes 3.2.7a [19,20]; nucleotide model testing with PartitionFinder2 as implemented in Phylosuite V1.2.2 [21,22]), checked for congruence and support values over 70% (bootstrap, bs) and 0.95 (posterior probability, pp) mapped onto the ML tree. The sequences generated in this study were deposited in GenBank (Table 1). The alignments used in the phylogenetic analysis are included in the Supplementary Material.

Table 1.
Isolates and reference strains of Diaporthe spp. included in the phylogenetic study. GenBank accession numbers in bold were newly generated in this study. Taxonomic novelty is indicated in bold italic.
The herbarium-type material and the ex-type strain of the new species were maintained at the collection of the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands.
2.3. Fermentation and Extraction
Diaporthe africana was cultivated in both, liquid and solid media. The liquid fermentation was conducted in a yeast malt glucose medium (YMG, 10 g malt extract, 4 g yeast extract, 4 g D-glucose in 1 L of deionized water with pH adjusted to 6.3 before sterilization). In detail, five plugs (obtained with a 7 mm cork borer) of the mycelial culture grown on YMG agar (composition as reported above, with 20 g/L of agar and without adjusting the pH) plates were used to inoculate 200 mL of liquid YMG medium contained in 5 × 500 mL Erlenmeyer flasks. The flasks were incubated at 23 °C under shaking conditions at 140 rpm on a rotary shaker. The growth of the fungus was monitored by checking the amount of free glucose daily using Medi-Test glucose strips (Macherey-Nagel, Düren, Germany). On the 6th day, the glucose was completely depleted, and fermentation was aborted 3 days afterward. The fungal mycelia were harvested by filtration, and the resulting supernatant was extracted three times with an equal amount of EtOAc using a separating funnel. After filtration over anhydrous sodium sulfate (Na2SO4), the ethyl acetate solution was evaporated to dryness in vacuo (40 °C) to afford 28 mg of the extract. The wet mycelia or biomass was extracted thrice with acetone in an ultrasonic bath (Sonorex Digital 10 P, Bandelin Electronic GmbH and Co. KG, Berlin, Germany) at 40 °C for 30 min. The obtained solvent was concentrated in vacuo using a rotary evaporator to yield an aqueous phase, which was extracted three times with the same amount of ethyl acetate. The ethyl acetate fraction was then filtered over anhydrous Na2SO4 and was evaporated to dryness to afford 198 mg of the mycelial extract.
The fungus was also cultivated in 10 × 500 mL flasks containing a solid autoclaved rice-based medium (BRFT: brown rice 28 g with 0.1 L base liquid (yeast extract 1 g/L, sodium tartrate 0.5 g/L, KH2PO4 0.5 g/L) per flask) [55]. Each flask was inoculated with 10 mL of the seed culture, which was prepared beforehand. To obtain the seed culture, three small pieces of mycelial culture grown on a YMG agar were used to inoculate a 250 mL Erlenmeyer flask containing 100 mL of the liquid YMG medium. The seed culture was incubated for 5 days at 23 °C under shaking conditions at 140 rpm, and was subsequently used to inoculate the flasks containing the solid rice medium. The flasks were incubated under static conditions at 23 °C for 28 days. The fermented cultures were extracted following the same previously described methodology as for the mycelia obtained from the liquid culture in YMG, to afford 3 g of orange-colored extract.
2.4. Isolation of Compounds 1–8
The EtOAc extracts obtained from liquid and solid fermentation were fractionated separately, as they contained different secondary metabolites. Following analytical HPLC-DAD profiling, the supernatant extract (28 mg) dissolved in methanol was purified using the preparative reverse phase HPLC (Büchi, Pure C-850, 2020, Flawil, Switzerland) with the following elution gradient: 20−45% solvent B in 20 min, 45−65% B in 30 min, 65−100% B in 10 min and thereafter isocratic conditions at 100% B for 10 min. A VP 250/21 Nucleodur C18 Htec 10 µm column (Macherey-Nagel) was used as a stationary phase with a flow rate of 15 mL/min. The mobile phase was constituted of two solvents, A (MilliQ H2O + 0.1% formic acid (FA)) and B (acetonitrile (ACN) + 0.1% FA). UV detection was set up to 210 and 350 nm. Four fractions (F1−F4) were collected according to the observed peaks, and their purity was examined using HPLC-DAD-MS. This resulted in the acquisition of two compounds: 3 (1.2 mg, tR = 40 min) and 6 (3.2 mg, tR = 57.9 min) from F2 to F4, respectively. The mycelial extract (198 mg) obtained from the liquid fermentation was discarded as it did not contain any metabolites of interest.
A portion of the extract (1 g) obtained from the solid rice medium was dissolved in a sufficient amount of methanol (100 mL) and was filtered using an SPME Strata™-X 33 u Polymeric RP cartridge (Phenomenex, Inc., Aschaffenburg, Germany). The resulting solution was evaporated in vacuo and re-dissolved in an adequate amount of methanol prior to purification using preparative RP-HPLC (VP 250/40 Nucleodur C18 Htec 10 µm column, flow rate of 30 mL/min, elution gradient from 5 to 80% solvent B in 70 min, 80−100% B in 20 min, and finally isocratic conditions at 100% B for 5 min). Three runs were performed on the RP-HPLC, and the obtained fractions (F1−F10) were combined according to UV absorption at 210 and 350 nm and concurrent HPLC-MS analysis. The fractionation yielded three compounds: 4 (1.5 mg, tR = 36.2 min), 5 (1.1 mg, tR = 45.4 min), and 8 (1.6 mg, tR = 86.5 min). F3 (11 mg), F8 (28 mg), and F9 (20 mg) were further purified using preparative RP-HPLC with an XBridgeTM Trifunctional C18, 250 × 19 mm, 135 Å, 5 μm column (Waters, Eschborn, Germany) at a flow rate of 15 mL/min. F3 (elution gradient: 5–10% solvent B in 10 min, then in an isocratic condition at 50% B for 30 min and finally 50–100% B in 10 min) yielded compound 1 (9.7 mg, tR = 16 min), whereas F8 (elution gradient: 5–60% solvent B in 10 min, then in an isocratic condition at 60% B for 30 min and finally 60–100% B in 5 min) and F9 (elution gradient: 5−65% solvent B in 10 min, followed by an isocratic condition at 65% B for 30 min and finally 65–100% B in 10 min) afforded compounds 2 (11.9 mg, tR = 17.6 min) and 7 (10 mg, tR = 27.5 min), respectively. A flow chart of the purification procedure can be found in the Supplementary Materials (Figure S76).
2.5. Determination of Amino Acid Stereochemistry
The stereochemistry of threonine in compound 1 was determined using Marfey’s analysis following the protocol described by Harms et al. [56] with slight modification. In detail, compound 1 (1 mg) was hydrolyzed in 6 N HCl (1 mL) at 90 °C for 18 h. The hydrolysate was evaporated to dryness using a speedVac vacuum connected to a chemistry hybrid pump (Wertheim, Germany), which was then dissolved in 400 μL Milli-Q H2O and divided into two individual vials. 1 M NaHCO3 (20 μL) and 1% N-(2,4-dinitro-5-fluorophenyl)-L-valinamide (L-FDVA, 100 μL in acetone) were added into one vial, and the other vial was supplemented with the same amount of 1 M NaHCO3 and 1% D-FDVA. Concurrently, 2 mg of the authentic amino acids: L-threonine, D/L-threonine, and L-allo-threonine (Sigma-Aldrich, Deisenhofen, Germany) were used as standards and were treated similarly to the hydrolysate of 1. The mixtures were incubated at 40 °C for 40 min. After cooling to room temperature, the solutions were neutralized with 2 N HCl (20 μL) and evaporated to dryness using the speedVac vacuum. Afterward, the residues were dissolved in 1 mL MeOH and analyzed using uHPLC connected to an amaZon speed ESI-Iontrap mass spectrometer (column and conditions were described in General Experimental Procedures). The stereochemistry of threonine in 1 was finally deduced as L-Thr by comparison with the retention times of the L-and D-FDVA-derivatized hydrolysate of 1 (L-Thr-L-FDVA tR 6.17 min; L-Thr-D-FDVA tR 7.30 min) with that of Marfey’s derivatized authentic amino acids (Tables S3 and S4 and Figures S72 and S73).
2.6. Antimicrobial and Cytotoxic Activities
The Minimum Inhibitory Concentration (MIC) of the isolated metabolites (1–8) was evaluated using serial dilution assays in a 96-well microtiter plate against a panel of Gram-positive bacteria, namely Bacillus subtilis DSM 10, Mycobacterium smegmatis ATCC 700084 and Staphylococcus aureus DSM 346; Gram-negative bacteria, including Acinetobacter baumannii DSM 30008, Chromobacterium violaceum DSM 30191, Escherichia coli DSM 1116 and Pseudomonas aeruginosa PA14 and fungal cultures of Candida albicans DSM 1665, Mucor hiemalis DSM 2656, Rhodotorula glutinis DSM 10134, Schizosaccharomyces pombe DSM 70572 and Wickerhamomyces anomalus DSM 6766, according to previously described protocols [57]. The MTT-based cytotoxicity assay was performed against several cancer cell lines (human endocervical adenocarcinoma KB 3.1, mouse fibroblasts L929, squamous cancer A431, breast cancer MCF-7, lung cancer A549, ovary cancer SK-OV-3, and prostate cancer PC-3) in accordance with our previously established protocols [57,58].
3. Results and Discussion
3.1. Molecular Phylogeny and Taxonomy
The generated sequences were checked for their phylogenetic placement first in a broad phylogenetic inference (see Supplementary Material for more details). Subsequently, we restricted the phylogenetic inference to a well-supported clade comprising 66 taxa for a more detailed study to save computing time. This new five-loci dataset was manually curated and yielded a data matrix that was used for the phylogenetic study comprising in total of 2449 (ITS: 554; cal: 423; his3: 359; tef1: 392; tub2: 721) sites. Both trees inferred by the Maximum-Likelihood criterion (l Ln = −14,653.6590) using IQTree and Bayesian methodology revealed a congruent tree (Figure 1). Here, the characterized strains formed a well-supported clade (100% bs/0.98 pp), and clustered with D. salinicola (78. bs/1 pp). The position of the newly formed clade consisting of D. salinicola and D. africana was not resolved. This was in agreement with single locus trees that were calculated for comparison.
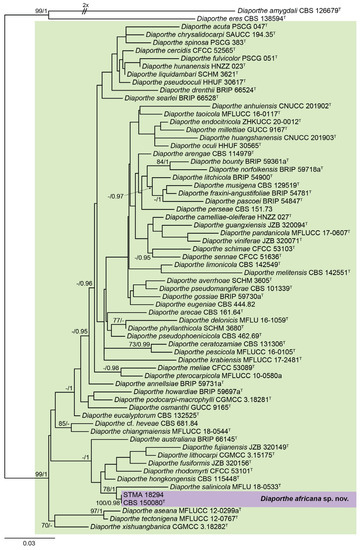
Figure 1.
ML phylogram obtained from a combined ITS, cal, his3, tef1, and tub2 dataset, including sequences of our strain and a selection of related Diaporthe spp., restricted according to the phylogenetic study of Supplementary Material (Figure S70). Diaporthe amygdali CBS 126679T and D. eres CBS 138594T were used as outgroup. Bootstrap support values ≥ 70/Bayesian posterior probability scores ≥ 0.95 are indicated along the branches. Branch lengths are proportional to the distance. The new taxon is indicated in bold. Sequences derived from the type material of the different species are indicated with T.
Diaporthe africana Y. Marín & C. Lamb., sp. nov. MycoBank MB849047. (Figure 2A–H).
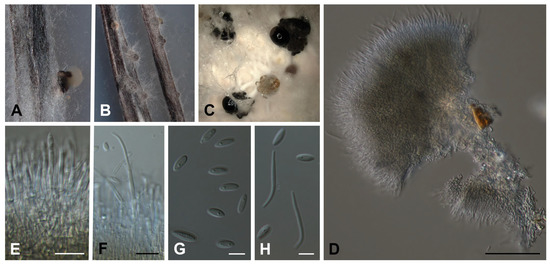
Figure 2.
Diaporthe africana CBS 150080T. (A,B) Conidiomata in PNA. (C) Conidiomata in OA. (D–F) Conidiophores, conidiogenous cells and conidia. (G) Alpha conidia. (H) Alpha and beta conidia. Scale bars: (D) = 50 µm; (E,F) = 10 µm; (G,H) = 5 µm.
Etymology: Refers to the continent from which the fungus has been isolated.
Type material: CAMEROON: Tonga, from the bark of Polyscias fulva, April 2019, S.F. Kouam (holotype CBS H-25264; ex-type cultures CBS 150080 = STMA 18293).
Additional material: CAMEROON: Kala Mountain, from the bark of Asystasia macrophylla, 3 January 2019, S.F. Kouam, A.M. Elodie Gisele and S.C.N. Wouamba, STMA 18294.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 190–390(–430) μm diam, white to yellow conidial drops exuded from ostioles; conidiomatal wall brown to dark brown, composed of 1–2 layers, textura angularis to epidermoidea. Conidiophores cylindrical to subcylindrical, base subhyaline to pale yellow, apex hyaline to subhyaline, straight, densely aggregated, smooth-walled, 1–2 septate, (12–)16–26(–28) × 1.5–3 μm. Conidiogenous cells phialidic, cylindrical, tapering towards the apex, hyaline to subhyaline, terminal, 7.5–16(–18) × 1.5–2.5 μm. Paraphyses not observed. Alpha conidia ovoid to ellipsoidal, hyaline, apex rounded, base acutate, multiguttulate, aseptate, (5.5–)6–7.5(–8) × 2–2.5 μm. Beta conidia filiform, straight to curved, apex rounded, tapering towards base, hyaline, not guttulate, aseptate, (10–)13.5–21(–22.5) × 1–2 μm. Gamma conidia not observed.
Culture characters: Colonies on PDA covering a 9 cm Petri dish in 2 weeks, white with margins transparent to pale yellow (11D), cottony, raised, margins fringed; reverse yellow (161B–D). Colonies on MEA reaching 85–90 in 2 weeks, white to greyed white (156A–D), cottony, crateriform, margins fringed; reverse greyed yellow (162B–D) with center and a ting surrounding it grey brown (199A–D) to black (202A–B). Colonies on OA covering all Petri dish in 2 weeks, white to yellow-white (158B–C) with center grey (201A), cottony, slightly crateriform, margins fringed; reverse transparent to greyed green (198C–D).
Notes: Diaporthe africana formed a well-supported basal clade (100% bs/0.98 pp) together with D. salinicola. For this latter species, only the sexual morph has been reported up to now. Our new species did not form the sexual morph in any of the media tested. Moreover, D. salinicola has been only reported from Thailand, Asia [42].
Diaporthe africana is also phylogenetically related to D. hongkongensis. Both species were so far characterized to form alpha and beta conidia. However, D. hongkongensis could be easily distinguished by the production of gamma conidia [24], which were not observed in D. africana. Diaporthe hongkongensis has been reported only from Asia, including China, Japan, Taiwan, and Turkey [24,59,60,61,62].
3.2. Structural Elucidation of Compounds 1–8
Purification of the EtOAc extracts of D. africana obtained from cultures in liquid YMG and solid rice media yielded 8 compounds, including two cyclic depsipeptides (1, 8), five tenellone derivatives (2–6), and one azaphilone (7) (Figure 3). The known compounds 5, 6, and 8 were identified as the mixture of diaporisoindoles A and B [63,64], tenellone B [65], and beauvericin [66,67], respectively. This identification was achieved through an analysis of their spectral data (ESI-MS, HR-ESIMS, 1D/2D NMR, Figures S54–S71) and specific rotation as well as a comparison with previously published data. Interestingly, this is the first NMR-confirmed report of the occurrence of beauvericin (8) in endophytes belonging to the genus Diaporthe. This cyclic hexadepsipeptide (8) was initially obtained from the entomopathogenic fungus Beauveria bassiana [66] but was also found to be produced by many species belonging to the genus Fusarium and some Isaria spp. [68].
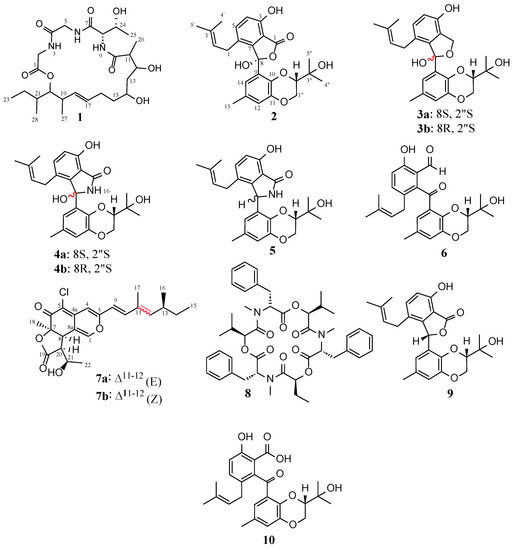
Figure 3.
Chemical structures of compounds 1–8 isolated from Diaporthe africana and the known compounds isoprenylisobenzofuran A (9) and tenellone C (10).
Compounds 2–4, 7 were isolated for the first time in the present study, and their structures were established after an exhaustive examination of their HR-ESIMS, NMR, and ECD spectra. It should be mentioned that although compound 1 had been previously characterized, the data regarding its structure elucidation have been included in this manuscript as well.
Compound 1 was obtained as a colorless oil. The molecular formula of C25H43N3O8 (6 degrees of unsaturation) was established based on (+) HR-ESIMS m/z 514.3124 [M + H]+ (calcd for 514.3123 C25H44N3O8+). An analysis of the 1H-NMR and 1H-13C HSQC spectra recorded in DMSO-d6 of 1 revealed clear signals corresponding to five methyl groups resonating at δH 0.83 (H3-28), δH 0.88 (H3-27), δH 1.20 (H3-26), δH 1.02 (H3-25), and δH 0.84 (H3-23). On the same spectrum, signals attributable to four methylene groups (δH 1.65, H2-13; δH 1.38, Ha-15; δH 1.48, Hb-15; δH 1.88, Ha-16; δH 1.99, Hb-16; δH 1.15, H-22), two N-bearing methylenes (δH 3.94, Ha-2; δH 3.69, Hb-2; δH 3.77, H2-5), three methines (δH 2.36, H-11; δH 2.32, H-19; δH 1.62, H-21), one N-bearing methine (δH 3.98, H-8), four O-bearing methines (δH 3.83, H-12; δH 3.57, H-14; δH 4.67, H-20, δH 4.18, H-24), two olefinic protons (δH 5.45, H-17; δH 5.09, H-18) and three amide hydrogens (δH 8.36, NH-3; δH 7.43, NH-6; δH 7.85, NH-9,) were observed. The signal-to-signal comparison of 13C NMR with 1H-13C HSQC spectrum of 1 confirmed 25 carbon signals which were classified into four carbonyl groups (δC 169.3, C-1; 168.6, C-4; 170.4, C-7 and 175.3, C-10), included two olefinic carbons (δC 130.7, C-17; δc 131.6, C-18), eight methines (including one N-bearing and four O-bearing ones), six methylenes (including two N-bearing ones) and five methyl groups (Table 2).

Table 2.
NMR spectroscopic data for compound 1 (1H 700 MHz, 13C 175 MHz, in DMSO-d6).
The careful interpretation of 2D NMR data and, more importantly, of the 1H-1H COSY and 1H-13C HMBC spectra enabled the construction of the gross structure of 1. On the COSY and HMBC spectra, three fragments (A–C) were unambiguously determined based on the observed correlations, as depicted in Figure 4. The HMBC correlations of NH-3 to C-2/C-4, NH-6 to C-4/C-5, and H2-2 to C-1/C-4 suggested that fragment A comprised two glycine residues. Similarly, a second amino acid residue (fragment B) was deduced as threonine based on key HMBC correlations of the amide proton NH-9 with C-8/C-24, alongside H-8 with carbonyl C-7, oxymethine C-24 and methyl C-25 (Figure 4). The downfield shifted chemical value of C-11, which resonated at δC 45.1, suggested its connection to a carbonyl group. This was further supported by a strong HMBC correlation between H-11 and the carbonyl C-10. Furthermore, the HMBC cross peaks of H-11, H-12, and H-26 to C-10 enabled the incorporation of the carbonyl group C-10 into the fatty acid residue (fragment C). On the HMBC spectrum, the key correlations to determine the linkage of moieties A and B were those of NH-6 and H-5 with C-7. Correlations of H-8 and NH-9 with C-10 validated the linkage of moieties B and C. The presence of four carbonyl groups and one olefinic bond, which accounted for five out of the six indices of hydrogen deficiency, as depicted by the molecular formula, suggested the existence of a ring in compound 1. The strong HMBC correlation observed between the oxymethine proton H-20 and the carbonyl C-1 corroborated the connection of moieties C and A, thus leading to the formation of a 21-membered macrocyclic core ring. Additionally, we investigated the absolute stereochemistry of fragment B. By using Marfey’s method (as described in the experimental section), the L-configuration (8S,24R) was assigned to the threonine residue by a comparison of the retention times of the L-and D-FDVA-derivatized hydrolysate of 1 (L-Thr-L-FDVA tR 6.17 min; L-Thr-D-FDVA tR 7.30 min) with that of Marfey’s derivatized authentic amino acids (Tables S3 and S4). In addition, the large coupling constant (J = 15.4 Hz) indicated an E-configuration for the ∆17–18 double bond. This was also confirmed by the absence of the ROESY correlation between protons H-17 and H-18, providing further evidence for their trans-relationship. It is worth mentioning that despite some important correlations observed on the ROESY spectrum of 1, as depicted in Figure 4, the determination of the relative stereochemistry at positions C-11, C-12, C-14, C-19, C-20, and C-21 remained inconclusive. This could be attributed to the inherent flexibility of the 21-membered macrolide ring, which could adopt various conformations and orientations, making it challenging to establish definitive stereochemical relationships at these positions. Based on the available evidence, the structure of compound 1 was successfully deduced, although its stereochemistry could only be partially determined. Curiously, while the current study has been under review, we became aware of a publication by Gao et al. [69], who reported a similar planar structure. Subsequently, a signals-to-signals comparison of the 1H NMR spectrum of 1 (recorded in CDCl3, Figure S9) with that of eucalactam B isolated from Diaporthe eucalyptorum showed a perfect match of chemical shifts (Table S1), suggesting that compound 1 was identical to eucalactam B [69]. However, while the relative configuration at positions C-12 and C-14 in eucalactam B were determined through a semisynthetic approach, revealing a syn stereochemistry [69], the absolute configuration of molecule 1 remains unresolved and requires further investigation. Previous attempts to employ X-ray crystallography and crystalline sponge methods to establish the absolute stereochemistry of this compound have proven unsuccessful, as reported by Gao et al. [69]. Therefore, other alternative methodologies must be explored, including chemical synthesis or computational approaches such as molecular modeling and density functional theory (DFT) calculations.

Figure 4.
Fragments A−C: key 1H-1H COSY, 1H-13C HMBC and 1H-1H ROESY correlations of compound 1.
Compound 2 was obtained as pale yellow oil. Its molecular formula was assigned as C25H28O7 based on (+) HR-ESIMS m/z 441.1905 [M + H]+ (calcd for 441.1908 C25H29O7+), indicating 12 degrees of unsaturation. An analysis of its 1D (Table 3) and 2D NMR spectroscopic data revealed similar features to those of the known isoprenylisobenzofuran A (9) [70]. In comparison to 9 (molecular formula: C25H28O6), the HR-ESIMS of compound 2 showed an increase in 16 Da, and the obtained molecular formula for 2 suggested that a hydrogen atom in 9 was possibly replaced by a hydroxyl group. The 1H NMR data of 2 recorded in CDCl3 showed broad signals, which led to the absence of important correlations in the 1H-13C HMBC spectrum. To solve this issue, a new set of NMR data were recorded in CDCl3 + 1%TFA, which facilitated the acquisition of well-defined 1H signals. The 1H−1H COSY and 1H-13C HMBC spectra supported the existence of these fragments: 1,2,3,4-tetrasubstituted benzene ring, isoprenyl, and a 2,3-dihydrobenzo[b][1, 4]dioxane moiety (Figure 5). The characteristics of HMBC correlations observed in compound 2 exhibited striking similarities to those reported for compound 9. However, the absence of the key signal C-8 in the 13C NMR spectrum, as well as the absence of the important HMBC correlation of H-14 to C-8 (to confirm the linkage point of units A and B), made establishing the structure of 2 ambiguous. In light of these observations, it was also conceivable to propose the structure of the previously known compound tenellone C (10) for compound 2. To further investigate this possibility, the NMR spectrum of compound 2 was recorded in deuterated methanol (MeOH-d4) for a more comprehensive comparison with tenellone C, which has been previously analyzed in that same solvent [63]. Interestingly, there were some significant differences in their NMR spectra (Figures S20–S22). Remarkably, compound 2 lacked some signals observed in tenellone C, such as the 13C signal of the carbonyl group at C-8. Additionally, the HMBC correlation between H-14 and C-8, which was evident in tenellone C, was not observed in compound 2. These findings effectively ruled out the hypothesis that compound 2 was identical to tenellone C. Conversely, the absence of a chemical shift at C-8 and the lack of an HMBC correlation between H-14 and C-8 have been observed in various instances where the isobenzofuran ring or indole rings were formed. This was notably observed in compounds such as isoprenylisobenzofuran A, as well as diaporisoindoles A and B [63,70]. However, it is important to mention that the structures and absolute configurations of these compounds (isoprenylisobenzofuran A and diaporisoindoles A and B) were confirmed through X-ray diffraction experiments. In the present study, an X-ray diffraction analysis was not pursued, and thus, the proposed planar structure of compound 2 was established based on the very close similarities of its spectral data (HR-ESIMS, 1D, and 2D NMR) with that of compound 9 [70]. As suggested by their molecular formula, the only remarkable difference was the incorporation of a hydroxyl group at C-8 in the structure of 2 instead of the hydrogen atom, as in 9. To determine the absolute configuration of 2, its ECD spectrum (Figure S24) was recorded and compared with that of 8-epi-isoprenylisobenzofuran A (lithocarol F) [71]. The Cotton effect pattern observed for compound 2 matched with that reported for lithocarol F [71] and enabled the assignment of the stereochemistry of 2 as 8R,2″S. The structure of compound 2 was therefore proposed and named isoprenylisobenzofuran B.

Table 3.
NMR data for compounds 2 (1H 700 MHz, 13C 175 MHz, in CDCl3 + 1% TFA) and 3 (1H 700 MHz, 13C 175 MHz, in DMSO-d6).
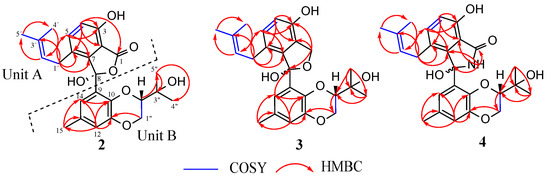
Figure 5.
Key 1H-1H COSY and 1H-13C HMBC correlations of compounds 2–4.
Compound 3 was isolated as pale yellow oil. Its purity was examined using HPLC-DAD-HR-ESIMS data (DAD detection at 210 nm/200–640 nm) which indicated the presence of a single compound (Figure S26). Its unique molecular formula C25H30O6 (indicating an index of hydrogen deficiency of 11) was, therefore, determined based on (+) HR-ESIMS m/z 449.1938 [M + Na]+ (calcd for C25H30NaO6+ 449.1935) and (−) HR-ESIMS m/z 425.1972 [M − H]− (calcd for C25H29O6− 425.1970). However, the features of the 1H and 13C-NMR spectra, which showed duplicated signals in a ratio of about 1.2:1, revealed that 3 was isolated as a mixture of stereoisomers. A detailed analysis of the 1D and 2D NMR spectroscopic data of 3 enabled the assignment of all protons and carbons (Table 3) corresponding to both isomers (3a and 3b). The careful interpretation of 1D and 2D NMR data of compound 3 displayed close similarities to that of 2. The main difference was the appearance of an O-bearing methylene group C-1 (δH 4.86 (Ha-1), δH 4.97 (Hb-1) and δC 69.1/69.2 (C-1)) in the structure of 3 (3a and 3b) instead of the C-1 carbonyl group (δC 171.9), as observed in 2. The structure of compound 3 was finally validated by the comprehensive analysis of its 1H-13C HMBC spectrum, which displayed key correlations with the methylene protons Ha-1 and Hb-1 with C-2/C-3/C-7/C-8 (Figure 5). The clear HMBC correlation of H-14 with C-8 that was observed in 3 further supports the linkage points of units A and B in its structure but also in that of its biogenetically related analog 2 described above, thus clarifying any ambiguity in the proposed structure of 2. Furthermore, the stereochemistry of each isomer in the mixture was tentatively determined. Among the hitherto reported tenellone derivatives isolated from Diaporthe spp., the stereogenic carbon at C-2″ in the [1, 4] dioxane core was found to be S-configured [63,65,70,71,72,73,74,75]. Taking into account biosynthetic considerations, which suggested that the formation of the [1, 4] dioxane ring occurred stereo-specifically, an S-configuration at C-2″ for both isomers of compound 3 was therefore proposed. This implies that 3 is a mixture of epimers differing only in the stereochemistry of the O-bearing quaternary carbon C-8. The configuration of 8S,2″S was thus arbitrarily assigned to the major isomer 3a, and the stereochemistry of the minor isomer 3b (or 8-epi-3a) was deduced to be 8R,2″S. The structures of compound 3 were therefore characterized and assigned the trivial names of isoprenylisobenzofuran C1 (3a) and isoprenylisobenzofuran C2 (3b).
Compound 4 was obtained as a pale yellow oil with the molecular formula of C25H29NO6 (12 degrees of unsaturation), which was assigned based on (+) HR-ESIMS m/z 440.2064 [M + H]+ (calcd for 440.2068 C25H30NO6+). Its purity was ascertained by using HPLC-DAD-HR-ESIMS data (DAD detection at 210 nm/200–640 nm) which showed ion clusters corresponding to a single molecular formula, and the single peak observed on the UV chromatogram suggested the presence of a pure compound (Figure S35). However, the characteristics of its 1H and 13C NMR data were similar to those of compound 3 and equally displayed duplicated signals in a ratio of 3:2, suggesting that 4 was also isolated as an epimeric mixture (4a + 4b). The interpretation of HR-ESIMS data of 4 (C25H29NO6) compared to that of 2 (C25H28O7) led to the hypothesis that the five-membered lactonic ring in 2 was possibly replaced by a five-membered lactam ring in 4. This assumption was verified by a detailed analysis of the 1H-13C HMBC spectrum. The HMBC cross peaks of NH-16 with C-1/C-2/C-7/C-8 formed the key correlations, which validated the presence of the lactam ring in 4 (Figure 5). The full assignment of all 1H and 13C’s chemical shifts corresponding to both isomers of 4 was finally accomplished by a comprehensive examination of 1D and 2D NMR spectroscopic data (Table 4). As stipulated above, C-2″ was suggested to be biogenetically S-configured [63,65,70,71,72,73,74,75], thus indicating arbitrarily an 8S,2″S stereochemistry for the major isomer 4a and 8R,2″S for the minor 4b. The structures of 4 were thus determined and given the trivial names diaporisoindole F1 (4a) and diaporisoindole F2 (4b).

Table 4.
NMR data for compound 4 (1H 700 MHz, 13C 175 MHz, in DMSO-d6).
Compound 7 was isolated as a yellow amorphous solid. The molecular formula C23H27ClO5 (10 degrees of unsaturation) was deduced from its (+) HR-ESIMS, which showed a cluster of the sodium adduct [M + Na]+ at m/z 441.1437/443.1415 (Calcd for C23H27ClNaO5+ 441.1439) with a ratio of 3:1, which is indicative of a monochlorinated compound. Although an examination of its HPLC-DAD-MS chromatogram confirmed the purity of compound 7, the 1H and 13C NMR spectra (recorded in DMSO-d6) rather exhibited duplicated signals, revealing that it was a mixture of isomers. The UV-Vis and 1H NMR spectra of 7 (Figures S45 and S52) displayed a typical pattern of an azaphilone skeleton [76]. A close inspection of its 1D NMR and 1H-13C HSQC spectra revealed characteristics that perfectly matched those of isochromophilonol: an azaphilone previously isolated from Arcopilus cupreus (syn. Chaetomium cupreum) RY202 [77]. The planar structure of this metabolite was further confirmed to be identical to that of isochromophinol by a careful analysis of its 1H-1H COSY and 1H-13C HMBC correlations (Figure 6). To make a proper comparison between compound 7 and the known isochromophilonol, the 1H NMR spectrum of compound 7 was additionally recorded in CDCl3 (Figure S51). Upon comparing the proton chemical shifts of compound 7 with those of isochromophilonol, noteworthy discrepancies, particularly for protons H-20, H-18, and H-22 (Table S2), were observed. This strongly suggests that compound 7 and isochromophinol did not have the same stereochemistry. In addition, the slight difference recorded between both isomers of 7 was rapidly detected on 1D NMR and ROESY spectra. A careful analysis of these spectra revealed a clear case of cis/trans isomerism through the observed variations in chemical shifts and correlations (1H and 13C NMR data of each isomer are summarized in Table 5). For the major isomer 7a, an E-geometry for the ∆11–12 double bond was established based on ROESY correlations of H-16 with H-17 and H-10 with H-12. Concurrently, ROESY cross-peaks between H-12 and H-17, H-10 and H-16 were in favor of a Z-geometry for the ∆11–12 double bond in the minor isomer 7b (Figure 6) [78]. Additionally, a large coupling constant J9,10 = 15.8 Hz indicated an E-geometry for the ∆9–10 double bond in both isomers. Furthermore, an investigative effort into the stereochemistry of compound 7 was completed by the detailed interpretation of its 1H-1H ROESY data and by comparison with its ECD spectra and its specific rotation with that of isochromophilonol [77]. On the ROESY spectrum of 7, important correlations were observed (for both isomers) between H-18 and H-8/H-20 and H-8 and H-21, indicating that those protons were located on the same side (Figure 6). Noteworthy, the stereochemistry at position C-7 of the azaphilones has already been firmly established by optical rotations, circular dichroism, and X-ray analysis [79,80,81]. On the basis of the ECD spectra and the known 7S configuration of isochromophilonol [77], the 7R configuration was assigned to compound 7. Explicitly, the ECD spectrum of 7 (Figure S53) showed a positive Cotton effect at ~315 nm (∆ε + 6.75), contrary to that of isochromophilonol, whose spectrum showed a negative cotton effect at the same wavelength [77]. Moreover, previous research by Whalley and co-workers has demonstrated that the sign of the specific rotation of azaphilones is seemingly influenced by the absolute configuration at the C-7 position [80]. Against this background, the opposite signs of specific rotation observed for 7 ( + 839) compared to that of isochromophilonol (− 141.6) further confirmed the 7R configuration assigned to this compound. Using C-7 as a reference, the absolute configuration at C-8, C-20, and C-21 was deduced as 8R, 20S, and 21R based on the aforementioned ROESY correlations. Taking into account biogenetic considerations, the absolute configuration at C-13 was proposed to be S since stereogenic carbon at C-13 in the aliphatic side chain was S-configured among the hitherto reported azaphilones; this harbored a branched C-7 side chain anchored at C-3 [64]. The structures of compound 7 were thus fully characterized and turned out to be unprecedented stereoisomers of isochromophilonol for which the trivial names isochromophilonol A1 (7a) and isochromophilonol A2 (7b) were assigned.
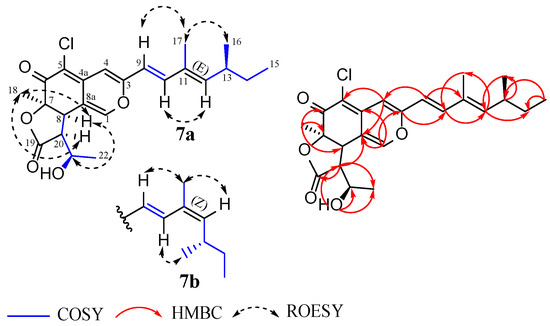
Figure 6.
Key 1H-1H COSY, 1H-13C HMBC and 1H-1H ROESY correlations of compound 7.

Table 5.
NMR data for compound 7 (1H 500 MHz, 13C 125 MHz, in DMSO-d6).
3.3. Physicochemical Data for Compounds 1–4, 7
Eucalactam B (1): colorless oil. 7 (c 0.35, MeOH), UV (MeOH, c = 0.02 mg/mL) λmax (log ε) 201 (4.1) nm. CD (c = 1.9 × 10−3 M, MeOH) λmax (Δε) 200 (−6.03) nm. (+) HR-ESIMS m/z 536.2935 [M + Na]+, m/z 1049.5973 [2 M + Na]+, m/z 1027.6173 [2 M + H]+, m/z 496.3018 [M + H − H2O]+, m/z 514.3124 [M + H]+ (calcd for C25H44N3O8+ 514.3123). tR = 8.48 min (HR-LC-ESIMS). For NMR data (1H: 700 MHz, 13C: 175 MHz, DMSO-d6), see Table 2.
Isoprenylisobenzofuran B (2): pale yellow oil.+ 34 (c 0.25, MeOH), UV (MeOH, c = 0.01 mg/mL) λmax (log ε) 312 (3.8) 269 (3.9) 208 (4.6) nm. CD (c = 2.3 × 10−3 M, MeOH) λmax (Δε) 225 (−6.3) 208 (+2.3) nm. (+) HR-ESIMS m/z 463.1726 [M + Na]+, m/z 903.3537 [2 M + Na]+, m/z 423.1797 [M + H − H2O]+, m/z 441.1905 [M + H]+ (calcd for C25H29O7+ 441.1908). tR = 10.99 min (HR-LC-ESIMS). For NMR data (1H: 700 MHz, 13C: 175 MHz, CDCl3 + 1%TFA), see Table 3.
Isoprenylisobenzofuran C1 (3a)/C2 (3b): pale yellow oil. UV (MeOH, c = 0.01 mg/mL) λmax (log ε) 284 (3.7) 204 (4.6) nm. (+) HR-ESIMS m/z 875.3981 [2 M + Na]+, m/z 409.2015 [M + H − H2O]+, m/z 449.1936 [M + Na]+ (calcd for C25H30NaO6+ 449.1935). (−) HR-ESIMS m/z 425.1972 [M − H]− (calcd for C25H29O6− 425.1970). tR = 10.65 min (HR-LC-ESIMS). For NMR data (1H: 500 MHz, 13C: 125 MHz, DMSO-d6), see Table 3.
Diaporisoindole F1 (4a)/F2 (4b): pale yellow oil. UV (MeOH, c = 0.01 mg/mL) λmax (log ε) 294 (3.7) 206 (4.6) nm. (+) HR-ESIMS m/z 462.1882 [M + Na]+, m/z 901.3876 [2 M + Na]+, m/z 422.1959 [M + H − H2O]+, m/z 440.2065 [M + H]+ (calcd for C25H30NO6+ 440.2068). tR = 10.70 min (HR-LC-ESIMS). For NMR data (1H: 700 MHz, 13C: 175 MHz, DMSO-d6), see Table 4.
Isochromophilonol A1 (7a)/A2 (7b): yellow amorphous solid.+ 839 (c 0.1, MeOH), UV (MeOH, c = 0.01 mg/mL) λmax (log ε) 409 (4.4), 363 (4.2) 252 (4.2), 209 (4.0) nm. CD (c = 2.4 × 10−3 M, MeOH) λmax (Δε) 411 (+7.76) 311 (+6.75) 262 (+0.93) 229 (+3.54) nm. HR-ESIMS m/z 441.1437 [M + Na]+, m/z 859.2989 [2 M + Na]+, m/z 419.1623 [M + H]+ (calcd for C23H28ClO5+ 419.1620), tR = 13.49 min (HR-LC-ESIMS). For NMR data (1H: 500 MHz, 13C: 125 MHz, DMSO-d6), see Table 5.
3.4. Biological Activities of Compounds 1–8
The inhibitory potentials of compounds 1–8 against a panel of bacteria and fungi were evaluated. Compound 1 was found to be devoid of any activity against all the tested microorganisms. Except for compound 2, which showed no activity, the tenellone derivatives (3–6) exhibited weak to moderate activity against certain microorganisms that were tested, namely B. subtilis, M. hiemalis, R. glutinis, S. pombe, and W. anomalus with MIC values in the range of 16.6–66.6 µg/mL (Table 6). Tenellone derivatives were previously found to exhibit interesting biological effects, and their main reported properties involved anti-tumor and anti-inflammatory activities [70,71,73,75,82]. However, to the best of our knowledge, their antimicrobial activities have hitherto not been reported. This study is thus the first to outline the antimicrobial effects of this class of compounds. In addition, compound 7 also showed weak to moderate activity against the Gram-positive bacteria B. subtilis (MIC 16.6 µg/mL), M. smegmatis (MIC 16.6 µg/mL), and S. aureus (MIC 66.6 µg/mL). Remarkably, its activity against the Mucoromycota fungus M. hiemalis was significant, reaching an MIC of 8.3 µg/mL, which is equal to that of nystatin used as a positive control. However, further investigations need to be conducted to delineate whether only one of the isomers is active or if the reported activity is the result of a synergistic effect. Furthermore, the cyclic hexadepsipeptide (8) was also active in the current assay, with MICs ranging from 8.3 to 66.6 µg/mL against some fungi and Gram-positive bacteria (Table 6).

Table 6.
Antimicrobial activities of compounds 1–8.
Different mammalian cell lines were used to assess the cytotoxicity of the isolated metabolites 1–8. In this assay, compound 1 did not show any cytotoxic effect under the tested conditions. Amidst the tenellone derivatives (2–6), compound 2 only demonstrated a slight inhibition of KB3.1 cell proliferation, whereas the other compounds (3–6) inhibited the growth of KB3.1 and L929 cell lines with IC50 values in the range of 20–59.2 µM (Table 7). Among these, compound 6 exhibited the strongest cytotoxic activity against the cancer cell line KB3.1 and the murine fibroblasts of line L929. This compound was thus further evaluated against five other mammalian cell lines, namely A431, MCF-7, A549, SKOV-3, and PC-3, and exhibited cytotoxic effects with IC50 values ranging from 17.7 to 42.5 µM. As stipulated in the previous paragraph, several tenellone-derived representatives have proven to possess interesting antitumor properties. Therefore, the cytotoxic activity of compounds 3–6 presented in this study corroborates previously published data [71,72,73,75]. This encourages further investigations into the cytotoxic effects of novel target molecules in this class for the discovery of potential antitumor agents. Interestingly, during the evaluation of the biological properties of diaporisoindoles A and B by Cui and their collaborators [63], diaporisoindole A with a configuration of 8S,2″S showed potent inhibitory activity against MptpB. However, diaporisoindole B (8R,2″S or 8-epi-diaporisoindole A) failed to show any activity under the test conditions, suggesting that the S-configuration at C-8 in these tenellone analogs probably promoted their inhibitory effect. The latter could explain the lack of activity observed for compound 2 (8R,2″S-configured) in comparison to the epimeric mixtures 3–5. In addition, compounds 7 and 8 exhibited significant growth inhibitory effects against the six cancer and the murine fibroblast cell lines assessed with IC50 values in the range of 0.9−12.9 µM and 1.9−3.3 µM, respectively (Table 7). Interestingly, the so-called emerging mycotoxin beauvericin (8), which belongs to the enniatin antibiotic family, has attracted a lot of attention over recent years due to its multifaceted nature. This compound has recently been recognized as a promising candidate for anticancer therapy, as reported by Sood et al. [83] and Wu et al. [84]. However, its potential as an emerging mycotoxin and its impact on animals, humans, and the environment still require clarification and further investigation [68]. In addition, beauvericin also displays other biological effects, including antibacterial, antiviral, antifungal, insecticidal, and nematicidal activities, to mention a few [68,84]. Therefore, the herein-reported antimicrobial and cytotoxic activities of metabolite 8 are consistent with previous research findings.

Table 7.
Cytotoxic activities of compounds 1–8.
4. Conclusions
In our continuous search for new therapeutic molecules, we investigated the secondary metabolism of endophytic fungal strains associated with Cameroonian medicinal plants. Through our extensive screening efforts, we were able to identify a new fungal species named Diaporthe africana, from which four novel polyketides were obtained alongside four known compounds. The isolation, characterization, and biological evaluation of these compounds not only demonstrated the potential of the Diaporthe species as a source of novel and structurally diverse bioactive compounds but also emphasized the importance of investigating new taxa of even well-studied phylogenetic groups such as Diaporthe for discovery of new forms of chemical diversity. In this study, two rationales were utilized to maximize the chances of expanding chemical diversity within the widely explored genus Diaporthe. On the one hand, we focused on new endophytic fungal strains from underexplored geographic areas like the tropical rainforests of Cameroon, which are known to possess a rich diversity of plant species that remain widely untapped in terms of the plant–endophyte interaction as well as the metabolites produced by these endophytes. On the other hand, traditional medicinal plants or plants with an ethnopharmacological background were selected for the isolation of these endophytes. These research findings complement previous results in relation to investigating chemical and biological diversity of the genus Diaporthe and some of the metabolites herein reported add to the growing pool of bioactive compounds that could have therapeutic applications. In addition, this work provides further evidence of the potential of endophytes as a promising source of new biologically active compounds and underscores the importance of further exploring these microorganisms for the discovery of potentially leading drug candidates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9070781/s1, Figure S1: HPLC-DAD chromatogram and ESI-MS data for eucalactam B (1), Figure S2: HPLC-DAD chromatogram and HR-ESI (+) MS data for eucalactam B (1), Figure S3: 1H NMR spectrum (DMSO-d6, 700 MHz) of eucalactam B (1), Figure S4: 13C NMR spectrum (DMSO-d6, 175 MHz) of eucalactam B (1), Figure S5: 1H-1H COSY NMR spectrum (DMSO-d6, 700 MHz) of eucalactam B (1), Figure S6: 1H-13C HSQC NMR spectrum (DMSO-d6, 700 MHz) of eucalactam B (1), Figure S7: 1H-13C HMBC NMR spectrum (DMSO-d6, 700 MHz) of eucalactam B (1), Figure S8: 1H-1H ROESY NMR spectrum (DMSO-d6, 700 MHz) of eucalactam B (1), Figure S9: 1H NMR spectrum (CDCl3, 500 MHz) of eucalactam B (1), Figure S10: UV-Vis spectrum (MeOH) of eucalactam B (1), Figure S11: ECD spectrum (MeOH) of eucalactam B (1), Figure S12: HPLC-DAD chromatogram and ESI-MS data for isoprenylisobenzofuran B (2), Figure S13: HPLC-DAD chromatogram and HR-ESI (+) MS data for isoprenylisobenzofuran B (2), Figure S14: 1H NMR spectrum (CDCl3 + 1%TFA, 700 MHz) of isoprenylisobenzofuran B (2), Figure S15: 13C NMR spectrum (CDCl3 + 1%TFA, 175 MHz) of isoprenylisobenzofuran B (2), Figure S16: 1H-1H COSY NMR spectrum (CDCl3 + 1%TFA, 700 MHz) of isoprenylisobenzofuran B (2), Figure S17: 1H-13C HSQC NMR spectrum (CDCl3 + 1%TFA, 700 MHz) of isoprenylisobenzofuran B (2), Figure S18: 1H-13C HMBC NMR spectrum (CDCl3 + 1%TFA, 700 MHz) of isoprenylisobenzofuran B (2), Figure S19: 1H-1H ROESY NMR spectrum (CDCl3 + 1%TFA, 700 MHz) of isoprenylisobenzofuran B (2), Figure S20: 1H NMR spectrum (MeOH-d4, 500 MHz) of compound 2, Figure S21: 13C NMR spectrum (MeOH-d4, 125 MHz) of compound 2, Figure S22: 1H-13C HMBC NMR spectrum (MeOH-d4, 500 MHz) of compound 2, Figure S23: UV-Vis spectrum (MeOH) of isoprenylisobenzofuran B (2), Figure S24: ECD spectrum (MeOH) of isoprenylisobenzofuran B (2), Figure S25: HPLC-DAD chromatogram and ESI-MS data for isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S26: HPLC-DAD chromatogram and (+)/(−) HR-ESIMS data for isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S27: 1H NMR spectrum (DMSO-d6, 700 MHz) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S28: 13C NMR spectrum (DMSO-d6, 175 MHz) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S29: 1H-1H COSY NMR spectrum (DMSO-d6, 700 MHz) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S30: 1H-13C HSQC NMR spectrum (DMSO-d6, 700 MHz) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S31: 1H-13C HMBC NMR spectrum (DMSO-d6, 700 MHz) isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S32: 1H-1H ROESY NMR spectrum (DMSO-d6, 700 MHz) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S33: UV-Vis spectrum (MeOH) of isoprenylisobenzofuran C1/C2 (3a + 3b), Figure S34: HPLC-DAD chromatogram and ESI-MS data for diaporisoindole F1/F2 (4a + 4b), Figure S35: HPLC-DAD chromatogram and HR-ESI (+) MS data for diaporisoindole F1/F2 (4a + 4b), Figure S36: 1H NMR spectrum (DMSO-d6, 700 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S37: 13C NMR spectrum (DMSO-d6, 175 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S38: 1H-1H COSY NMR spectrum (DMSO-d6, 700 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S39: 1H-13C HSQC NMR spectrum (DMSO-d6, 700 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S40: 1H-13C HMBC NMR spectrum (DMSO-d6, 700 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S41: 1H-1H ROESY NMR spectrum (DMSO-d6, 700 MHz) of diaporisoindole F1/F2 (4a + 4b), Figure S42: UV-Vis spectrum (MeOH) of diaporisoindole F1/F2 (4a + 4b), Figure S43: HPLC-DAD chromatogram and ESI-MS data for isochromophilonol A1/A2 (7a + 7b), Figure S44: HPLC-DAD chromatogram and HR-ESI (+) MS data for isochromophilonol A1/A2 (7a + 7b), Figure S45: 1H NMR spectrum (DMSO-d6, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S46: 13C NMR spectrum (DMSO-d6, 125 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S47: 1H-1H COSY NMR spectrum (DMSO-d6, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S48: 1H-13C HSQC NMR spectrum (DMSO-d6, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S49: 1H-13C HMBC NMR spectrum (DMSO-d6, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S50: 1H-1H ROESY NMR spectrum (DMSO-d6, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S51: 1H NMR spectrum (CDCl3, 500 MHz) of isochromophilonol A1/A2 (7a + 7b), Figure S52: UV-Vis spectrum (MeOH) of isochromophilonol A1/A2 (7a + 7b), Figure S53: ECD spectrum (MeOH) of isochromophilonol A1/A2 (7), Figure S54: HPLC-DAD chromatogram and ESI-MS data for diaporisoindole A/B (5), Figure S55: HPLC-DAD chromatogram and HR-ESI (+) MS data for diaporisoindole A/B (5), Figure S56: 1H NMR spectrum (CDCl3, 700 MHz) of diaporisoindole A/B (5), Figure S57: 1H-1H COSY NMR spectrum (CDCl3, 700 MHz) of diaporisoindole A/B (5), Figure S58: 1H-13C HSQC NMR spectrum (CDCl3, 700 MHz) of diaporisoindole A/B (5), Figure S59: 1H-13C HMBC NMR spectrum (CDCl3, 700 MHz) of diaporisoindole A/B (5), Figure S60: HPLC-DAD chromatogram and ESI-MS data for tenellone B (6), Figure S61: HPLC-DAD chromatogram and HR-ESI (+) MS data for tenellone B (6), Figure S62: 1H NMR spectrum (DMSO-d6, 700 MHz) of tenellone B (6), Figure S63: 1H-1H COSY NMR spectrum (DMSO-d6, 700 MHz) of tenellone B (6), Figure S64: 1H-13C HSQC NMR spectrum (DMSO-d6, 700 MHz) of tenellone B (6), Figure S65: 1H-13C HMBC NMR spectrum (DMSO-d6, 700 MHz) of tenellone B (6), Figure S66: HPLC-DAD chromatogram and ESI-MS data for beauvericin (8), Figure S67: HPLC-DAD chromatogram and HR-ESI (+) MS data for beauvericin (8), Figure S68: 1H NMR spectrum (DMSO-d6, 700 MHz) of beauvericin (8), Figure S69: 1H-1H COSY NMR spectrum (DMSO-d6, 700 MHz) of beauvericin (8), Figure S70: 1H-13C HSQC NMR spectrum (DMSO-d6, 700 MHz) of beauvericin (8), Figure S71: 1H-13C HMBC NMR spectrum (DMSO-d6, 700 MHz) of beauvericin (8), Figure S72: HPLC-DAD chromatograms of eucalactam B derived D-FDVA (D)/authentic amino acid derived D-FDVA. (A) DL-threonine, (B) L-Allothreonine, (C) L-threonine, Figure S73: HPLC-DAD chromatograms of eucalactam B derived L-FDVA (D)/authentic amino acid derived L-FDVA. (A) DL-threonine, (B) L-Allothreonine, (C) L-threonine, Figure S74: General Marfey’s reaction with threonine, Figure S75: RAxML phylogram including our strains and type and reference strains of Diaporthe spp., Figure S76: Flow chart of the purification procedure, Table S1: Comparison of 1H NMR data of compound 1 with that of eucalactam B in CDCl3, Table S2: Comparison of 1H NMR data of compound 7 with that of isochromophilonol in CDCl3, Table S3: Retention time of L or D authentic amino acid derived D-FDVA. Table S4: Retention time of L or D authentic amino acid derived L-FDVA. Table S5: GenBank accession numbers of the strains included in the broad phylogenetic study. Table S6: Selected edge-linked proportional partition substitution models subjected to IQTree2 calculated with ModelTest as implemented in IQTree using Bayesian information criterion (BIC). Table S7: Characteristics of the restricted MAFFT alignments following the first phylogenetic analysis using IQTree 2 for phylogenetic inference. Table S8: Selected unlinked partition substitution model subjected to MrBayes calculated with ModelFinder as implemented in the Phylosuite program package using Bayesian information criterion (BIC). Alignments of the ITS, cal, his3, tef1, and tub2 sequences used in the phylogenetic study.
Author Contributions
B.M.K., conceptualization, screening, large scale fermentation, isolation of compounds, structure elucidation and preparing original draft; C.L., conceptualization, identification of the strain and editing the draft; M.S., supervision, funding acquisition, and editing the draft. S.K.F., funding acquisition and correcting the draft. Y.M.-F., conceptualization, fungal identification, supervision, preparing and polishing the draft. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided by a personal PhD stipend from the German Academic Exchange Service (DAAD) to B.M.K., who is gratefully acknowledged (programme ID-57440921). Y.M.F was supported by the Deutsche Forschungsgemeinschaft (DFG)—Project-ID 490821847. We are also grateful to The World Academy of Sciences (TWAS) (grant 18-178 RG/CHE/AF/AC_G-FR 3240303654), the AvH Foundation through the equipment subsidies (Ref 3.4-8151/20 002), the Research Group Linkage (grant IP-CMR-1121341) and the AvH Research hub project CECANAPROF (Ref 3.4-CMR-Hub). C.L. is thankful for a stipend from the Life Science Foundation, Braunschweig, Germany.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequences are deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and all other relevant data are included in the Supplementary Information.
Acknowledgments
We are grateful to Kirsten Harmrolfs and Christel Kakoschke for conducting NMR spectroscopic measurements, Wera Collisi for conducting cytotoxicity assays, and Aileen Gollasch and Esther Surges for their assistance in recording HR-ESIMS and HPLC-DAD/MS spectra, respectively. We also acknowledge Elodie Gisele Anoumedem Mouafo for providing the fungal specimens and Lena Schweizer for their assistance in the laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu, T.C.; Lu, Y.H.; Wang, J.F.; Song, Z.Q.; Hou, Y.G.; Liu, S.S.; Liu, C.S.; Wu, S.H. Bioactive secondary metabolites of the genus Diaporthe and anamorph Phomopsis from terrestrial and marine habitats and endophytes: 2010–2019. Microorganisms 2021, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug resistance (MDR): A widespread phenomenon in pharmacological therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef] [PubMed]
- Hilário, S.; Gonçalves, M.F.M. Endophytic Diaporthe as promising leads for the development of biopesticides and biofertilizers for a sustainable agriculture. Microorganisms 2022, 10, 2453. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Khan, B.; Dai, Q.; Lin, J.; Kang, L.; Rajput, N.A.; Yan, W.; Liu, G. Potential of secondary metabolites of Diaporthe species associated with terrestrial and marine origins. J. Fungi 2023, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Riga, R.; Happyana, N.; Quentmeier, A.; Zammarelli, C.; Kayser, O.; Hakim, E.H. Secondary metabolites from Diaporthe lithocarpus isolated from Artocarpus heterophyllus. Nat. Prod. Res. 2021, 35, 2324–2328. [Google Scholar] [CrossRef]
- Niaz, S.; Khan, D.; Naz, R.; Safdar, K.; Abidin, S.Z.U.; Khan, I.U.; Gul, R.; Khan, W.U.; Khan, M.A.U.; Lan, L. Antimicrobial and antioxidant chlorinated azaphilones from mangrove Diaporthe perseae sp. isolated from the stem of Chinese mangrove Pongamia pinnata. J. Asian Nat. Prod. Res. 2021, 23, 1077–1084. [Google Scholar] [CrossRef]
- Yang, X.-W.; Salome, C.; Rongbiao Pi, S.; Zhou, X.; Liu, S.; Luo, X.; Yang, J.; Chen, F.; Lin, X.; Chen, C.; et al. Structurally diverse polyketides from the mangrove-derived fungus Diaporthe sp. SCSIO 41011 with their anti-influenza A virus activities. Front. Chem. 2018, 1, 282. [Google Scholar] [CrossRef]
- Matio Kemkuignou, B.; Schweizer, L.; Lambert, C.; Gisèle, E.; Anoumedem, M.; Kouam, S.F.; Stadler, M.; Marin-Felix, Y.; Felix, Y.M. New polyketides from the liquid culture of Diaporthe breyniae sp. nov. (Diaporthales, Diaporthaceae). MycoKeys 2022, 90, 85–118. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; Crous, P.W.; Coutinho, I.A. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. S. Afr. J. Bot. 1996, 62, 86–88. [Google Scholar] [CrossRef]
- Crous, P.W.; Verkley, G.J.M.; Groenewald, J.Z.; Samson, R.A. Fungal Biodiversity. CBS Laboratory Manual Series 1; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2009. [Google Scholar]
- The Royal Horticultural Society London. R.H.S. Colour Chart; The Royal Horticultural Society London: London, UK, 1966. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Guo, Y.S.; Crous, P.W.; Bai, Q.; Fu, M.; Yang, M.M.; Wang, X.H.; Du, Y.M.; Hong, N.; Xu, W.X.; Wang, G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 2020, 45, 132–162. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Hou, C. Three new species of Diaporthe from China based on morphological characters and DNA sequence data analyses. Phytotaxa 2019, 422, 157–174. [Google Scholar] [CrossRef]
- Tan, Y.P.; Shivas, R.G. Nomenclatural novelties. Index Aust. Fungi 2022, 2, 1–12. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Wrona, C.J.; Mohankumar, V.; Schoeman, M.H.; Tan, Y.P.; Shivas, R.G.; Jeff-Ego, O.S.; Akinsanmi, O.A. Phomopsis husk rot of macadamia in Australia and South Africa caused by novel Diaporthe species. Plant Pathol. 2020, 69, 911–921. [Google Scholar] [CrossRef]
- Chang, C.Q.; Cheng, Y.H.; Xiang, M.M.; Jiang, Z.D. New species of Phomopsis on woody plants in Fujian Province. Mycosystema 2005, 24, 6–11. [Google Scholar]
- Yang, Q.; Tang, J.; Zhou, G.Y. Characterization of Diaporthe species on Camellia oleifera in Hunan Province, with descriptions of two new species. MycoKeys 2021, 84, 15–33. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Romberg, M.; Mel’nik, V.A.; Verkley, G.J.M.; Groenewald, J.Z. Fungal planet description sheets: 92–106. Persoonia 2011, 27, 130–162. [Google Scholar] [CrossRef]
- Yang, Q.; Fan, X.L.; Guarnaccia, V.; Tian, C.M. High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 2018, 39, 97–149. [Google Scholar] [CrossRef]
- de Silva, N.; Hyde, K.D.; Lumyong, S.; Phillips, A.J.L.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Tennakoon, D.S.; Suwannarach, N.; Karunarathna, S.C. Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere 2022, 13, 955–1076. [Google Scholar] [CrossRef]
- Huang, S.; Xia, J.; Zhang, X.; Sun, W. Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 2021, 78, 49–77. [Google Scholar] [CrossRef]
- Perera, R.H.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Jones, E.B.G.; McKenzie, E.H.C.; Stadler, M.; Lee, H.B.; Samarakoon, M.C.; Ekanayaka, A.H.; Camporesi, E.; et al. Fungi on wild seeds and fruits. Mycosphere 2020, 11, 2108–2480. [Google Scholar] [CrossRef]
- Dong, Z.; Manawasinghe, I.S.; Huang, Y.; Shu, Y.; Phillips, A.J.L.; Dissanayake, A.J.; Hyde, K.D.; Xiang, M.; Luo, M. Endophytic Diaporthe associated with Citrus grandis cv. Tomentosa in China. Front. Microbiol. 2021, 11, 3621. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Burgess, T.I.; Decock, C.A.; Dreyer, L.L.; Granke, L.L.; Guest, D.I.; Hardy, G.E.S.J.; Hausbeck, M.K.; et al. Fungal Planet description sheets: 107–127. Persoonia 2012, 28, 138–182. [Google Scholar] [CrossRef]
- Tan, Y.P.; Edwards, J.; Grice, K.R.E.; Shivas, R.G. Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Divers. 2013, 61, 251–260. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Jayawardena, R.S.; Li, H.L.; Zhou, Y.Y.; Zhang, W.; Phillips, A.J.L.; Wanasinghe, D.N.; Dissanayake, A.J.; Li, X.H.; Li, Y.H.; et al. Microfungi associated with Camellia sinensis: A case study of leaf and shoot necrosis on Tea in Fujian, China. Mycosphere 2021, 12, 430–518. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Dissanayake, A.J.; Li, X.; Liu, M.; Wanasinghe, D.N.; Xu, J.; Zhao, W.; Zhang, W.; Zhou, Y.; Hyde, K.D.; et al. High genetic diversity and species complexity of Diaporthe Associated with grapevine dieback in China. Front. Microbiol. 2019, 10, 1936. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef]
- Gao, Y.H.; Sun, W.; Su, Y.Y.; Cai, L. Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycol. Prog. 2014, 13, 111–121. [Google Scholar] [CrossRef]
- Cao, L.; Luo, D.; Lin, W.; Yang, Q.; Deng, X. Four new species of Diaporthe (Diaporthaceae, Diaporthales) from forest plants in China. MycoKeys 2022, 91, 25–47. [Google Scholar] [CrossRef]
- Long, H.; Zhang, Q.; Hao, Y.-Y.; Shao, X.-Q.; Wei, X.-X.; Hyde, K.D.; Wang, Y.; Zhao, D.-G. Diaporthe species in south-western China. MycoKeys 2019, 57, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K.; Mochizuki, K.; Takagi, D.; Ishida, K.; Sunada, A.; Ohkusu, K.; Kamei, K.; Hashimoto, A.; Tanaka, K. Identification and antifungal sensitivity of two new species of Diaporthe isolated. J. Infect. Chemother. 2019, 25, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.B.; Tang, A.M.C.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Zhang, W.; Liu, M.; Hyde, K.D.; Zhao, W.S.; Li, X.H.; Yan, J.Y. Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere 2017, 8, 533–549. [Google Scholar] [CrossRef]
- Gao, Y.; Duan, W.; Crous, P.W.; Cai, L. Diaporthe is paraphyletic. IMA Fungus 2017, 8, 153–187. [Google Scholar] [CrossRef]
- Udayanga, D.; Liu, X.; McKenzie, E.H.C.; Chukeatirote, E.; Hyde, K.D. Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogam. Mycol. 2012, 33, 295–309. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, N.; Tian, C.M. New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 2021, 77, 41–64. [Google Scholar] [CrossRef]
- Yang, Q.; Fan, X.L.; Du, Z.; Tian, C.M. Diaporthe species occurring on Senna bicapsularis in southern China, with descriptions of two new species. Phytotaxa 2017, 302, 145–155. [Google Scholar] [CrossRef]
- Doilom, M.; Dissanayake, A.J.; Wanasinghe, D.N.; Boonmee, S.; Liu, J.K.; Bhat, D.J.; Taylor, J.E.; Bahkali, A.H.; McKenzie, E.H.C.; Hyde, K.D. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2017, 82, 107–182. [Google Scholar] [CrossRef]
- Becker, K.; Wongkanoun, S.; Wessel, A.C.; Bills, G.F.; Stadler, M.; Luangsa-Ard, J.J. Phylogenetic and chemotaxonomic studies confirm the affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae. J. Fungi 2020, 6, 144. [Google Scholar] [CrossRef] [PubMed]
- Harms, K.; Surup, F.; Stadler, M.; Stchigel, A.M.; Marin-Felix, Y. Morinagadepsin, a depsipeptide from the fungus Morinagamyces vermicularis gen. et comb. nov. Microorganisms 2021, 9, 1191. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Wessel, A.C.; Luangsa-Ard, J.J.; Stadler, M. Viridistratins A–C, Antimicrobial and Cytotoxic Benzo[j]fluoranthenes from Stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef] [PubMed]
- Mulwa, L.S.; Jansen, R.; Praditya, D.F.; Mohr, K.I.; Okanya, P.W.; Wink, J.; Steinmann, E.; Stadler, M. Lanyamycin, a macrolide antibiotic from Sorangium cellulosum, strain Soce 481 (Myxobacteria). Beilstein J. Org. Chem. 2018, 14, 1554. [Google Scholar] [CrossRef]
- Huang, F.; Udayanga, D.; Wang, X.; Hou, X.; Mei, X.; Fu, Y.; Hyde, K.D.; Li, H. Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015, 119, 331–347. [Google Scholar] [CrossRef]
- Erper, I.; Turkkan, M.; Ozcan, M.; Luongo, L.; Belisario, A. Characterization of Diaporthe hongkongensis species causing stem-end rot on kiwifruit in Turkey. J. Plant Pathol. 2017, 99, 779–782. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Tsai, I.; Wang, J.Y.; Withee, P.; Tanjira, M.; Lin, S.R.; Suwannarach, N.; Kumla, J.; Elgorban, A.M.; Cheewangkoon, R. Molecular phylogenetic diversity and biological characterization of Diaporthe species associated with leaf spots of Camellia sinensis in Taiwan. Plants 2021, 10, 1434. [Google Scholar] [CrossRef]
- Du, Y.; Wang, X.; Guo, Y.; Xiao, F.; Peng, Y.; Hong, N.; Wang, G.; Du, Y.; Wang, X.; Guo, Y.; et al. Biological and molecular characterization of seven Diaporthe species associated with kiwifruit shoot blight and leaf spot in China. Phytopathol. Mediterr. 2021, 60, 177–198. [Google Scholar] [CrossRef]
- Cui, H.; Lin, Y.; Luo, M.; Lu, Y.; Huang, X.; She, Z. Diaporisoindoles A–C: Three isoprenylisoindole alkaloid derivatives from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. Org. Lett. 2017, 19, 5621–5624. [Google Scholar] [CrossRef]
- Luo, X.; Lin, X.; Tao, H.; Wang, J.; Li, J.; Yang, B.; Zhou, X.; Liu, Y. Isochromophilones A–F, cytotoxic chloroazaphilones from the marine mangrove endophytic fungus Diaporthe sp. SCSIO 41011. J. Nat. Prod. 2018, 81, 934–941. [Google Scholar] [CrossRef]
- Zhang, C.; Ondeyka, J.G.; Herath, K.B.; Guan, Z.; Collado, J.; Platas, G.; Pelaez, F.; Leavitt, P.S.; Gurnett, A.; Nare, B.; et al. Tenellones A and B from a Diaporthe sp.: Two highly substituted benzophenone inhibitors of parasite cGMP-dependent protein kinase activity. J. Nat. Prod. 2005, 68, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Hamill, R.L.; Higgens, C.E.; Boaz, H.E.; Gorman, M. The structure of beauvericin, a new depsipeptide antibiotic toxic to Artemia salina. Tetrahedron Lett. 1969, 10, 4255–4258. [Google Scholar] [CrossRef]
- Ola, A.R.B.; Aly, A.H.; Lin, W.; Wray, V.; Debbab, A. Structural revision and absolute configuration of lateritin. Tetrahedron Lett. 2014, 55, 6184–6187. [Google Scholar] [CrossRef]
- Caloni, F.; Fossati, P.; Anadón, A.; Bertero, A. Beauvericin: The beauty and the beast. Environ. Toxicol. Pharmacol. 2020, 75, 103349. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.Q.; Du, S.T.; Xiao, J.; Wang, D.C.; Han, W.B.; Zhang, Q.; Gao, J.M. Isolation and characterization of antifungal metabolites from the Melia azedarach-Associated Fungus Diaporthe eucalyptorum. J. Agric. Food Chem. 2020, 68, 2418–2425. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.; Li, J.; Huang, X.; Yan, T.; Cao, W.; Liu, H.; Long, Y.; She, Z. Diaporindenes A–D: Four unusual 2,3-dihydro-1H-indene analogues with anti-inflammatory activities from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. J. Org. Chem. 2018, 83, 11804–11813. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Z.; Chen, Y.; Tan, H.; Li, H.; Li, S.; Guo, H.; Huang, Z.; Gao, X.; Liu, H.; et al. Lithocarols A–F, six tenellone derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508. Bioorg. Chem. 2019, 87, 728–735. [Google Scholar] [CrossRef]
- Xu, J.; Tan, H.; Chen, Y.; Li, S.; Guo, H.; Huang, Z.; Li, H.; Gao, X.; Liu, H.; Zhang, W. Lithocarpinols A and B, a pair of diastereomeric antineoplastic tenellone derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508. Chin. Chem. Lett. 2019, 30, 439–442. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Y.; Liu, Z.; Li, S.; Wang, Y.; Ren, Y.; Liu, H.; Zhang, W. Lithocarpins E–G, potent anti-tumor tenellone-macrolides from the deep-sea fungus Phomopsis lithocarpus FS508. Chin. J. Chem. 2021, 39, 1104–1112. [Google Scholar] [CrossRef]
- Xu, J.; Tan, H.; Chen, Y.; Li, S.; Huang, Z.; Guo, H.; Li, H.; Gao, X.; Liu, H.; Zhang, W. Lithocarpins A–D: Four tenellone-macrolide conjugated [4 + 2] hetero-adducts from the deep-sea derived fungus Phomopsis lithocarpus FS508. Org. Chem. Front. 2018, 5, 1792–1797. [Google Scholar] [CrossRef]
- Liu, H.B.; Liu, Z.M.; Chen, Y.C.; Tan, H.B.; Li, S.N.; Li, D.L.; Liu, H.X.; Zhang, W.M. Cytotoxic diaporindene and tenellone derivatives from the fungus Phomopsis lithocarpus. Chin. J. Nat. Med. 2021, 19, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.N.; Stadler, M.; Fournier, J.; Tomita, A.; Hashimoto, T. Cohaerins C–F, four azaphilones from the xylariaceous fungus Annulohypoxylon cohaerens. Tetrahedron 2006, 62, 6349–6354. [Google Scholar] [CrossRef]
- Panthama, N.; Kanokmedhakul, S.; Kanokmedhakul, K.; Soytong, K. Chemical constituents from the fungus Chaetomium cupreum RY202. Arch. Pharm. Res. 2015, 38, 585–590. [Google Scholar] [CrossRef]
- Omura, S.; Tanaka, H.; Matsuzaki, K.; Ikeda, H.; Masuma, R. Isochromophilones I and II, novel inhibitors against gp120-CD4 binding from Penicillium sp. J. Antibiot. 1993, 46, 1908–1911. [Google Scholar] [CrossRef]
- Whalley, W.B.; Ferguson, G.; Marsh, W.C.; Restivo, R.J. The Chemistry of Fungi. Part LXVIII. The Absolute Configuration of (+)-Sclerotiorin and of the Azaphilones. J. Chem. Soc. Perkin Trans. 1 1976, 13, 1366–1369. [Google Scholar] [CrossRef]
- Chen, C.; Manchand, S.; Gray, R.W.; Whalley, W.B. The chemistry of fungi. Part LXIV. The structure of monascin: The relative stereochemistry of the azaphilones. J. Chem. Soc. C Org. 1969, 3577–3579. [Google Scholar] [CrossRef]
- Steyn, P.S.; Vleggaar, R. The structure of dihydrodeoxy-8-epi-austdiol and the absolute configuration of the azaphilones. J. Chem. Soc. Perkin Trans. 1 1974, 2, 204–206. [Google Scholar] [CrossRef]
- Liu, H.; Xie, H.; Li, C.; Wang, L.; Chen, Q.; Ouyang, X.; Yan, C. Diaporisoindole B reduces lipid accumulation in THP-1 macrophage cells via MAPKS and PPARΓ-LXRᾳ pathways and promotes the reverse cholesterol transport by upregulating SR-B1 and LDLR in HepG2 cells. J. Nat. Prod. 2022, 85, 2769–2778. [Google Scholar] [CrossRef]
- Sood, S.; Sandhu, S.S.; Mukherjee, T.K. Pharmacological and therapeutic potential of beauvericin: A Short Review. J. Proteom. Bioinform. 2017, 10, 18–23. [Google Scholar] [CrossRef]
- Wu, Q.; Patocka, J.; Nepovimova, E.; Kuca, K. A review on the synthesis and bioactivity aspects of beauvericin, a Fusarium mycotoxin. Front. Pharmacol. 2018, 9, 1338. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).