Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Antifungal Susceptibility Testing
2.3. Cell Labeling
2.4. Imaging Flow Data Acquisition
2.5. Chemical Library and Drug Treatment for Imaging Flow Analysis
2.6. Data Analysis
3. Results
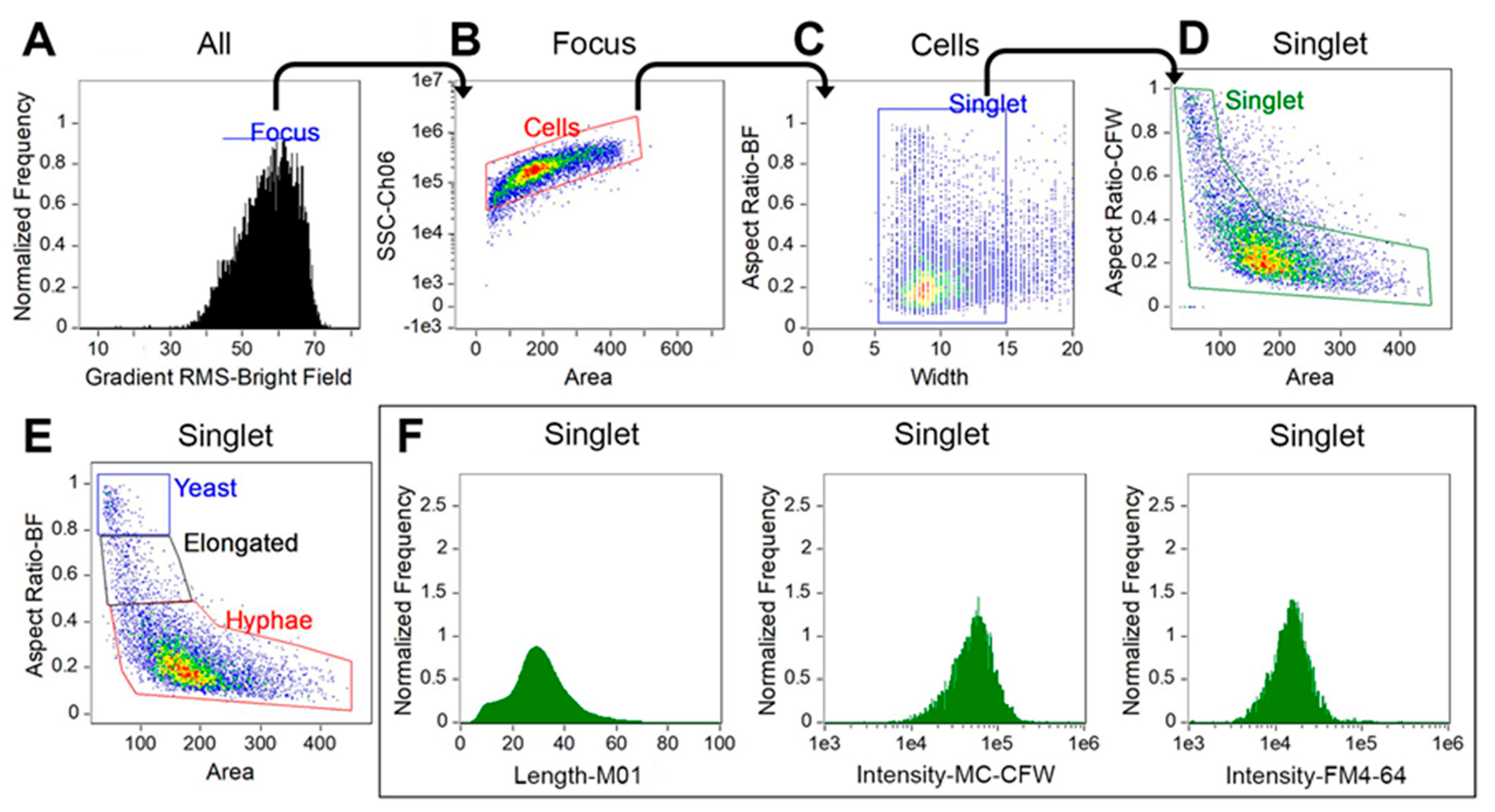
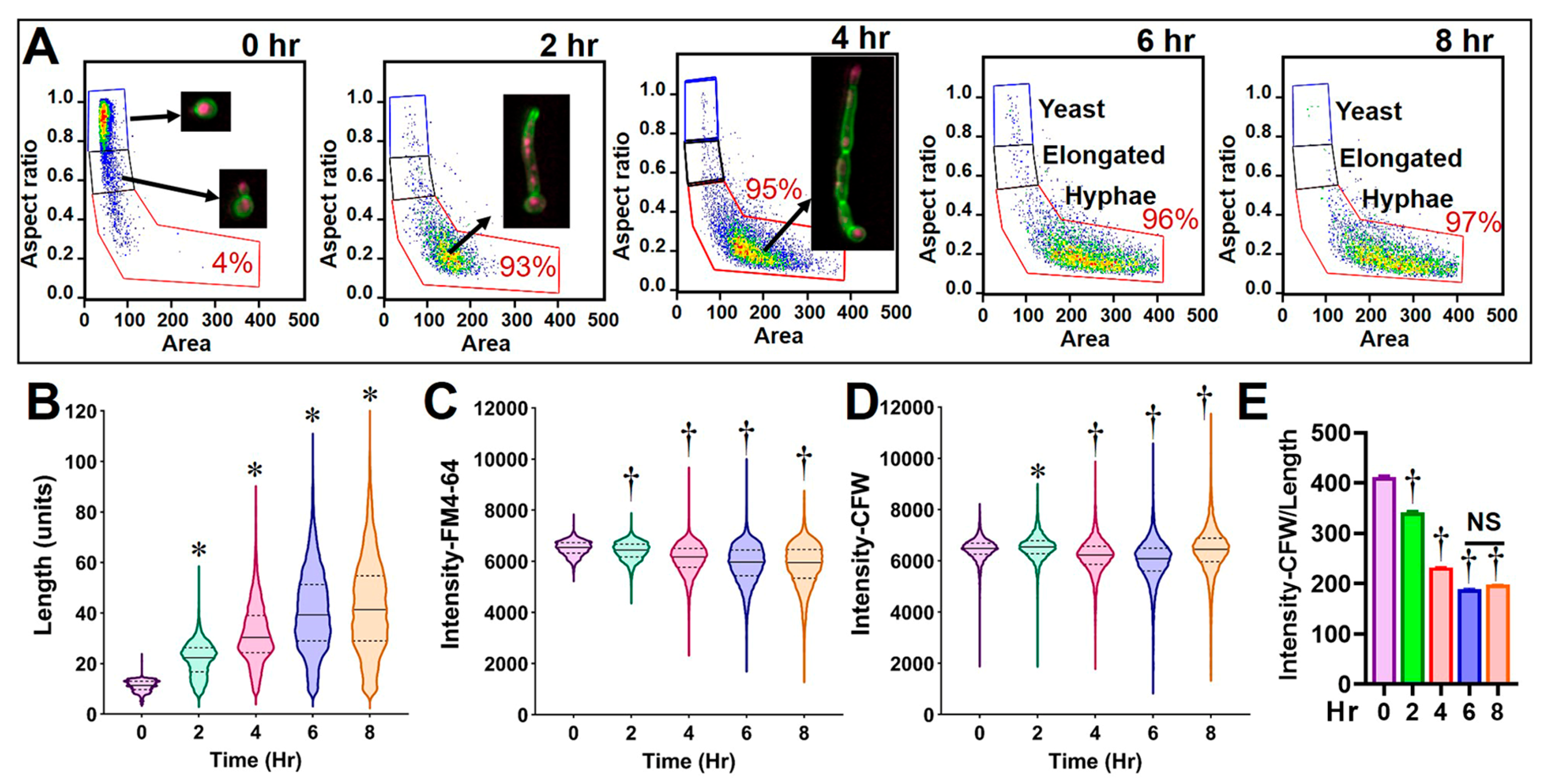
3.1. Optimization of Culture Conditions and FCP Analysis for C. albicans
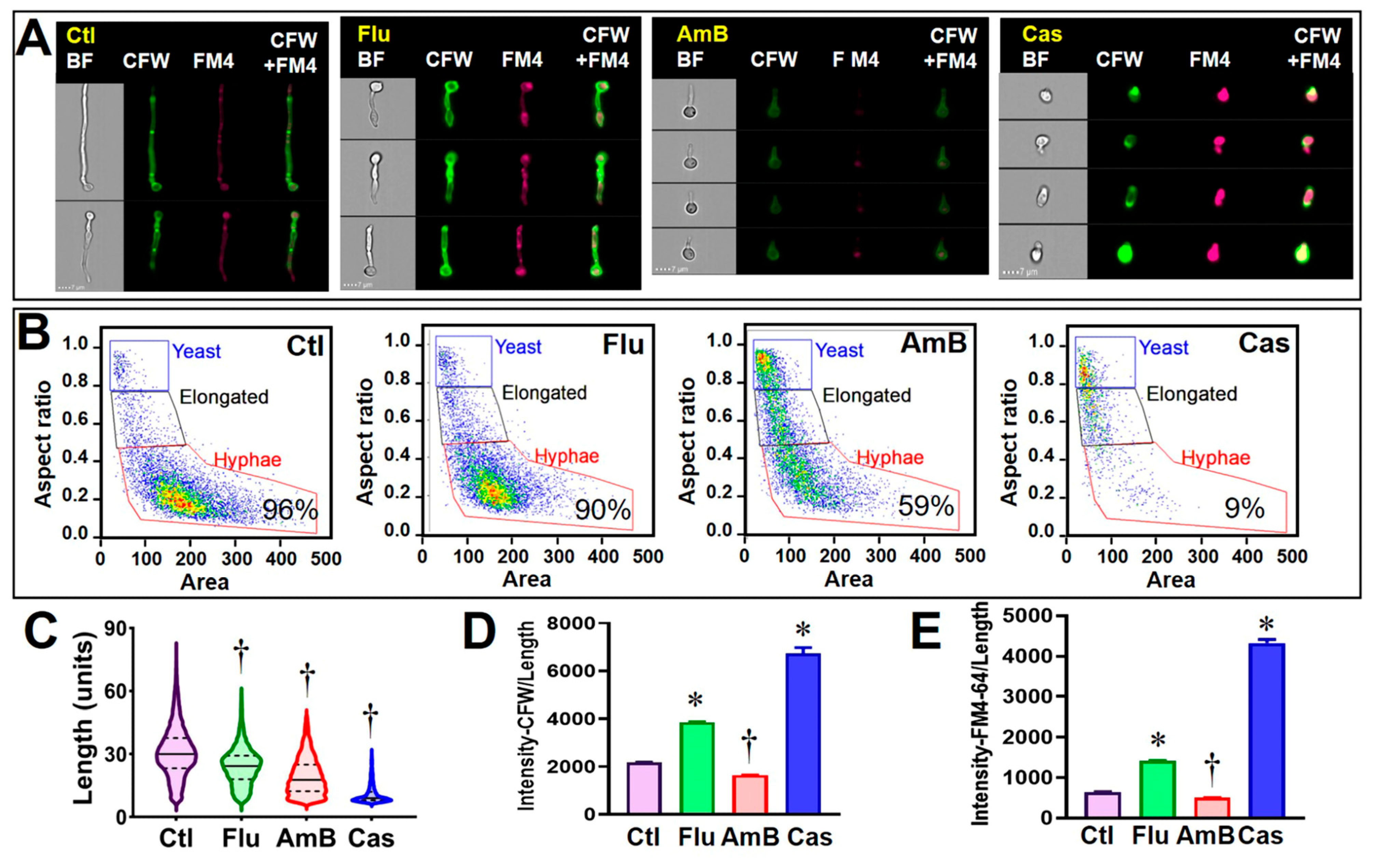
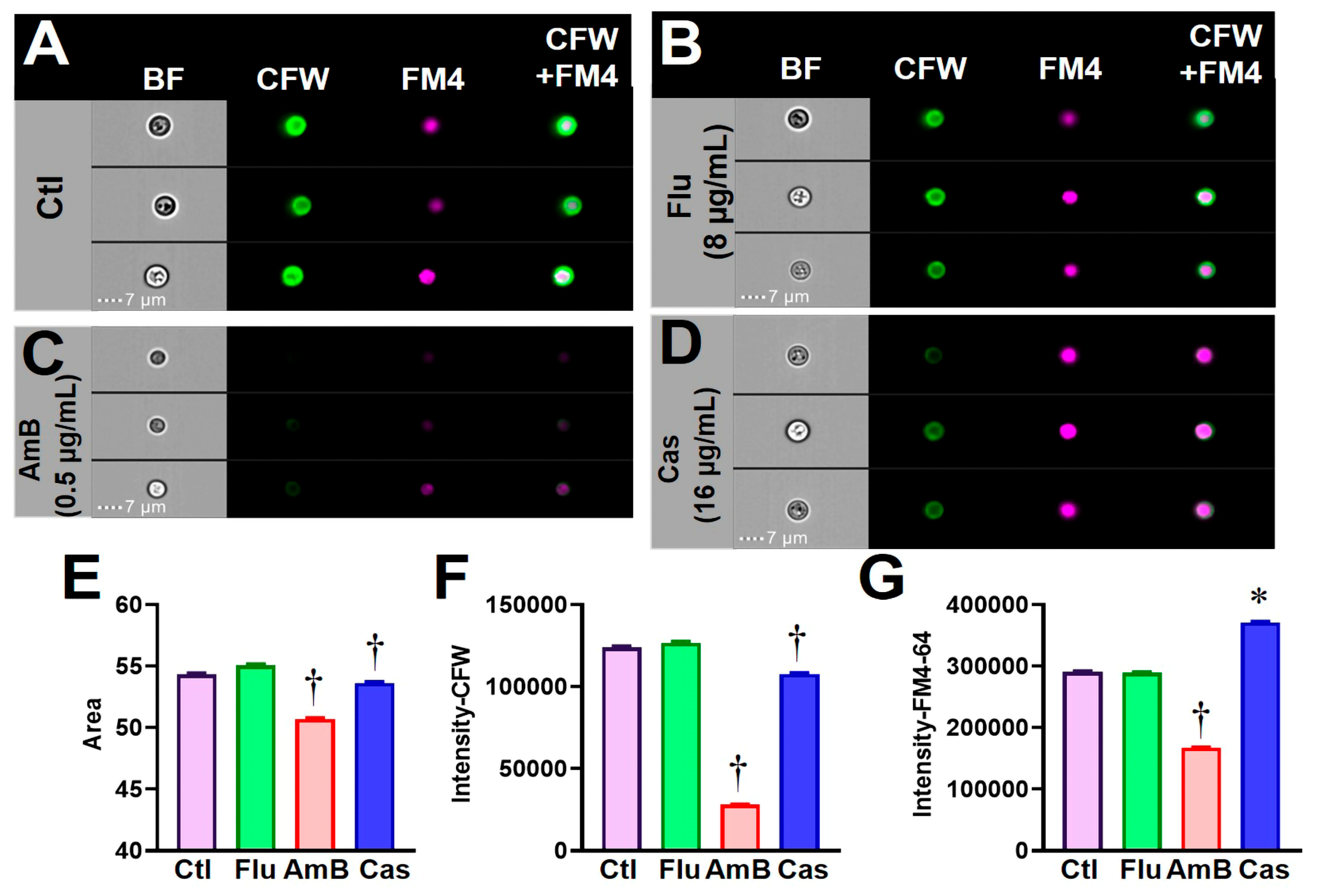
3.2. Validating C. albicans FCP after Exposure to Clinical Antifungal Drugs
3.3. The Newly Established FCP Method Is Useful for Profiling Fungal Cellular States
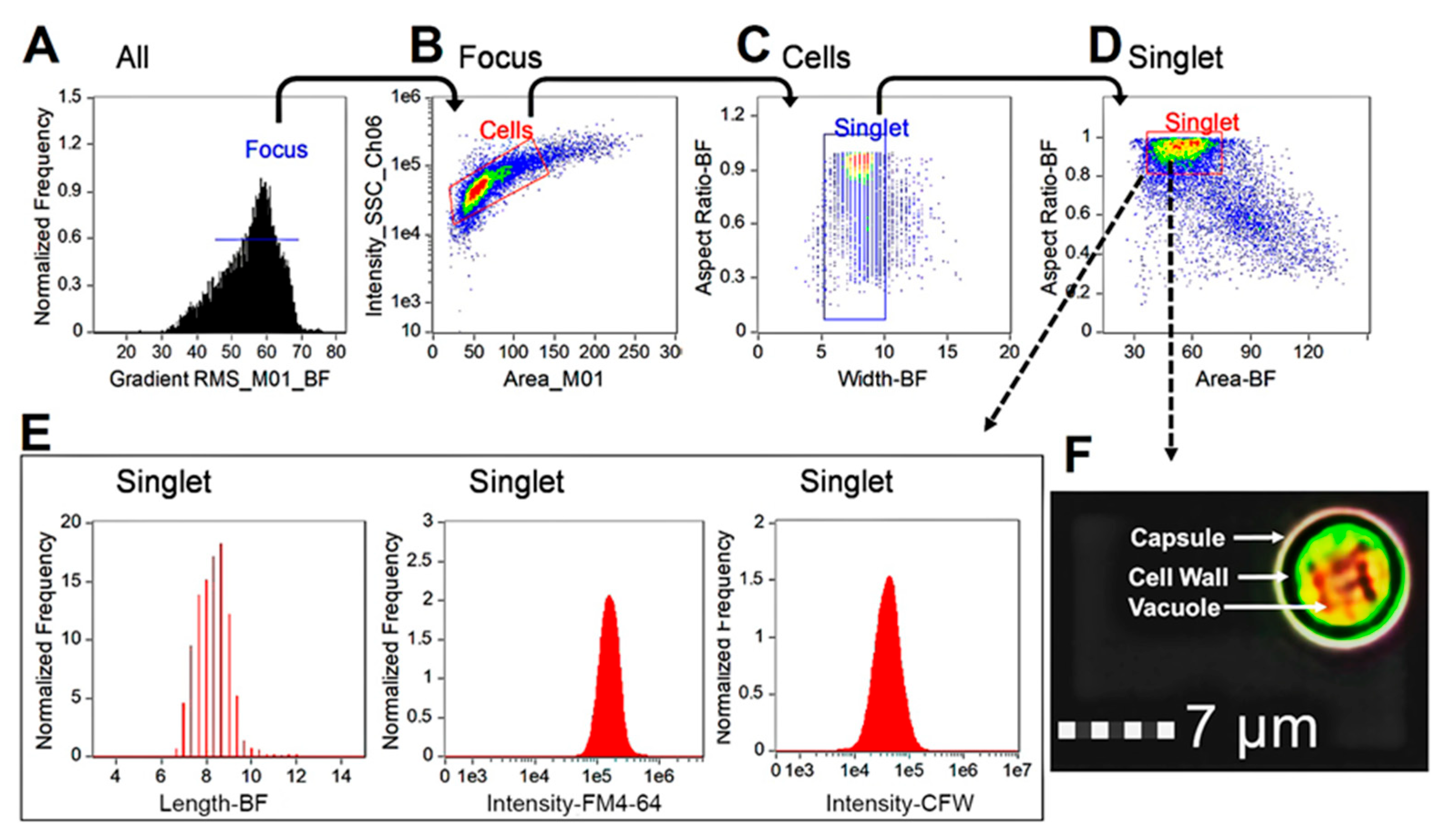
3.4. Profiling Cytological States of C. neoformans after Exposure to Clinical Antifungal Drugs
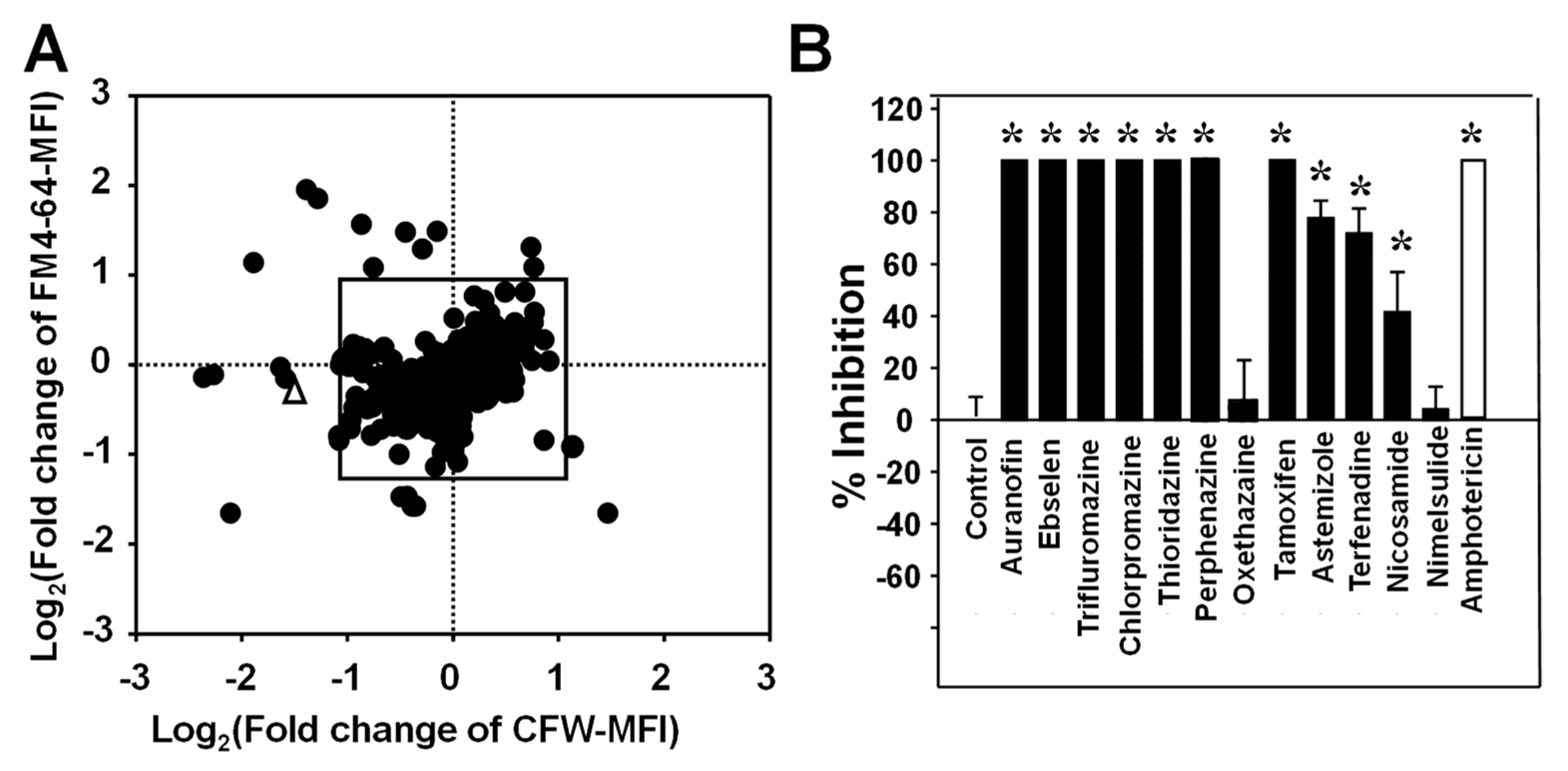
3.5. Potential Application of the FCP Method for Screening a Drug Library to Identify Compounds with Antifungal Activity
| Drug Type | Drug Name | MFI Fold Change | MICs (µg/mL) | Butts et al. [19] Reported Hits | Reported Drugs against Other Fungal Species and Cited References | ||
|---|---|---|---|---|---|---|---|
| CFW | FM4-64 | ||||||
| Phenothiazine antipsychotic | Thioridazine | −3.79 | 2.27 | 1.77 (50%) | Y | Paracoccidioides Cryptococcus | [20] [19] |
| Chlorpromazine | −2.49 | 3.72 | 2.28 (50%) | N | Aspergillus Candida Cryptococcus | [21] [22] [23] | |
| Trifluoperazine | −2.68 | 3.99 | 3.36 (50%) | Y | Aspergillus Candida Cryptococcus | [22] [23] [24] | |
| Perphenazine | −1.40 | 2.87 | 1.20 (50%) 3.50 (90%) | N | Cryptococcus | This study | |
| Anti-inflammatory | Auranofin | −4.57 | −2.05 | 2.41 (50%) | N | Aspergillus Blastomyces Candida Cryptococcus | [25] [26] [27] |
| Ebselen | −5.40 | −3.95 | 0.88 (50%) | N | Aspergillus Candida Cryptococcus | [28] [29] [26] | |
| Nimesulide | −1.38 | −2.68 | 9.83 (50%) | N | Aspergillus, Candida Cryptococcus | [30] | |
| Antihistamine | Astemizole | 1.66 | 2.19 | 0.43 (50%) | N | Cryptococcus | [31] |
| Terfenadine | 1.63 | 2.56 | 7.78 (50%) | N | Candida | [32] | |
| Antihelmintic | Niclosamide | −2.16 | −1.74 | 6.42 (50%) | N | Candida | [33] |
| Antineoplastic | Tamoxifen | −1.14 | 2.90 | 1.21 (50%) | Y | Candida Cryptococcus | [34] [35] |
| Anesthetic | Oxethazaine | −1.87 | 3.05 | 11.1 (50%) >20.0 (90%) | N | Cryptococcus | This study |
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56 (Suppl. S1), i5–i11. [Google Scholar] [CrossRef] [PubMed]
- Gintjee, T.J.; Donnelley, M.A.; Thompson, G.R., 3rd. Aspiring Antifungals: Review of Current Antifungal Pipeline Developments. J. Fungi 2020, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Rauseo, A.M.; Coler-Reilly, A.; Larson, L.; Spec, A. Hope on the Horizon: Novel Fungal Treatments in Development. Open Forum Infect. Dis. 2020, 7, ofaa016. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.I.; Villmann, M. Echinocandins in the management of invasive fungal infections, Part 2. Am. J. Health Syst. Pharm. 2006, 63, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.I.; Villmann, M. Echinocandins in the management of invasive fungal infections, Part 1. Am. J. Health Syst. Pharm. 2006, 63, 1693–1703. [Google Scholar] [CrossRef]
- Hamill, R.J. Amphotericin B formulations: A comparative review of efficacy and toxicity. Drugs 2013, 73, 919–934. [Google Scholar] [CrossRef]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jimenez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef]
- Sanglard, D. Emerging Threats in Antifungal-Resistant Fungal Pathogens. Front. Med. 2016, 3, 11. [Google Scholar] [CrossRef]
- Mitchison, T.J. Small-molecule screening and profiling by using automated microscopy. Chembiochem 2005, 6, 33–39. [Google Scholar] [CrossRef]
- Perlman, Z.E.; Slack, M.D.; Feng, Y.; Mitchison, T.J.; Wu, L.F.; Altschuler, S.J. Multidimensional drug profiling by automated microscopy. Science 2004, 306, 1194–1198. [Google Scholar] [CrossRef]
- Bray, M.-A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef]
- Willis, C.; Nyffeler, J.; Harrill, J. Phenotypic Profiling of Reference Chemicals across Biologically Diverse Cell Types Using the Cell Painting Assay. SLAS Discov. Adv. Sci. Drug Discov. 2020, 25, 755–769. [Google Scholar] [CrossRef]
- Gustafsdottir, S.M.; Ljosa, V.; Sokolnicki, K.L.; Anthony Wilson, J.; Walpita, D.; Kemp, M.M.; Petri Seiler, K.; Carrel, H.A.; Golub, T.R.; Schreiber, S.L.; et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS ONE 2013, 8, e80999. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Machado, M.-J.; Benahmed, F.H.; Conville, P.; Shawar, R.M.; Patel, J.; Brown, A.C. FDA-CDC Antimicrobial Resistance Isolate Bank: A Publicly Available Resource To Support Research, Development, and Regulatory Requirements. J. Clin. Microbiol. 2018, 56, e01415–e01417. [Google Scholar] [CrossRef]
- Wayne, P. CLSI, Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi. In Approved Standard: CLSI Document M38–A2; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Harrington, B.J.; Hageage, J.G.J. Calcofluor White: A Review of its Uses and Applications in Clinical Mycology and Parasitology. Lab. Med. 2003, 34, 361–367. [Google Scholar] [CrossRef]
- Zheng, B.; Wu, J.N.; Schober, W.; Lewis, D.E.; Vida, T. Isolation of yeast mutants defective for localization of vacuolar vital dyes. Proc. Natl. Acad. Sci. USA 1998, 95, 11721–11726. [Google Scholar] [CrossRef]
- Wall, G.; Chaturvedi, A.K.; Wormley, F.L.; Wiederhold, N.P.; Patterson, H.P.; Patterson, T.F.; Lopez-Ribot, J.L. Screening a Repurposing Library for Inhibitors of Multidrug-Resistant Candida auris Identifies Ebselen as a Repositionable Candidate for Antifungal Drug Development. Antimicrob. Agents Chemother. 2018, 62, e01084-18. [Google Scholar] [CrossRef]
- Butts, A.; DiDone, L.; Koselny, K.; Baxter, B.K.; Chabrier-Rosello, Y.; Wellington, M.; Krysan, D.J. A repurposing approach identifies off-patent drugs with fungicidal cryptococcal activity, a common structural chemotype, and pharmacological properties relevant to the treatment of cryptococcosis. Eukaryot. Cell 2013, 12, 278–287. [Google Scholar] [CrossRef]
- Jabes, D.L.; de Freitas Oliveira, A.C.; Alencar, V.C.; Menegidio, F.B.; Reno, D.L.; Santos, D.S.; Barbosa, D.A.; Vilas Boas, R.O.; de Oliveira Rodrigues Cunha, R.L.; Rodrigues, T.; et al. Thioridazine inhibits gene expression control of the cell wall signaling pathway (CWI) in the human pathogenic fungus Paracoccidioides brasiliensis. Mol. Genet. Genom. MGG 2016, 291, 1347–1362. [Google Scholar] [CrossRef]
- Vitale, R.G.; Afeltra, J.; Meis, J.F.; Verweij, P.E. Activity and post antifungal effect of chlorpromazine and trifluopherazine against Aspergillus, Scedosporium and zygomycetes. Mycoses 2007, 50, 270–276. [Google Scholar] [CrossRef]
- Galgoczy, L.; Bacsi, A.; Homa, M.; Viragh, M.; Papp, T.; Vagvolgyi, C. In vitro antifungal activity of phenothiazines and their combination with amphotericin B against different Candida species. Mycoses 2011, 54, e737–e743. [Google Scholar] [CrossRef] [PubMed]
- Eilam, Y.; Polacheck, I.; Ben-Gigi, G.; Chernichovsky, D. Activity of phenothiazines against medically important yeasts. Antimicrob. Agents Chemother. 1987, 31, 834–836. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Casadevall, A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to the melanin-binding compounds trifluoperazine and chloroquine. Antimicrob. Agents Chemother. 1996, 40, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.B.; RajaMuthiah, R.; Souza, A.C.; Eatemadpour, S.; Rossoni, R.D.; Santos, D.A.; Junqueira, J.C.; Rice, L.B.; Mylonakis, E. Inhibition of bacterial and fungal pathogens by the orphaned drug auranofin. Future Med. Chem. 2016, 8, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Eldesouky, H.E.; Mohammad, H.; Pascuzzi, P.E.; Avramova, L.; Hazbun, T.R.; Seleem, M.N. Ebselen exerts antifungal activity by regulating glutathione (GSH) and reactive oxygen species (ROS) production in fungal cells. Biochim. Biophys. Acta 2017, 1861, 3002–3010. [Google Scholar] [CrossRef]
- Wiederhold, N.P.; Patterson, T.F.; Srinivasan, A.; Chaturvedi, A.K.; Fothergill, A.W.; Wormley, F.L.; Ramasubramanian, A.K.; Lopez-Ribot, J.L. Repurposing auranofin as an antifungal: In vitro activity against a variety of medically important fungi. Virulence 2017, 8, 138–142. [Google Scholar] [CrossRef]
- Orie, N.N.; Warren, A.R.; Basaric, J.; Lau-Cam, C.; Pietka-Ottlik, M.; Mlochowski, J.; Billack, B. In vitro assessment of the growth and plasma membrane H+-ATPase inhibitory activity of ebselen and structurally related selenium- and sulfur-containing compounds in Candida albicans. J. Biochem. Mol. Toxicol. 2017, 31, e21892. [Google Scholar] [CrossRef]
- Wall, G.; Herrera, N.; Lopez-Ribot, J.L. Repositionable Compounds with Antifungal Activity against Multidrug Resistant Candida auris Identified in the Medicines for Malaria Venture’s Pathogen Box. J. Fungi 2019, 5, 92. [Google Scholar] [CrossRef]
- de Matos, R.F.; Mendonca, L.C.V.; da Silva Souza, K.G.; Fonseca, A.A.D.; Costa, E.M.S.; de Lima, M.V.D.; Vieira, J.; de Brito, M.; Monteiro, M.C. Nimesulide inhibits pathogenic fungi: PGE2-dependent mechanisms. Folia Microbiol. 2017, 62, 169–174. [Google Scholar] [CrossRef]
- Vu, K.; Gelli, A. Astemizole and an analogue promote fungicidal activity of fluconazole against Cryptococcus neoformans var. grubii and Cryptococcus gattii. Med. Mycol. 2010, 48, 255–262. [Google Scholar] [CrossRef]
- Dennis, E.K.; Garneau-Tsodikova, S. Synergistic combinations of azoles and antihistamines against Candida species in vitro. Med. Mycol. 2018, 57, 874–884. [Google Scholar] [CrossRef]
- Garcia, C.; Burgain, A.; Chaillot, J.; Pic, E.; Khemiri, I.; Sellam, A. A phenotypic small-molecule screen identifies halogenated salicylanilides as inhibitors of fungal morphogenesis, biofilm formation and host cell invasion. Sci. Rep. 2018, 8, 11559. [Google Scholar] [CrossRef]
- Beggs, W.H. Anti-Candida activity of the anti-cancer drug tamoxifen. Res. Commun. Chem. Pathol. Pharmacol. 1993, 80, 125–128. [Google Scholar]
- Dolan, K.; Montgomery, S.; Buchheit, B.; Didone, L.; Wellington, M.; Krysan, D.J. Antifungal activity of tamoxifen: In vitro and in vivo activities and mechanistic characterization. Antimicrob. Agents Chemother. 2009, 53, 3337–3346. [Google Scholar] [CrossRef]
- Maguire, O.; Collins, C.; O’Loughlin, K.; Miecznikowski, J.; Minderman, H. Quantifying nuclear p65 as a parameter for NF-κB activation: Correlation between ImageStream cytometry, microscopy, and Western blot. Cytom. Part A 2011, 79 Pt A, 461–469. [Google Scholar] [CrossRef]
- George, T.C.; Fanning, S.L.; Fitzgeral-Bocarsly, P.; Medeiros, R.B.; Highfill, S.; Shimizu, Y.; Hall, B.E.; Frost, K.; Basiji, D.; Ortyn, W.E.; et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J. Immunol. Methods 2006, 311, 117–129. [Google Scholar] [CrossRef]
- Cerveira, J.; Begum, J.; Di Marco Barros, R.; van der Veen, A.G.; Filby, A. An imaging flow cytometry-based approach to measuring the spatiotemporal calcium mobilisation in activated T cells. J. Immunol. Methods 2015, 423, 120–130. [Google Scholar] [CrossRef]
- Filby, A.; Perucha, E.; Summers, H.; Rees, P.; Chana, P.; Heck, S.; Lord, G.M.; Davies, D. An imaging flow cytometric method for measuring cell division history and molecular symmetry during mitosis. Cytom. Part A 2011, 79 Pt A, 496–506. [Google Scholar] [CrossRef]
- Patterson, J.O.; Swaffer, M.; Filby, A. An Imaging Flow Cytometry-based approach to analyse the fission yeast cell cycle in fixed cells. Methods 2015, 82, 74–84. [Google Scholar] [CrossRef]
- Chia, W.N.; Lee, Y.Q.; Tan, K.S.-W. Imaging flow cytometry for the screening of compounds that disrupt the Plasmodium falciparum digestive vacuole. Methods 2017, 112, 211–220. [Google Scholar] [CrossRef]
- Barteneva, N.S.; Fasler-Kan, E.; Vorobjev, I.A. Imaging flow cytometry: Coping with heterogeneity in biological systems. J. Histochem. Cytochem. 2012, 60, 723–733. [Google Scholar] [CrossRef] [PubMed]
- McFarlin, B.K.; Gary, M.A. Flow cytometry what you see matters: Enhanced clinical detection using image-based flow cytometry. Methods 2017, 112, 1–8. [Google Scholar] [CrossRef]
- Georgopapadakou, N.H. Antifungals: Mechanism of action and resistance, established and novel drugs. Curr. Opin. Microbiol. 1998, 1, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Demers, E.G.; Biermann, A.R.; Masonjones, S.; Crocker, A.W.; Ashare, A.; Stajich, J.E.; Hogan, D.A. Evolution of drug resistance in an antifungal-naive chronic Candida lusitaniae Infection. Proc. Natl. Acad. Sci. USA 2018, 115, 12040–12045. [Google Scholar] [CrossRef] [PubMed]
- Vanden Bossche, H.; Dromer, F.; Improvisi, I.; Lozano-Chiu, M.; Rex, J.H.; Sanglard, D. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 1998, 36 (Suppl. S1), 119–128. [Google Scholar]
- Fernandez-Silva, F.; Lackner, M.; Capilla, J.; Mayayo, E.; Sutton, D.; Castanheira, M.; Fothergill, A.W.; Lass-Florl, C.; Guarro, J. In vitro antifungal susceptibility of Candida glabrata to caspofungin and the presence of FKS mutations correlate with treatment response in an immunocompromised murine model of invasive infection. Antimicrob. Agents Chemother. 2014, 58, 3646–3649. [Google Scholar] [CrossRef]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017, 13, e1006290. [Google Scholar] [CrossRef]
- Klepser, M.E.; Wolfe, E.J.; Pfaller, M.A. Antifungal pharmacodynamic characteristics of fluconazole and amphotericin B against Cryptococcus neoformans. J. Antimicrob. Chemother. 1998, 41, 397–401. [Google Scholar] [CrossRef]
- Kordalewska, M.; Lee, A.; Park, S.; Berrio, I.; Chowdhary, A.; Zhao, Y.; Perlin, D.S. Understanding Echinocandin Resistance in the Emerging Pathogen Candida auris. Antimicrob. Agents Chemother. 2018, 62, e00238-18. [Google Scholar] [CrossRef]
- Maligie, M.A.; Selitrennikoff, C.P. Cryptococcus neoformans resistance to echinocandins:(1,3) β-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 2005, 49, 2851–2856. [Google Scholar] [CrossRef]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Kwon-Chung, K.J. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 2009, 53, 2804–2815. [Google Scholar] [CrossRef]
- Butts, A.; Martin, J.A.; DiDone, L.; Bradley, E.K.; Mutz, M.; Krysan, D.J. Structure-activity relationships for the antifungal activity of selective estrogen receptor antagonists related to tamoxifen. PLoS ONE 2015, 10, e0125927. [Google Scholar] [CrossRef]
- Ngan, N.T.T.; Mai, N.T.H.; Tung, N.L.N.; Lan, N.P.H.; Tai, L.T.H.; Phu, N.H.; Chau, N.V.V.; Binh, T.Q.; Hung, L.Q.; Beardsley, J.; et al. A randomized open label trial of tamoxifen combined with amphotericin B and fluconazole for cryptococcal meningitis. Wellcome Open Res. 2019, 4, 8. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McMahon, C.L.; Esqueda, M.; Yu, J.-J.; Wall, G.; Romo, J.A.; Vila, T.; Chaturvedi, A.; Lopez-Ribot, J.L.; Wormley, F.; Hung, C.-Y. Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development. J. Fungi 2023, 9, 722. https://doi.org/10.3390/jof9070722
McMahon CL, Esqueda M, Yu J-J, Wall G, Romo JA, Vila T, Chaturvedi A, Lopez-Ribot JL, Wormley F, Hung C-Y. Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development. Journal of Fungi. 2023; 9(7):722. https://doi.org/10.3390/jof9070722
Chicago/Turabian StyleMcMahon, Courtney L., Marisol Esqueda, Jieh-Juen Yu, Gina Wall, Jesus A. Romo, Taissa Vila, Ashok Chaturvedi, Jose L. Lopez-Ribot, Floyd Wormley, and Chiung-Yu Hung. 2023. "Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development" Journal of Fungi 9, no. 7: 722. https://doi.org/10.3390/jof9070722
APA StyleMcMahon, C. L., Esqueda, M., Yu, J.-J., Wall, G., Romo, J. A., Vila, T., Chaturvedi, A., Lopez-Ribot, J. L., Wormley, F., & Hung, C.-Y. (2023). Development of an Imaging Flow Cytometry Method for Fungal Cytological Profiling and Its Potential Application in Antifungal Drug Development. Journal of Fungi, 9(7), 722. https://doi.org/10.3390/jof9070722








