Candida auris in Dog Ears
Abstract
1. Introduction
2. Material and Methods
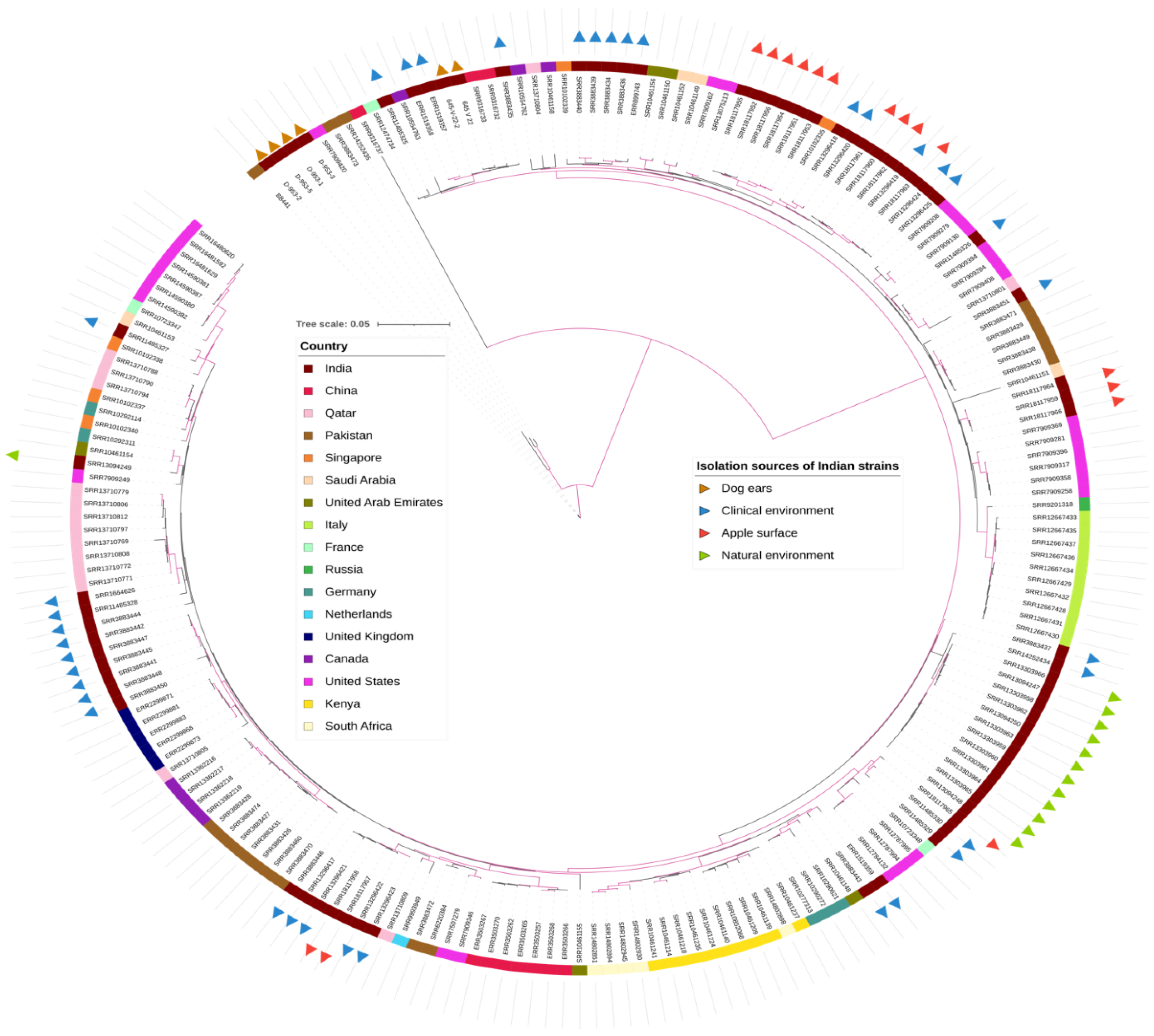
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J. Population genomic analyses reveal evidence for limited recombination in the superbug Candida auris in nature. Comput. Struct. Biotechnol. J. 2022, 20, 3030–3040. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: The World Health Organization (WHO) fungal priority pathogens list in response to emerging fungal pathogens during the COVID-19 pandemic. Med. Sci. Monit. 2022, 28, e939088. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Singh, P.; Wang, Y.; Yadav, A.; Pawar, K.; Singh, A.; Padmavati, G.; Xu, J.; Chowdhary, A. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio 2021, 12, e03181-20. [Google Scholar] [CrossRef] [PubMed]
- Escandón, P. Novel environmental niches for Candida auris: Isolation from a coastal habitat in Colombia. J. Fungi 2022, 8, 748. [Google Scholar] [CrossRef]
- Yadav, A.; Jain, K.; Wang, Y.; Pawar, K.; Kaur, H.; Sharma, K.K.; Tripathy, V.; Singh, A.; Xu, J.; Chowdhary, A. Candida auris on apples: Diversity and clinical significance. mBio 2022, 13, e0051822. [Google Scholar] [CrossRef]
- Chavez, J.; Iverson, T.; Hergert, J.; Oakeson, K.; LaCross, N.; Njoku, C.; Gorzalski, A.; Gerrity, D. Candida auris discovery through community wastewater surveillance during healthcare outbreak, Nevada, USA, 2022. Emerg. Infect. Dis. 2023, 29, 422–425. [Google Scholar]
- Irinyi, L.; Roper, M.; Malik, R.; Meyer, W. Finding a needle in a haystack—In silico search for environmental traces of Candida auris. Jpn. J. Infect. Dis. 2022, 75, 490–495. [Google Scholar] [CrossRef]
- Seyedmousavi, S.; Bosco, S.M.G.; de Hoog, S.; Ebel, F.; Elad, D.; Gomes, R.R.; Jacobsen, I.D.; Jensen, H.E.; Martel, A.; Mignon, B.; et al. Fungal infections in animals: A patchwork of different situations. Med. Mycol. 2018, 56 (Suppl. S1), S165–S187. [Google Scholar] [CrossRef]
- Yadav, A.; Singh, A.; Wang, Y.; Haren, M.H.V.; Singh, A.; de Groot, T.; Meis, J.F.; Xu, J.; Chowdhary, A. Colonisation and transmission dynamics of Candida auris among chronic respiratory diseases patients hospitalised in a chest hospital, Delhi, India: A comparative analysis of whole genome sequencing and microsatellite typing. J. Fungi 2021, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [PubMed]
- Gweon, H.S.; Oliver, A.; Taylor, J.; Booth, T.; Gibbs, M.; Read, D.S.; Griffiths, R.I.; Schonrogge, K. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 2015, 6, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- M27-A3; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 3rd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008.
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–2017) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef]
- M100; Performance Standards for Antimicrobial Disk Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Sharma, C.; Kumar, N.; Pandey, R.; Meis, J.F.; Chowdhary, A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016, 13, 77–82. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees, in 2010 Gateway Computing Environments Workshop (GCE); IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Abastabar, M.; Haghani, I.; Ahangarkani, F.; Rezai, M.S.; Taghizadeh Armaki, M.; Roodgari, S.; Kiakojuri, K.; Al-Hatmi, A.M.S.; Meis, J.F.; Badali, H. Candida auris otomycosis in Iran and review of recent literature. Mycoses 2019, 62, 101–105. [Google Scholar] [CrossRef]
- Chow, N.A.; de Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential Fifth Clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef] [PubMed]
- Spruijtenburg, B.; Badali, H.; Abastabar, M.; Mirhendi, H.; Khodavaisy, S.; Sharifisooraki, J.; Taghizadeh Armaki, M.; de Groot, T.; Meis, J.F. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes Infect. 2022, 11, 2405–2411. [Google Scholar] [CrossRef] [PubMed]
- Puigdemont, A.; D’Andreano, S.; Ramió-Lluch, L.; Cuscó, A.; Francino, O.; Brazis, P. Effect of an anti-inflammatory pomegranate otic treatment on the clinical evolution and microbiota profile of dogs with otitis externa. Vet. Dermatol. 2021, 32, 158-e37. [Google Scholar] [CrossRef] [PubMed]
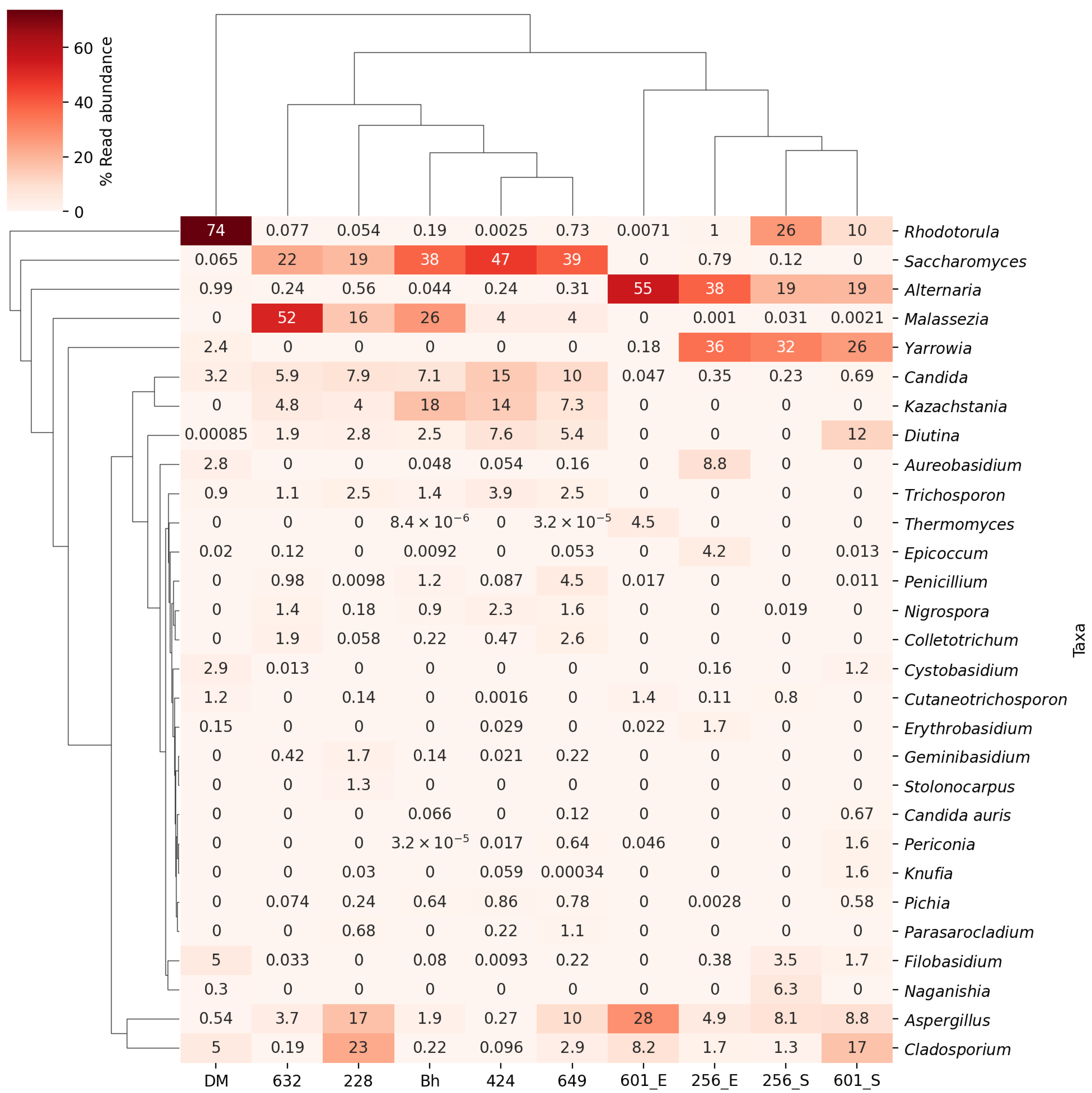
| Disease Categories in Dogs (Numbers) | Period of Sampling | Yeasts | Bacterial Isolation (Body Site, No. of Dogs Positive for the Given Species) | |
|---|---|---|---|---|
| Culture (Body Site, No. of Dogs Positive for the Given Species) | Amplicon Sequencing (ITS Reads for C. auris/Total Reads Matched to Fungal ITS; Percentage of ITS Reads) | |||
| Group IA Chronic skin disease with otitis externa (n = 8), | 27-09-22 to 17-04-23 | C. auris (E *, n = 1) (D1) @ C. rugosa (E, n = 2), C. krusei (E, n = 3) (D2) @ C. lusitaniae (E, n = 1) C. glabrata (S #, n = 1) (D3) @ | C. auris from ear swab of D1 dog (34394/27920633; 0.12%) C. auris from ear swab of D2 dog (39577/59853350; 0.06%) C. auris from skin swab of D3 dog (3231/479708; 0.67%) | A. baumanii (E, n = 1) S. pseudointermedius (S, n = 1) |
| Group IB Chronic skin disease (n = 44), | 14-08-22 to 17-04-23 | C. auris (E, n = 1) (D4) @ M. pachydermatis (E, n = 11; S, n = 3) C. tropicalis (E, n = 8; S, n = 3), T. asahii (E, n = 7), C. glabrata (E, n = 6, S; n = 1), C. rugosa (E, n =1), C. albicans (E, n = 1), | ND | A. baumanii (E, n =15; S, n = 2) S. pseudointermedius (E, n = 9; S, n = 2) S. intermedius, (E, n = 1; S, n = 1) K. pneumoniae (E, n = 1) S. simulans (S, n = 1) S. schleiferi (S, n = 1) P. aeruginosa (E, n = 1) |
| Group II Negative for skin and ear infections (n = 35), | 11-05-22 to 02-09-22 | C. tropicalis (E, n = 3; S, n = 3) M. pachydermatis (E, n = 3; S, n = 1) C. krusei (E, n = 2) C. lusitaniae (S, n = 2) C. parapsilosis (E, n = 1) | ND | A. baumanii (E, n = 1; S, n = 2), S. pseudointermedius (E, n = 6), P. aeruginosa (E, n = 3; S, n = 1) S. schleiferi (E, n = 1; S, n = 1) S. intermedius (S, n = 2), K. pneumoniae (S, n = 1), E. faecalis (S, n = 1), B. cereus (E, n = 1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, A.; Wang, Y.; Jain, K.; Panwar, V.A.R.; Kaur, H.; Kasana, V.; Xu, J.; Chowdhary, A. Candida auris in Dog Ears. J. Fungi 2023, 9, 720. https://doi.org/10.3390/jof9070720
Yadav A, Wang Y, Jain K, Panwar VAR, Kaur H, Kasana V, Xu J, Chowdhary A. Candida auris in Dog Ears. Journal of Fungi. 2023; 9(7):720. https://doi.org/10.3390/jof9070720
Chicago/Turabian StyleYadav, Anamika, Yue Wang, Kusum Jain, Vijay Amrit Raj Panwar, Hardeep Kaur, Vikas Kasana, Jianping Xu, and Anuradha Chowdhary. 2023. "Candida auris in Dog Ears" Journal of Fungi 9, no. 7: 720. https://doi.org/10.3390/jof9070720
APA StyleYadav, A., Wang, Y., Jain, K., Panwar, V. A. R., Kaur, H., Kasana, V., Xu, J., & Chowdhary, A. (2023). Candida auris in Dog Ears. Journal of Fungi, 9(7), 720. https://doi.org/10.3390/jof9070720







