Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Genotyping
2.2. PCR and Sequencing of the Group I Introns Present in the cob and cox1 Genes of Cryptococcus
2.3. Characterization of the Amplified Group I Introns and Search for Endonuclease Genes
2.4. Phylogenetic Analyses
2.5. Statistical Analyses
3. Results
3.1. Size and Sequence Polymorphism of the Group I Introns from Mitochondrial Genes cob and cox1 among Cryptococcus neoformans and Cryptococcus gattii Genotypes
3.2. Distribution of Group I Introns in the Mitogenome of Cryptococcus neoformans and Cryptococcus gattii
3.3. Phylogenetic Analyses
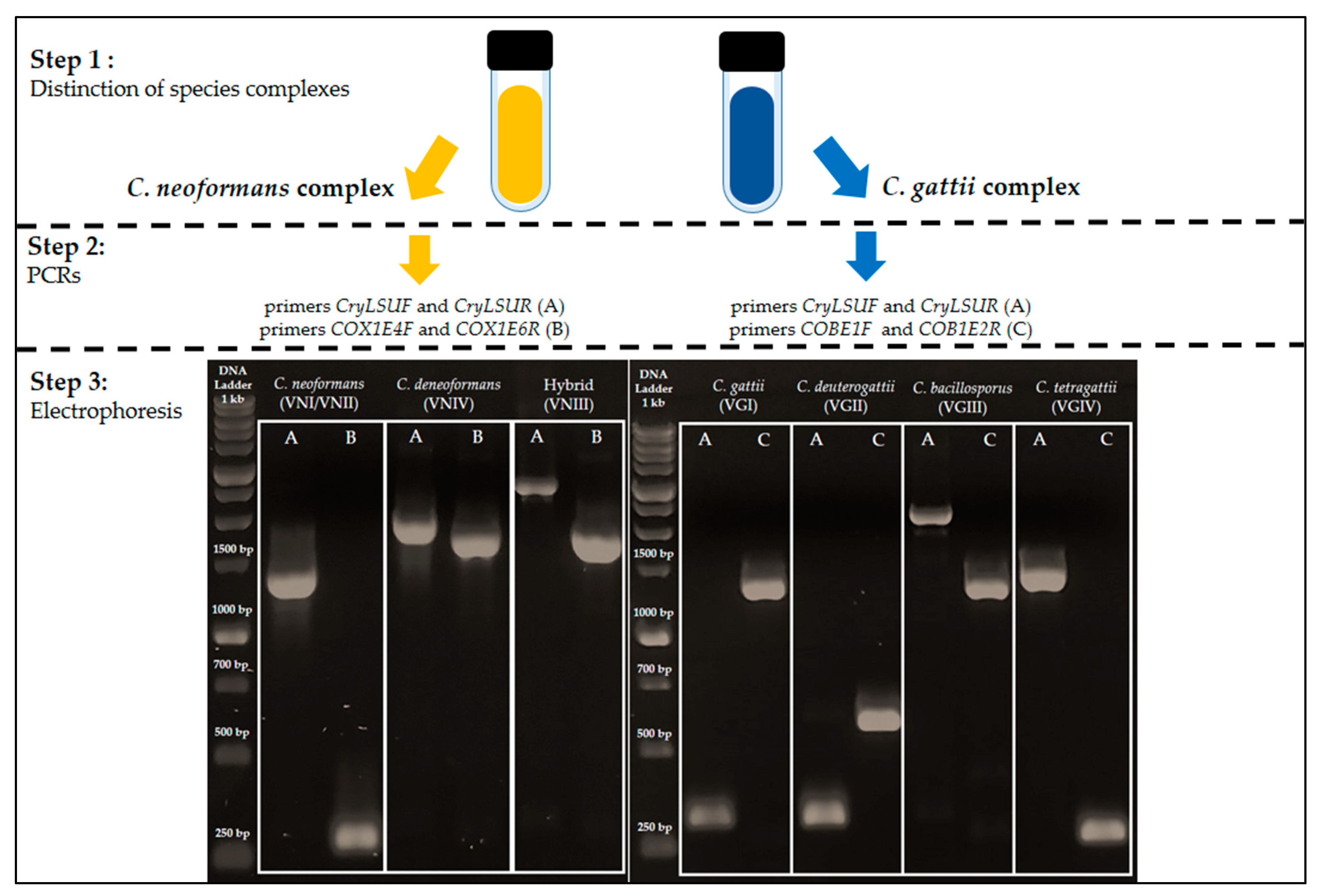
3.4. Proposal of PCR Reactions for Differentiation of Cryptococcus Species Based on Mitochondrial Group I Introns
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rathore, S.S.; Sathiyamoorthy, J.; Lalitha, C.; Ramakrishnan, J. A Holistic Review on Cryptococcus neoformans. Microb. Pathog. 2022, 166, 105521. [Google Scholar] [CrossRef] [PubMed]
- Beardsley, J.; Dao, A.; Keighley, C.; Garnham, K.; Halliday, C.; Chen, S.C.-A.; Sorrell, T.C. What’s New in Cryptococcus gattii: From Bench to Bedside and Beyond. J. Fungi 2022, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Rajasingham, R.; Govender, N.P.; Jordan, A.; Loyse, A.; Shroufi, A.; Denning, D.W.; Meya, D.B.; Chiller, T.M.; Boulware, D.R. The Global Burden of HIV-Associated Cryptococcal Infection in Adults in 2020: A Modelling Analysis. Lancet Infect. Dis. 2022, 22, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Theelen, B.; Diaz, M.; Fell, J.W.; Hop, W.C.J.; Abeln, E.C.A.; Dromer, F.; Meyer, W. Hybrid Genotypes in the Pathogenic Yeast Cryptococcus neoformans. Microbiology 2001, 147, 891–907. [Google Scholar] [CrossRef]
- Meyer, W.; Castañeda, A.; Jackson, S.; Huynh, M.; Castañeda, E.; IberoAmerican Cryptococcal Study Group. Molecular Typing of IberoAmerican Cryptococcus neoformans Isolates. Emerg. Infect. Dis. 2003, 9, 189–195. [Google Scholar] [CrossRef]
- Hagen, F.; Khayhan, K.; Theelen, B.; Kolecka, A.; Polacheck, I.; Sionov, E.; Falk, R.; Parnmen, S.; Lumbsch, H.T.; Boekhout, T. Recognition of Seven Species in the Cryptococcus gattii/Cryptococcus neoformans Species Complex. Fungal Genet. Biol. 2015, 78, 16–48. [Google Scholar] [CrossRef]
- Farrer, R.A.; Chang, M.; Davis, M.J.; van Dorp, L.; Yang, D.-H.; Shea, T.; Sewell, T.R.; Meyer, W.; Balloux, F.; Edwards, H.M.; et al. A New Lineage of Cryptococcus gattii (VGV) Discovered in the Central Zambezian Miombo Woodlands. mBio 2019, 10, e02306–e02319. [Google Scholar] [CrossRef]
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.-H.; et al. Clinical Practice Guidelines for the Management of Cryptococcal Disease: 2010 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 291–322. [Google Scholar] [CrossRef]
- Sorrell, T.C. Cryptococcus neoformans Variety gattii. Med. Mycol. 2001, 39, 155–168. [Google Scholar] [CrossRef]
- Speed, B.; Dunt, D. Clinical and Host Differences between Infections with the Two Varieties of Cryptococcus neoformans. Clin. Infect. Dis. 1995, 21, 28–34; discussion 35–36. [Google Scholar] [CrossRef]
- Trilles, L.; Meyer, W.; Wanke, B.; Guarro, J.; Lazéra, M. Correlation of Antifungal Susceptibility and Molecular Type within the Cryptococcus neoformans/C. gattii Species Complex. Med. Mycol. 2012, 50, 328–332. [Google Scholar] [CrossRef]
- Chong, H.S.; Dagg, R.; Malik, R.; Chen, S.; Carter, D. In Vitro Susceptibility of the Yeast Pathogen Cryptococcus to Fluconazole and Other Azoles Varies with Molecular Genotype. J. Clin. Microbiol. 2010, 48, 4115–4120. [Google Scholar] [CrossRef]
- Hagen, F.; Illnait-Zaragozi, M.-T.; Bartlett, K.H.; Swinne, D.; Geertsen, E.; Klaassen, C.H.W.; Boekhout, T.; Meis, J.F. In Vitro Antifungal Susceptibilities and Amplified Fragment Length Polymorphism Genotyping of a Worldwide Collection of 350 Clinical, Veterinary, and Environmental Cryptococcus gattii Isolates. Antimicrob. Agents Chemother. 2010, 54, 5139–5145. [Google Scholar] [CrossRef]
- Iqbal, N.; DeBess, E.E.; Wohrle, R.; Sun, B.; Nett, R.J.; Ahlquist, A.M.; Chiller, T.; Lockhart, S.R.; Cryptococcus gattii Public Health Working Group. Correlation of Genotype and in Vitro Susceptibilities of Cryptococcus gattii Strains from the Pacific Northwest of the United States. J. Clin. Microbiol. 2010, 48, 539–544. [Google Scholar] [CrossRef]
- Karahan, Z.C.; Saran, B.; Yenice, S.; Ağırbaşlı, H.; Arıkan Akan, O.; Tekeli, A. [25S intron analysis followed by restriction enzyme digestion performed for genotyping Candida albicans isolates]. Mikrobiyol. Bul. 2012, 46, 257–265. [Google Scholar]
- Haugen, P.; Simon, D.M.; Bhattacharya, D. The Natural History of Group I Introns. Trends Genet. 2005, 21, 111–119. [Google Scholar] [CrossRef]
- Lang, B.F.; Laforest, M.-J.; Burger, G. Mitochondrial Introns: A Critical View. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef]
- Robbins, J.B.; Smith, D.; Belfort, M. Redox-Responsive Zinc Finger Fidelity Switch in Homing Endonuclease and Intron Promiscuity in Oxidative Stress. Curr. Biol. 2011, 21, 243–248. [Google Scholar] [CrossRef]
- Mercure, S.; Montplaisir, S.; Lemay, G. Correlation between the Presence of a Self-Splicing Intron in the 25S RDNA of C. albicans and Strains Susceptibility to 5-Fluorocytosine. Nucleic Acids Res. 1993, 21, 6020–6027. [Google Scholar] [CrossRef]
- Liu, Y.; Tidwell, R.R.; Leibowitz, M.J. Inhibition of in Vitro Splicing of a Group I Intron of Pneumocystis carinii. J. Eukaryot. Microbiol. 1994, 41, 31–38. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Pilch, D.S.; Leibowitz, M.J. Pentamidine Inhibits Catalytic Activity of Group I Intron Ca.LSU by Altering RNA Folding. Nucleic Acids Res. 2002, 30, 2961–2971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bell, A.; Perlman, P.S.; Leibowitz, M.J. Pentamidine Inhibits Mitochondrial Intron Splicing and Translation in Saccharomyces cerevisiae. RNA 2000, 6, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Miletti, K.E.; Leibowitz, M.J. Pentamidine Inhibition of Group I Intron Splicing in Candida albicans Correlates with Growth Inhibition. Antimicrob. Agents Chemother. 2000, 44, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the Emerging Threat of Antifungal Resistance to Human Health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.E.E.S.; Arantes, T.D.; Fernandes, J.A.L.; Ferreira, L.C.; Romero, H.; Bosco, S.M.G.; Oliveira, M.T.B.; Del Negro, G.M.B.; Theodoro, R.C. Polymorphism in Mitochondrial Group I Introns among Cryptococcus neoformans and Cryptococcus gattii Genotypes and Its Association with Drug Susceptibility. Front. Microbiol. 2018, 9, 86. [Google Scholar] [CrossRef]
- Megarioti, A.H.; Kouvelis, V.N. The Coevolution of Fungal Mitochondrial Introns and Their Homing Endonucleases (GIY-YIG and LAGLIDADG). Genome Biol. Evol. 2020, 12, 1337–1354. [Google Scholar] [CrossRef]
- Trilles, L.; dos Lazéra, M.S.; Wanke, B.; Oliveira, R.V.; Barbosa, G.G.; Nishikawa, M.M.; Morales, B.P.; Meyer, W. Regional Pattern of the Molecular Types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Memórias Do Inst. Oswaldo Cruz 2008, 103, 455–462. [Google Scholar] [CrossRef]
- Litter, J.; Keszthelyi, A.; Hamari, Z.; Pfeiffer, I.; Kucsera, J. Differences in Mitochondrial Genome Organization of Cryptococcus neoformans Strains. Antonie Van Leeuwenhoek 2005, 88, 249–255. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Eddy, S.R.; Durbin, R. RNA Sequence Analysis Using Covariance Models. Nucleic Acids Res. 1994, 22, 2079–2088. [Google Scholar] [CrossRef]
- Mullineux, S.T.; Willows, K.; Hausner, G. Evolutionary dynamics of the mS952 intron: A novel mitochondrial group II intron encoding a LAGLIDADG homing endonuclease gene. J. Mol. Evol. 2011, 72, 433–449. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Altamirano, S.; Jackson, K.M.; Nielsen, K. The interplay of phenotype and genotype in Cryptococcus neoformans disease. Biosci. Rep. 2020, 40, BSR20190337. [Google Scholar] [CrossRef]
- Min, K.H.; Kwon-Chung, K.J. The Biochemical Basis for the Distinction between the Two Cryptococcus neoformans varieties with CGB Medium. Zentralbl Bakteriol. Mikrobiol. Hyg. A 1986, 261, 471–480. [Google Scholar] [CrossRef]
- Feng, X.; Fu, X.; Ling, B.; Wang, L.; Liao, W.; Yao, Z. Development of a Singleplex PCR Assay for Rapid Identification and Differentiation of Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. neoformans, Cryptococcus gattii, and Hybrids. J. Clin. Microbiol. 2013, 51, 1920–1923. [Google Scholar] [CrossRef]
- McTaggart, L.; Richardson, S.E.; Seah, C.; Hoang, L.; Fothergill, A.; Zhang, S.X. Rapid Identification of Cryptococcus neoformans var. grubii, C. neoformans var. neoformans, and C. gattii by Use of Rapid Biochemical Tests, Differential Media, and DNA Sequencing. J. Clin. Microbiol. 2011, 49, 2522–2527. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J. Mitochondrial Genome Polymorphisms in the Human Pathogenic Fungus Cryptococcus neoformans. Front. Microbiol. 2020, 11, 706. [Google Scholar] [CrossRef]
- Wang, Z.; Wilson, A.; Xu, J. Mitochondrial DNA Inheritance in the Human Fungal Pathogen Cryptococcus gattii. Fungal Genet. Biol. 2015, 75, 1–10. [Google Scholar] [CrossRef]
- Yan, Z.; Hull, C.M.; Heitman, J.; Sun, S.; Xu, J. SXI1alpha Controls Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Curr. Biol. 2004, 14, R743–R744. [Google Scholar] [CrossRef]
- Yan, Z.; Li, Z.; Yan, L.; Yu, Y.; Cheng, Y.; Chen, J.; Liu, Y.; Gao, C.; Zeng, L.; Sun, X.; et al. Deletion of the Sex-Determining Gene SXI1α Enhances the Spread of Mitochondrial Introns in Cryptococcus neoformans. Mob. DNA 2018, 9, 24. [Google Scholar] [CrossRef]
- Birky, C.W. Uniparental Inheritance of Mitochondrial and Chloroplast Genes: Mechanisms and Evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 11331–11338. [Google Scholar] [CrossRef] [PubMed]
- Law, R.; Hutson, V. Intracellular Symbionts and the Evolution of Uniparental Cytoplasmic Inheritance. Proc. Biol. Sci. 1992, 248, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Sex Is Dangerous in a World of Potential Symbionts or the Basis of Selection for Uniparental Inheritance. J. Theor. Biol. 1982, 97, 367–369. [Google Scholar] [CrossRef]
- Lambowitz, A.M.; Belfort, M. Introns as Mobile Genetic Elements. Annu. Rev. Biochem. 1993, 62, 587–622. [Google Scholar] [CrossRef]
- Belfort, M. Two for the Price of One: A Bifunctional Intron-Encoded DNA Endonuclease-RNA Maturase. Genes. Dev. 2003, 17, 2860–2863. [Google Scholar] [CrossRef]
- Edgell, D.R.; Chalamcharla, V.R.; Belfort, M. Learning to Live Together: Mutualism between Self-Splicing Introns and Their Hosts. BMC Biol. 2011, 9, 22. [Google Scholar] [CrossRef]
- Novo, M.; Bigey, F.; Beyne, E.; Galeote, V.; Gavory, F.; Mallet, S.; Cambon, B.; Legras, J.-L.; Wincker, P.; Casaregola, S.; et al. Eukaryote-to-Eukaryote Gene Transfer Events Revealed by the Genome Sequence of the Wine Yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 2009, 106, 16333–16338. [Google Scholar] [CrossRef]
- Fox, T.D. Mitochondrial Protein Synthesis, Import, and Assembly. Genetics 2012, 192, 1203–1234. [Google Scholar] [CrossRef]
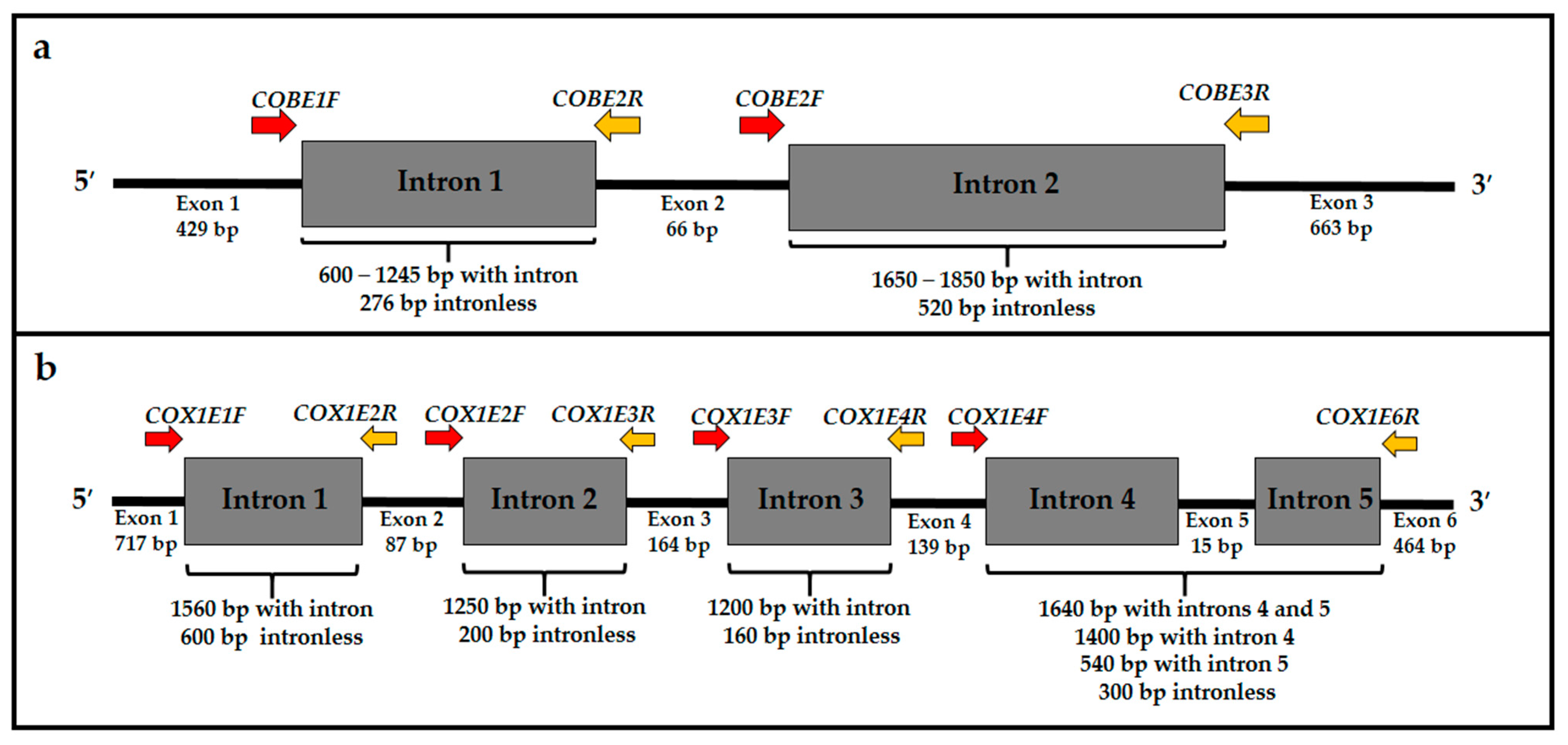

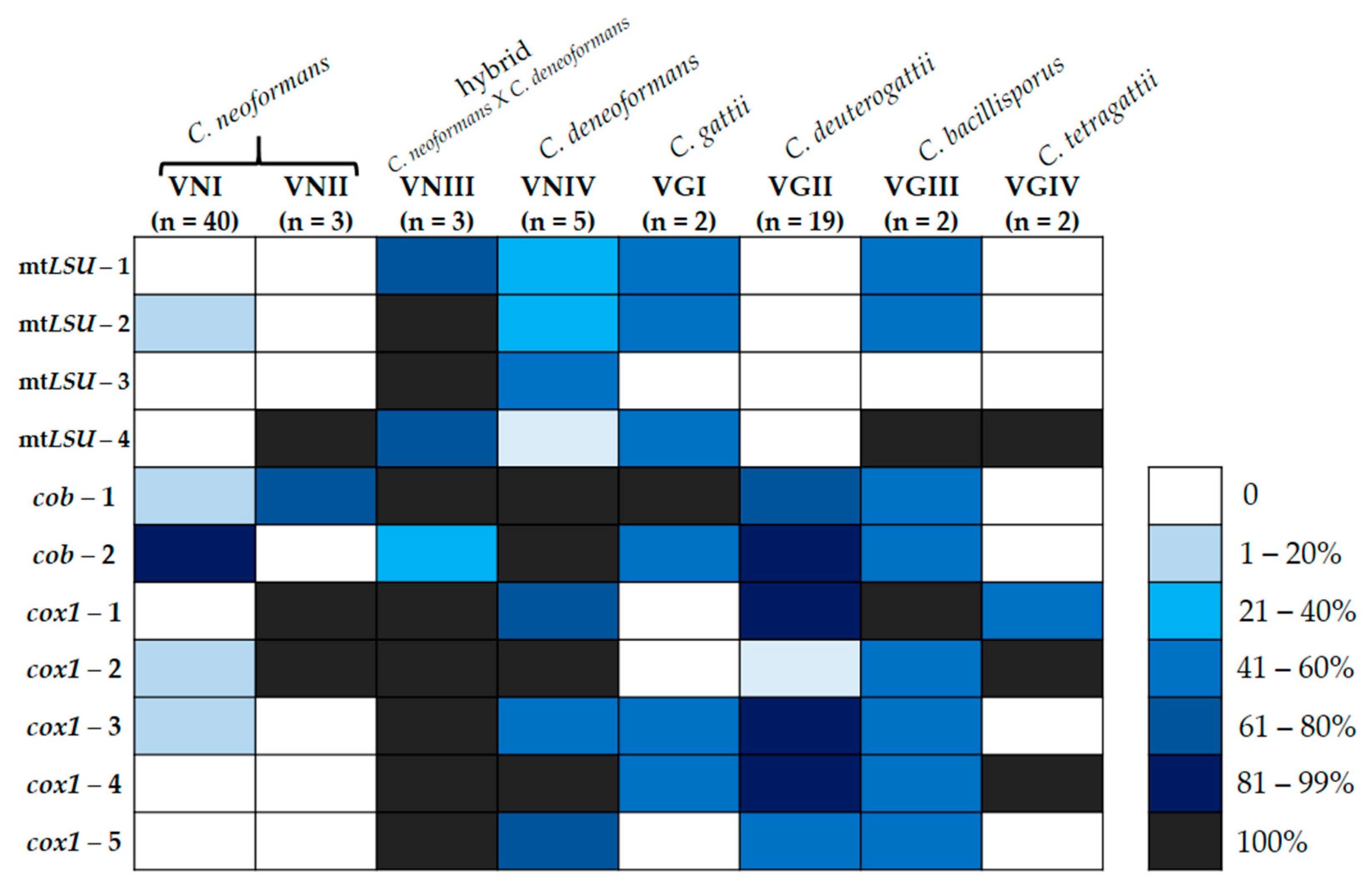

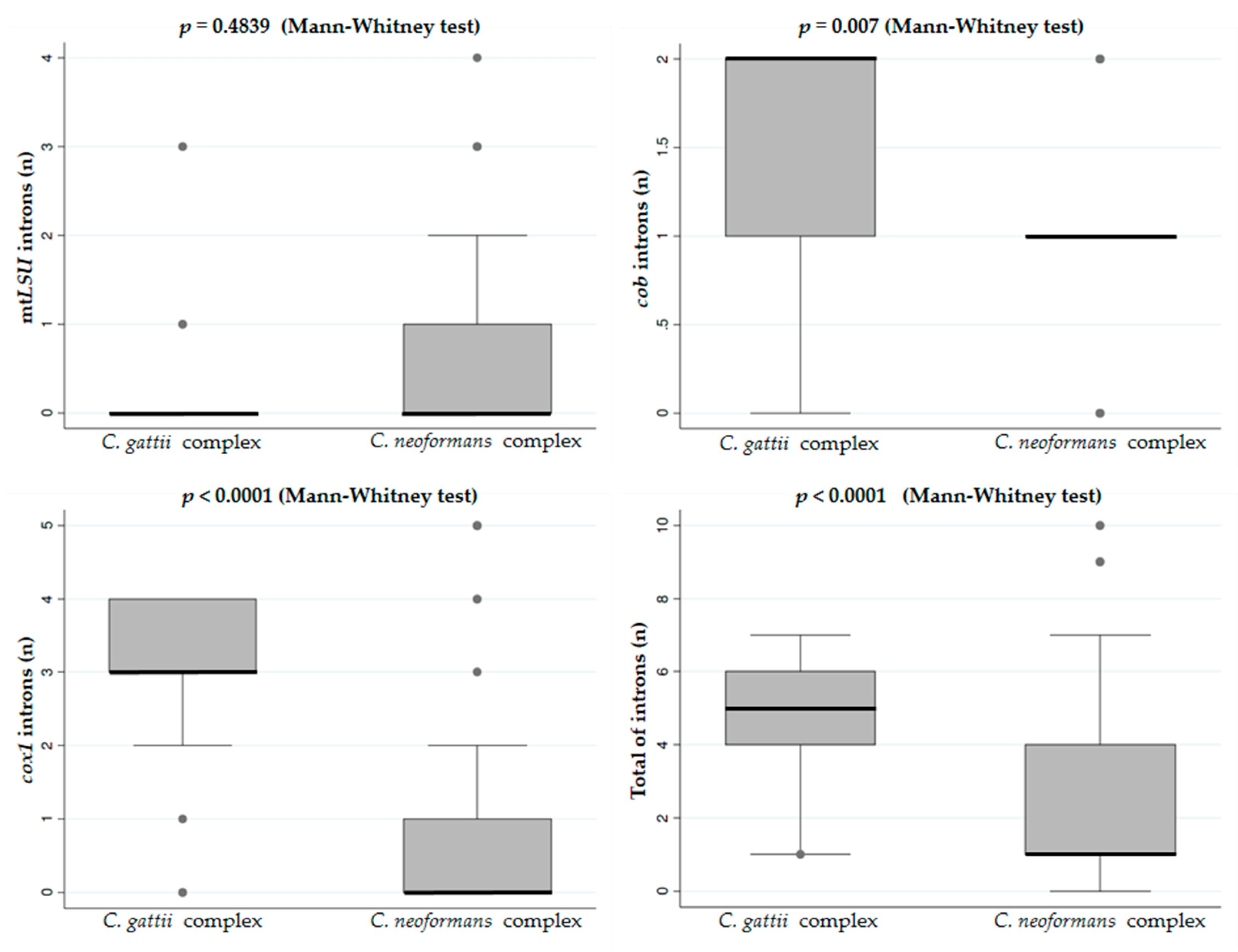

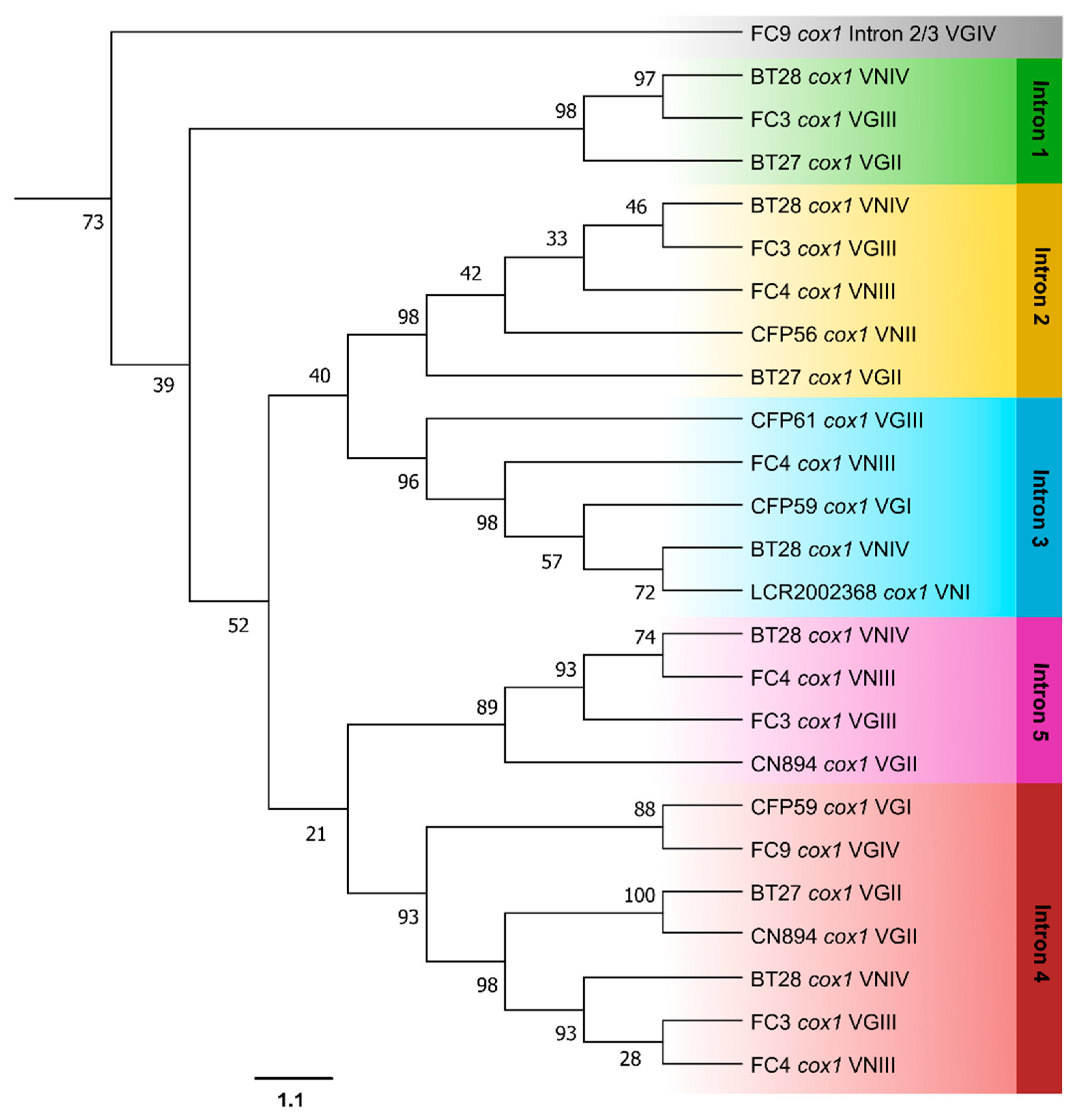

| Gene | Intron | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| cob | 1 | COBE1F ATGTTAACTACGGATGGATG | COB1E2R GTACCAATCCAAGGAATAGC |
| 2 | COB1E2F GCTATTCCTTGGATTGGTAC | COB1E3R CAGAAGGCAAATCGTGTTAG | |
| cox1 | 1 | COX1E1F CCAACTATACAATGTAATCGC | COX1E2R TGAAATCATACCRAAACCTGG |
| 2 | COX1E2F CAGGTTTYGGTATGATTTCAC | COX1E3R GCTGCTGTAAAGTAAGCTCG | |
| 3 | COX1E3F CGAGCTTACTTTACAGCAGC | COX1E4R ATTGTAAATAGAGCTACAAATCC | |
| 4 and 5 | COX1E4F GGATTTGTAGCTCTATTTACAAT | COX1E6R GAGGCATTCCAGCTAGTCC |
| Sequenced Introns | Classification (RNAweasel) | Homing Endonuclease (Conserved Domain Database) | |||
|---|---|---|---|---|---|
| Gene | Strain/Genotype | Family | e-Value | Family | HEG Accession/e-Value |
| cob | BT8/VG2 (1) | ID | 8.02 × 10−36 | – | – |
| CFP61/VGIII (1) | ID | 9.18 × 10−38 | LAGLIDADG | Accession cl03916/E-value 4.32 × 10−14 | |
| CFP56/VNII (1) | ID | 4.29 × 10−36 | LAGLIDADG | Accession pfam03161/E-value 6.51 × 10−58 | |
| CFP58/VNIV (1) | ID | 4.29 × 10−36 | LAGLIDADG | Accession pfam03161/E-value 7.64 × 10−56 | |
| FC4/VNIII (1) | ID | 3.16 × 10−36 | LAGLIDADG | Accession cl03916/E-value 5.31 × 10−12 | |
| BT8/VG2 (2) | IA | 1.49 × 10−22 | LAGLIDADG | Accession cl24136/E-value 4.06 × 10−3 | |
| CFP58/VNIV (2) | IA | 2.06 × 10−22 | LAGLIDADG | Accession cl24136/E-value 4.09 × 10−3 | |
| BT26/VNI (2) | IA | 3.10 × 10−22 | LAGLIDADG | Accession cl24136/E-value 5.12 × 10−3 | |
| CFP59/VGI (2) | IA | 3.68 × 10−20 | – | – | |
| HGT17/VNI (2) | IA | 1.38 × 10−18 | LAGLIDADG | Accession cl24136/E-value 5.13 × 10−3 | |
| cox1 | FC4/VNIII (1) | IB | 2.70 × 10−25 | LAGLIDADG | Accession pfam00961/E-value 2.92 × 10−6 |
| BT28/VNIV (1) | IB | 2.70 × 10−25 | LAGLIDADG | Accession pfam00961/E-value 2.92 × 10−6 | |
| BT27/VGII (1) | IB | 1.41 × 10−24 | LAGLIDADG | Accession cl08299/E-value 3.83 × 10−7 | |
| FC3/VGIII (1) | IB | 2.54 × 10−25 | LAGLIDADG | Accession cl08299/E-value 2.92 × 10−6 | |
| CFP56/VNII (2) | IB | 1.48 × 10−22 | LAGLIDADG | Accession pfam14528/E-value 4.26 × 10−4 | |
| Fc4/VNIII (2) | IB | 1.48 × 10−22 | LAGLIDADG | Accession pfam14528/E-value 3.20 × 10−4 | |
| BT28/VNIV (2) | IB | 9.30 × 10−23 | LAGLIDADG | Accession pfam14528/E-value 1.56 × 10−4 | |
| BT27/VGII (2) | IB | 8.97 × 10−20 | LAGLIDADG | Accession pfam14528/E-value 2.75 × 10−4 | |
| FC3/VGIII (2) | IB | 8.65 × 10−23 | LAGLIDADG | Accession pfam14528/E-value 1.55 × 10−4 | |
| FC9/VGIV (2) | IB | 3.61 × 10−29 | LAGLIDADG LAGLIDADG | Accession pfam00961/E-value 1.67 × 10−12 Accession cl24136/E-value 9.98 × 10−3 | |
| LCR2002368/VNI (3) | Similar to IB | 1.52 × 10−29 | LAGLIDADG | Accession cl03916/E-value 1.25 × 10−28 | |
| FC4/VNIII (3) | Similar to IB | 5.06 × 10−29 | LAGLIDADG | Accession pfam03161/E-value 1.31 × 10−49 | |
| BT28/VNIV (3) | Similar to IB | 5.80 × 10−30 | LAGLIDADG | Accession pfam03161/E-value 5.30 × 10−53 | |
| CFP59/VGI (3) | Similar to IB | 5.80 × 10−30 | LAGLIDADG | Accession pfam03161/E-value 2.42 × 10−51 | |
| CFP61/VGIII (3) | Similar to IB | 5.68 × 10−31 | – | – | |
| FC4/VNIII (4) | Similar to IB | 1.12 × 10−9 | LAGLIDADG | Accession pfam00961/E-value 4.68 × 10−12 | |
| BT28/VNIV (4) | Similar to IB | 1.30 × 10−9 | LAGLIDADG | Accession pfam00961/E-value 4.68 × 10−12 | |
| CFP59/VGI (4) | Similar to IB | 1.30 × 10−9 | LAGLIDADG LAGLIDADG | Accession pfam00961/E-value 5.67 × 10−13 Accession cl24136/E-value 8.57 × 10−3 | |
| BT27/VGII (4) | Similar to IB | 1.98 × 10−7 | LAGLIDADG | Accession pfam00961/E-value 1.07 × 10−11 | |
| CN894/VGII (4) | NI | NI | LAGLIDADG | Accession pfam00961/E-value 7.76 × 10−12 | |
| FC3/VGIII (4) | NI | NI | LAGLIDADG | Accession pfam00961/E-value 4.80 × 10−12 | |
| FC9/VGIV (4) | Similar to IB | 1.37 × 10−9 | LAGLIDADG LAGLIDADG | Accession pfam00961/E-value 5.59 × 10−13 Accession cl24136/E-value 8.49 × 10−3 | |
| Fc4/VNIII (5) | IA | 7.78 × 10−17 | – | – | |
| BT28/VNIV (5) | IA | 8.84 × 10−17 | – | – | |
| CN894/VGII (5) | IA | 2.98 × 10−19 | – | – | |
| FC3/VGIII (5) | IA | 9.06 × 10−17 | – | – | |
| PCR Target | Primer’s Pair | C. neoformans Complex Expected Product Sizes (bp) | |||
| C. neoformans | Hybrid | C. deneoformans | |||
| VNI | VNII | VNIII | VNIV | ||
| mtLSU * | CryLSUF/CryLSUR | 300 or 1300 | 1300 | 2000 or 2500 | 300 or 1900 |
| introns 4 and 5 of cox1 | COX1E4F/COX1E6R | 300 | 300 | 1600 | 1600 |
| PCR Target | Primer’s Pair | C. gattii Complex Expected Product Sizes (bp) | |||
| C. gattii | C. deuterogattii | C. bacillosporus | C. tetragattii | ||
| VGI | VGII | VGIII | VGIV | ||
| mtLSU * | CryLSUF/CryLSUR | 300 or 1100 | 300 | 1800 | 1300 |
| intron 1 of cob | COBE1F/COB1E2R | 1200 | 250 or 600 | 250 or 1200 | 250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, R.M.O.d.S.; da Silva, K.J.G.; Ferreira, L.C.; Arantes, T.D.; Theodoro, R.C. Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii. J. Fungi 2023, 9, 629. https://doi.org/10.3390/jof9060629
Gomes RMOdS, da Silva KJG, Ferreira LC, Arantes TD, Theodoro RC. Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii. Journal of Fungi. 2023; 9(6):629. https://doi.org/10.3390/jof9060629
Chicago/Turabian StyleGomes, Ronald Muryellison Oliveira da Silva, Kássia Jéssica Galdino da Silva, Leonardo Capistrano Ferreira, Thales Domingos Arantes, and Raquel Cordeiro Theodoro. 2023. "Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii" Journal of Fungi 9, no. 6: 629. https://doi.org/10.3390/jof9060629
APA StyleGomes, R. M. O. d. S., da Silva, K. J. G., Ferreira, L. C., Arantes, T. D., & Theodoro, R. C. (2023). Distribution and Polymorphisms of Group I Introns in Mitochondrial Genes from Cryptococcus neoformans and Cryptococcus gattii. Journal of Fungi, 9(6), 629. https://doi.org/10.3390/jof9060629







