Abstract
An altered gut microbiota is a possible contributing pathogenic factor in myasthenia gravis (MG), an autoimmune neuromuscular disease. However, the significance of the fungal microbiome is an understudied and neglected part of the intestinal microbiome in MG. We performed a sub-analysis of the MYBIOM study including faecal samples from patients with MG (n = 41), non-inflammatory neurological disorder (NIND, n = 18), chronic inflammatory demyelinating polyradiculoneuropathy (CIDP, n = 6) and healthy volunteers (n = 12) by sequencing the internal transcribed spacer 2 (ITS2). Fungal reads were obtained in 51 out of 77 samples. No differences were found in alpha-diversity indices computed between the MG, NIND, CIDP and HV groups, indicating an unaltered fungal diversity and structure. Overall, four mould species (Penicillium aurantiogriseum, Mycosphaerella tassiana, Cladosporium ramonetellum and Alternaria betae-kenyensis) and five yeast species (Candida. albicans, Candida. sake, Candida. dubliniensis, Pichia deserticola and Kregervanrija delftensis) were identified. Besides one MG patient with abundant Ca. albicans, no prominent dysbiosis in the MG group of the mycobiome was found. Not all fungal sequences within all groups were successfully assigned, so further sub-analysis was withdrawn, limiting robust conclusions.
1. Introduction
Myasthenia gravis (MG) is an autoimmune-mediated disorder with autoantibodies targeting different proteins of the neuromuscular junction [1]. Classification of different autoantibody types, particularly antibodies against the acetylcholine receptor (AChR), helped to clarify the mechanisms of symptoms such as exercise-induced muscle weakness. However, the pathogenic trigger of MG is yet not fully understood. Altered gut microbiota is a possible predisposing factor for MG [2,3]. Bacterial gut microbiota profiles of MG patients differ from those of healthy volunteers [3,4,5]. In the last few years, several cohort studies, as well as rat models, have implicated the bacterial gut microbiota in MG induction and severity [3,4,5,6]. However, sufficient data on fungal gut microbiota profiles of MG patients compared with those of patients with other inflammatory or non-inflammatory neurological diseases is still lacking. Specific compositions of the gut mycobiome are associated with autoimmune conditions such as multiple sclerosis (MS) [7,8], and an increase in Candida spp. is linked to neurological disorders such as autism spectrum disorders [9], schizophrenia [10] and Rett syndrome [11]. Although it is not fully understood how C. albicans contributes to the pathogenesis of these diseases, one well-established mechanism for the increase in C. albicans abundance seen in disease is that colonisation of C. albicans can drive T-helper-17 cell-mediated immune responses, leading to exacerbation [12]. Through the activation of pattern recognition receptors, mannans, fungal cell wall constituents from C. spp., induced Th17 and IL-23 responses, leading to the worsening of intestinal graft-versus-host disease and colitis in mice [13,14,15]. We present here a sub-analysis of the previous MYBIOM study, a single-centre observational study on bacterial gut microbiota with 77 participants, to determine whether the fungal gut microbiota is altered in MG patients compared with non-inflammatory neurological disorder (NIND), patients with chronic inflammatory demyelinating polyradiculoneuropathy (CIDP) and healthy volunteers (HV) without neurological disorders [3].
2. Materials and Methods
2.1. Study Design and Patients
MYBIOM was a single-centre observational study conducted at the Department of Neurology at the University Hospital of Essen in Germany from July 2017 to March 2018. The study design and the bacterial microbiome results have been published elsewhere [3]. In brief, MG patients were included based on their clinical characteristics, such as fluctuating fatigability and muscle weakness, a positive response to cholinesterase inhibitors and, optionally, a recorded decrease in repetitive motor nerve stimulation [16]. Three different groups of patients were recruited for comparison: subjects with NIND, those with CIDP diagnosis based on the European Federation of Neurological Societies/Peripheral Nerve Society diagnostic criteria [17] and HV without any underlying neurological or systemic inflammatory disease were recruited as controls. Study participants were 18 years or older and had no history of treatment with antibiotics within the previous 4 weeks, and no intentional consumption of probiotics or anti-obesity agents within 3 months prior to study participation. Exclusion criteria were chronic inflammatory bowel disease, short bowel syndrome, irritable bowel syndrome, pregnancy and recent treatment with chemotherapeutics or monoclonal antibodies.
2.2. Sample Collection, DNA Extraction and Sequencing
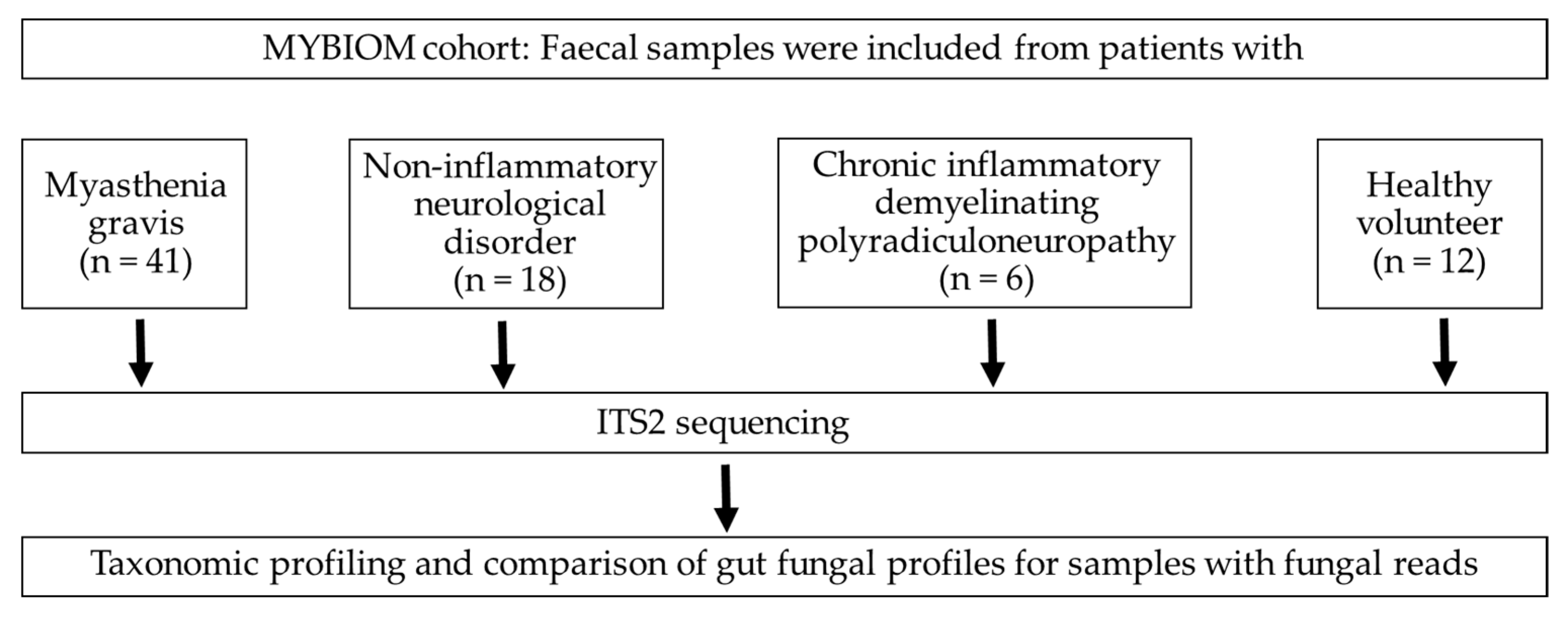
Fresh faecal samples were collected from participants and were transported to the Institute of Medical Microbiology at 4°C within 12 h of specimen collection. A total of 77 faecal samples (MG (n = 41), CIDP (n = 6), NIND (n = 18) and HV (n = 12), Figure 1), as well as 6 negative controls (RNA-free water) and 2 positive controls (Escherichia coli and a mix of E. coli, Staphylococcus aureus and Corynebacterium striatum), were stored at −80°C until DNA extraction [3] Using a QIAmp Fast DNA Stool Mini kit (Qiagen). DNA aliquots were stored at −80°C until use. The internal transcribed spacer 2 (ITS2) region was amplified from faecal DNA using primers ITS3 and ITS4 (ITS3 F (5′- GCATCGATGAAGAACGCAGC-3′) and ITS4 R (5′- TCCTCCGCTTATTGATATGC-3′)) [18]. BaseClear B.V. (Leiden, the Netherlands) performed library preparation and sequencing on the Illumina MiSeq platform (10 k paired-end reads) as well as Illumina raw data processing.
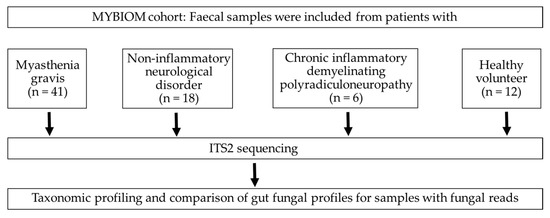
Figure 1.
Flow chart and enrolled participants in the study.
2.3. Illumina Demultiplexing
Paired-end (2 × 300 bp) sequence reads were generated using the Illumina MiSeq system. The sequences generated performed under accreditation according to the scope of BaseClear B.V. (L457; NEN-EN-ISO/IEC 17025). FASTQ read sequence files were generated using bcl2fastq version 2.20 (Illumina). Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing PhiX control signal were removed using an inhouse filtering protocol. In addition, reads containing (partial) adapters were clipped (up to a minimum read length of 50 bp). The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.11.8. A total of 1,579,359 read-pairs were obtained from sequencing the ITS2 region after demultiplexing and filtering. Samples without fungal reads were excluded from further analysis.
2.4. Processing and Statistical Analysis of Metataxonomic Data
Sequence processing was performed using DADA2 to cluster sequences into ASV and to provide taxonomic annotations. The DADA2 ITS Pipeline Workflow (1.8) was used to process the reads (https://benjjneb.github.io/dada2/ITS_workflow.html, accessed on 4 July 2022). Fungal taxonomy was assigned using the UNITE database (https://unite.ut.ee, accessed on 4 July 2022). Statistical analyses were performed in MicrobiomeAnalyst [19] and the data exported to CSVs for further analysis. Within-sample alpha-diversity indices [Chao1, Simpson, and Shannon] were calculated in the MicrobiomeAnalyst. For comparison between groups, the nonparametric Kruskal–Wallis and Mann–Whitney tests were calculated in GraphPad Prism v7.05 (GraphPad Software, San Diego, CA, USA, www.graphpad.com, accessed on 4 July 2022). Statistical tests with p ≤ 0.05 were considered significant.
3. Results
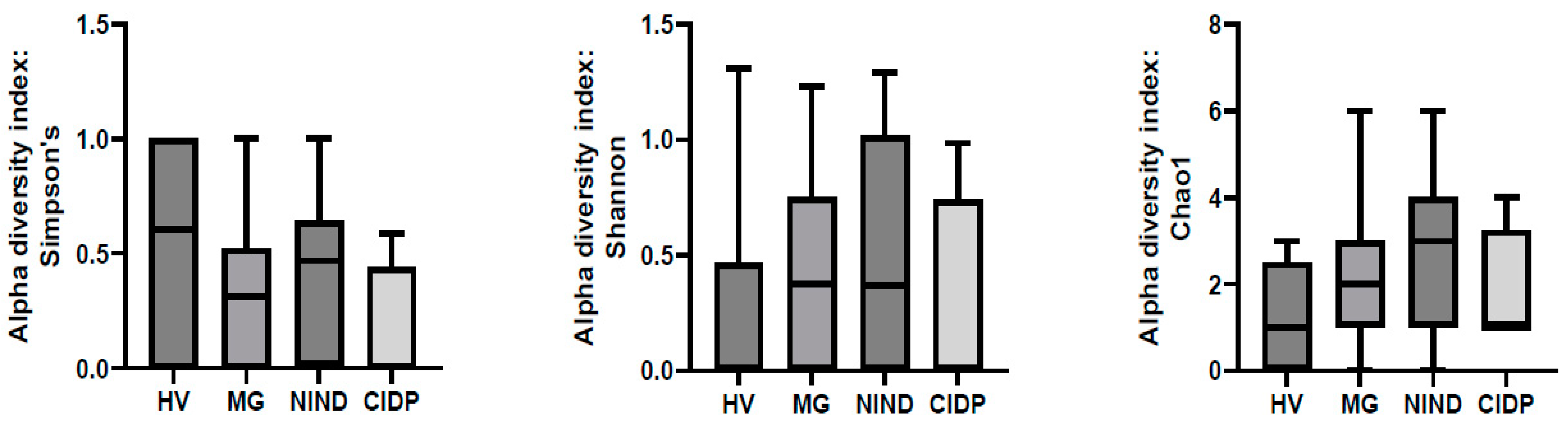
After exclusion of samples without fungal reads, 51 samples were further analysed ((MG (n = 27), CIDP (n = 4), NIND (n = 11) and HV (n = 9)). To assess differences in the gut microbiota of patients with MG, CIDP, NIND and HV, ecological features of the faecal fungal communities were evaluated using the alpha diversity indices Simpson’s, Shannon and Chao1. None of the indices were significantly different between groups (p > 0.05) (Figure 2).
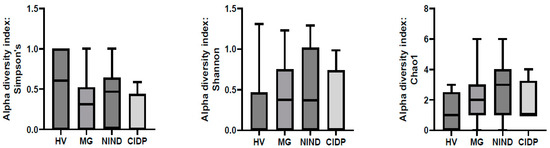
Figure 2.
Comparison of alpha-diversity indices of the different study groups MG, NIND and CIDP using the Kruskal–Wallis test. Data are presented as mean ± standard error of the mean. HV, healthy volunteer; MG, myasthenia gravis; NIND, non-inflammatory neurological disorder; CIDP, chronic inflammatory demyelinating polyradiculoneuropathy.
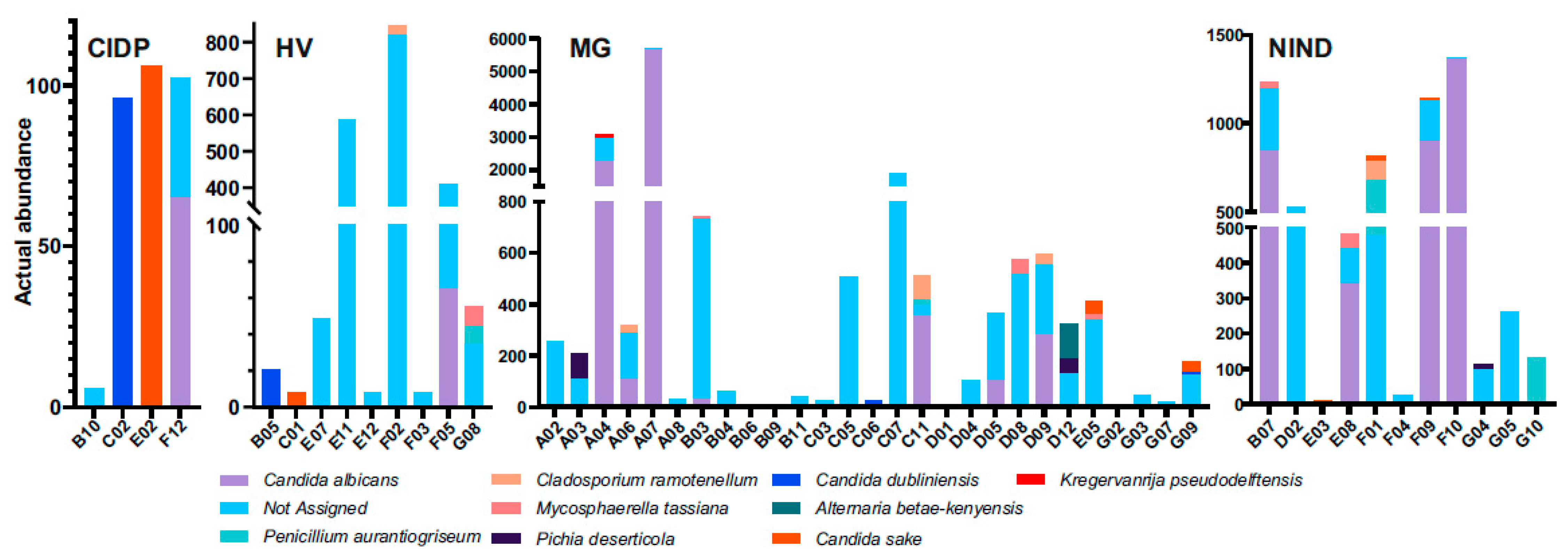
The composition of fungal microbiota from faeces was characterized by the presence of yeasts and moulds. Subjects with failed assignment to fungal species belonged to all groups (CIDP 2/4, HV 7/9, MG 22/27 and NIND 8/11).
Failed assignment affected either all fungal sequences per subject (CIDP 1/2, HV 4/7, MG 10/22 and NIND 3/8) or a portion. Overall, four mould species (Penicillium aurantiogriseum, Mycosphaerella tassiana, Cladosporium ramonetellum and Alternaria betae-kenyensis) and five yeast species (C. albicans, C. sake, C. dubliniensis, Pichia deserticola and Kregervanrija delftensis) were identified (Figure 3). In subject A07 (MG) C. albicans abundance dominated within the fungal community. Furthermore, no prominent dysbiosis of the mycobiome was found. Due to the limited numbers of successfully assigned fungal sequences, we refrained from further data analysis and representations.
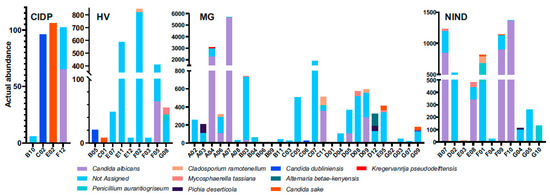
Figure 3.
Differentially abundant fungal species in faecal samples from patients. HV, healthy volunteer; MG, myasthenia gravis; NIND, non-inflammatory neurological disorder; CIDP, chronic inflammatory demyelinating polyradiculoneuropathy.
4. Discussion
In this study, extracted DNA from faecal samples from MYBIOM study participants was examined to assess the gut mycobiome. No difference was found comparing the alpha-diversity indices of the different study groups. Our findings include Penicillium aurantiogriseum, Mycosphaerella tassiana, Cladosporium ramonetellum and Alternaria betae-kenyensis among filamentous fungi. Mycosphaerella tassiana, the heterotypic synonym of Cladosporium herbarum, is a common fungus found worldwide, and its spores are highly prevalent in the air [20,21]. Together with Cladosporium herbarum, Alternaria spp. are common allergens, and the latter are known as major plant pathogens ubiquitously distributed in the environment. The genus Alternaria can also be found on normal human skin is part of the oral mycobiome in healthy individuals [22,23]. In vitro, Alternaria and Cladosporium species have been found to reveal acetylcholinesterase inhibitory activities [24,25]. Interestingly, acetylcholinesterase inhibitors are common therapeutics in MG, but further studies need to clarify potential benefits of these fungal genera in MG. As stool samples for this study were processed in a Class II safety cabinet, fungal spores possibly occurred during defecation and sampling or may have been swallowed by patients.
Among yeast, the species Kregervanrija pseudodelftensis and Pichia deserticola were found, both belonging to the Pichiaceae family, and together with C. sake they are associated with the consumption of fruits [26,27,28]. C. albicans causes serious infections in hospitalized patients, associated with high morbidity and mortality rates, among others [29]. In our study, C. albicans was detected in all groups. However, in one MG patient (A07), C. albicans was dominantly present. According to the Human Microbiome Project and their study on gut mycobiomes of healthy subjects, fungal communities were characterized by a high prevalence of yeast including C. albicans, with operational taxonomic units (OTUs) present in 80.8% of samples [30]. Although there was a high degree of inter-subject variability in fungal communities, C. albicans amplicon sequence variants (ASV) were found in 63.6% of subjects. In contrast to bacteria in the digestive tract, C. albicans colonizes all segments from the oral cavity to the anus [31]. We hypothesize that subject A07 might have had an intestinal fungal overgrowth or an oral candidiasis. Both often occur after long-term antibiotic treatment or in short bowel syndrome, a malabsorptive disorder, as a result of the loss of bowel mass mostly secondary to surgical resection of the small intestine. Both conditions were excluded by study inclusion criteria (no history of treatment with antibiotics within the previous 4 weeks and no short bowel syndrome).
However, some fungal sequences could not be assigned to fungal genera or species, which reduced the impact of the mycobiome analysis. Therefore, comparison between groups was not reasonable based on the obtained data. To extract meaningful community profiles for fungi, challenges must be overcome [22] which are in parts different from those of bacteria, e.g., recognition of process-induced sequencing errors [32], accurate taxonomic assignments to define community members and structure [33] as well as binary naming and phylogenetic classifications [34]. In our study, limited content and annotation most likely led to non-robust fungal identification or failure to achieve a genus-level assignment. The applied DNA extraction protocol was appropriate for PCR amplification of fungal regions regarding DNA yield and quality [35]. However, the best extraction protocol for analysis of mycobiome data was achieved using the standardized IHMS Protocol Q with additional repeated beat-beating steps [36]. Furthermore, sequencing of the ITS2 region provides greater resolution of the relatively low-abundance mycobiome species in comparison to 18S rRNA gene sequencing [30]. In contrast to the former bacterial analysis, taxonomic classification and interpretation of fungal results was more challenging [37], although the UNITE database, a well-curated, high quality database that is constantly updated, was utilised [38]. To discard non-target sequences and to increase taxonomic resolution, Nilsson et al. recommend pipelines such as PipeCraft, PIPITS, LotuS and AMPtk for Illumina data without erasing all errors that occurred during sample preparation and sequencing [39]. The parallel use of different taxonomy assignment tools for comparison and combination of results may be helpful [40].
As a sub-analysis from the initial study performed between July 2017 and August 2018, DNA aliquots were stored at −80°C after DNA extraction and were twice thawed, namely once for the bacterial microbiome analysis and again for the present mycobiome analysis. This might have had an impact on the sequencing yield and quality but should be only marginal.
In MS, the gut mycobiome differs from that of healthy individuals, with enrichment of C. and Epicoccum as well as a lower relative abundance of Saccharomyces [8] and over-representation of Saccharomyces and Aspergillus, respectively [7]. Different mycobiome profiles could be defined with different immune cell subsets in the blood [7]. Due to the above stated reasons, a distinct gut mycobiome signature could not be generated and, therefore, comparisons with other autoimmune neurological diseases such as MS are difficult. If compared to the recently described high prevalence of Saccharomyces, Malassezia, and C. within the gut mycobiome of the Human Microbiome Project healthy cohort, our findings within the healthy volunteer group HV bore a resemblance in part [30].
5. Conclusions
This study evaluated potential alterations of the gut mycobiome in patients with MG in comparison to subjects with other (autoimmune) neurological diseases and healthy subjects, and found no difference between the groups. However, due to the small groups, further investigations are required to assess associations of the mycobiome with MG and interactions between fungi and autoimmunity, including analysis of fungal functional profiles and immune responses as well as unravelling the significance of fungal species with acetylcholinesterase inhibitory activities in MG.
Author Contributions
Conceptualization, H.L.V., T.H. and A.T.; methodology, H.L.V. and J.R.M.; formal analysis, H.L.V., J.R.M. and A.T.; investigation and data curation, H.L.V., E.R., M.S., J.R.M. and A.T.; writing—original draft preparation, H.L.V., J.R.M. and A.T.; writing—review and editing, J.B., T.H. and C.K.; visualization, H.L.V., A.T. and T.H.; supervision, J.B., T.H. and C.K.; project administration, H.L.V. and A.T.; funding acquisition, H.L.V. and T.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Medical Faculty at the University Duisburg-Essen, Germany (approval number: 17-7439-BO). The study was registered with the German Clinical Trials Register: DRKS00017134.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Sequenced reads generated in this study have been deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home) (accessed on 4 July 2022). under accession number PRJEB61825.
Acknowledgments
This work was supported by the University of Duisburg-Essen, Faculty of Medicine, program “Willkommen zurück”. The Division of Digestive Diseases and JRM received financial and infrastructure support from the NIHR Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London.
Conflicts of Interest
H.L.V., E.R., M.S. and J.B. declare no conflict of interest. J.R.M. has received consultancy fees from EnterBiotix and Cultech Ltd. C.K. received speaker's and or consultation honoraria from Alexion Pharmaceuticals and Biogen. A.T. and T.H. are members of the medical advisory board of the German Myasthenia Society. A.T. received consultation honoraria from Argenx. T.H. received speaker's and or consultation honoraria from Alexion Pharmaceuticals, Argenx, Novartis, Hormosan Pharma, Biogen, Roche and Sanofi-Genzyme.
References
- Koneczny, I.; Herbst, R. Myasthenia Gravis: Pathogenic Effects of Autoantibodies on Neuromuscular Architecture. Cells 2019, 8, 671. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Li, L.; Kang, X.; Zhao, Y.; Cai, Y. Gut microbiota and metabolites in myasthenia gravis: Early diagnostic biomarkers and therapeutic strategies. Clin. Immunol. 2022, 245, 109173. [Google Scholar] [CrossRef]
- Totzeck, A.; Ramakrishnan, E.; Schlag, M.; Stolte, B.; Kizina, K.; Bolz, S.; Thimm, A.; Stettner, M.; Marchesi, J.R.; Buer, J.; et al. Gut bacterial microbiota in patients with myasthenia gravis: Results from the MYBIOM study. Ther. Adv. Neurol. Disord. 2021, 14, 17562864211035657. [Google Scholar] [CrossRef] [PubMed]
- Moris, G.; Arboleya, S.; Mancabelli, L.; Milani, C.; Ventura, M.; de Los Reyes-Gavilan, C.G.; Gueimonde, M. Fecal microbiota profile in a group of myasthenia gravis patients. Sci. Rep. 2018, 8, 14384. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xia, Z.; Jiao, X.; Deng, J.; Zhang, L.; Li, J. Altered Gut Microbiota in Myasthenia Gravis. Front. Microbiol. 2018, 9, 2627. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, E.; Consonni, A.; Cordiglieri, C.; Sacco, G.; Crasa, C.; Fontana, A.; Morelli, L.; Elli, M.; Mantegazza, R.; Baggi, F. Therapeutic Effect of Bifidobacterium Administration on Experimental Autoimmune Myasthenia Gravis in Lewis Rats. Front. Immunol. 2019, 10, 2949. [Google Scholar] [CrossRef]
- Shah, S.; Locca, A.; Dorsett, Y.; Cantoni, C.; Ghezzi, L.; Lin, Q.; Bokoliya, S.; Panier, H.; Suther, C.; Gormley, M.; et al. Alterations of the gut mycobiome in patients with MS. EBioMedicine 2021, 71, 103557. [Google Scholar] [CrossRef]
- Yadav, M.; Ali, S.; Shrode, R.L.; Shahi, S.K.; Jensen, S.N.; Hoang, J.; Cassidy, S.; Olalde, H.; Guseva, N.; Paullus, M.; et al. Multiple sclerosis patients have an altered gut mycobiome and increased fungal to bacterial richness. PLoS ONE 2022, 17, e0264556. [Google Scholar] [CrossRef]
- Zou, R.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zheng, H. Dysbiosis of Gut Fungal Microbiota in Children with Autism Spectrum Disorders. J. Autism Dev. Disord. 2021, 51, 267–275. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, L.Y.; Zhang, Z.; Zhou, Y.Y.; Jiang, H.Y.; Ruan, B. Analysis of gut mycobiota in first-episode, drug-naive Chinese patients with schizophrenia: A pilot study. Behav. Brain Res. 2020, 379, 112374. [Google Scholar] [CrossRef]
- Strati, F.; Calabro, A.; Donati, C.; De Felice, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Rizzetto, L.; De Filippo, C.; et al. Intestinal Candida parapsilosis isolates from Rett syndrome subjects bear potential virulent traits and capacity to persist within the host. BMC Gastroenterol. 2018, 18, 57. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Santos, N.; Gaffen, S.L. Th17 cells in immunity to Candida albicans. Cell Host Microbe 2012, 11, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef]
- van der Velden, W.J.; Netea, M.G.; de Haan, A.F.; Huls, G.A.; Donnelly, J.P.; Blijlevens, N.M. Role of the mycobiome in human acute graft-versus-host disease. Biol. Blood Marrow Transplant. 2013, 19, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Aschenbrenner, D.; Yoo, J.Y.; Zuo, T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 2022, 3, e969–e983. [Google Scholar] [CrossRef]
- Melzer, N.; Ruck, T.; Fuhr, P.; Gold, R.; Hohlfeld, R.; Marx, A.; Melms, A.; Tackenberg, B.; Schalke, B.; Schneider-Gold, C.; et al. Clinical features, pathogenesis, and treatment of myasthenia gravis: A supplement to the Guidelines of the German Neurological Society. J. Neurol. 2016, 263, 1473–1494. [Google Scholar] [CrossRef]
- Van den Bergh, P.Y.; Hadden, R.D.; Bouche, P.; Cornblath, D.R.; Hahn, A.; Illa, I.; Koski, C.L.; Leger, J.M.; Nobile-Orazio, E.; Pollard, J.; et al. European Federation of Neurological Societies/Peripheral Nerve Society guideline on management of chronic inflammatory demyelinating polyradiculoneuropathy: Report of a joint task force of the European Federation of Neurological Societies and the Peripheral Nerve Society—First revision. Eur. J. Neurol. 2010, 17, 356–363. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Meklin, T.; Reponen, T.; McKinstry, C.; Cho, S.H.; Grinshpun, S.A.; Nevalainen, A.; Vepsalainen, A.; Haugland, R.A.; Lemasters, G.; Vesper, S.J. Comparison of mold concentrations quantified by MSQPCR in indoor and outdoor air sampled simultaneously. Sci. Total. Environ. 2007, 382, 130–134. [Google Scholar] [CrossRef]
- Schubert, K.; Groenewald, J.Z.; Braun, U.; Dijksterhuis, J.; Starink, M.; Hill, C.F.; Zalar, P.; de Hoog, G.S.; Crous, P.W. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud. Mycol. 2007, 58, 105–156. [Google Scholar] [CrossRef]
- Dupuy, A.K.; David, M.S.; Li, L.; Heider, T.N.; Peterson, J.D.; Montano, E.A.; Dongari-Bagtzoglou, A.; Diaz, P.I.; Strausbaugh, L.D. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: Discovery of Malassezia as a prominent commensal. PLoS ONE 2014, 9, e90899. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F.J.; Guarro, J. Alternaria infections: Laboratory diagnosis and relevant clinical features. Clin. Microbiol. Infect. 2008, 14, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, J.; Kaur, A.; Kaur, R.; Yadav, A.K.; Sharma, V.; Chadha, B.S. Cholinesterase inhibitor (Altenuene) from an endophytic fungus Alternaria alternata: Optimization, purification and characterization. J. Appl. Microbiol. 2016, 121, 1015–1025. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, Q.G.; Zhang, Z.B.; Yan, R.M.; Wang, L.Y.; Zhu, D. Isolation and characterization of endophytic huperzine A-producing fungi from Huperzia serrata. J. Ind. Microbiol. Biotechnol. 2011, 38, 1267–1278. [Google Scholar] [CrossRef]
- Archer, D.L. Freezing: An underutilized food safety technology? Int. J. Food Microbiol. 2004, 90, 127–138. [Google Scholar] [CrossRef]
- Kurtzman, C.P. New species and new combinations in the yeast genera Kregervanrija gen. nov., Saturnispora and Candida. FEMS Yeast Res. 2006, 6, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.J.; Yao, T.; Ma, L.; Ren, X.; Li, L. Application of Pichia Deserticola in Prevention and Treatment of Postharvest Diseases of Fruits. Patent application filed by Beijing Technology and Business University, China CN107988088B, 2017. [Google Scholar]
- Kett, D.H.; Azoulay, E.; Echeverria, P.M.; Vincent, J.L.; FCCM for the Extended Prevalence of Infection in the ICU Study (EPIC II) Group of Investigators. Candida bloodstream infections in intensive care units: Analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 2011, 39, 665–670. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Poulain, D. Candida albicans, plasticity and pathogenesis. Crit. Rev. Microbiol. 2015, 41, 208–217. [Google Scholar] [CrossRef]
- Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Jairus, T.; Sadam, A.; Saar, I.; Bahram, M.; Bechem, E.; Chuyong, G.; Koljalg, U. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New. Phytol. 2010, 188, 291–301. [Google Scholar] [CrossRef]
- Lee, C.K.; Herbold, C.W.; Polson, S.W.; Wommack, K.E.; Williamson, S.J.; McDonald, I.R.; Cary, S.C. Groundtruthing next-gen sequencing for microbial ecology-biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS ONE 2012, 7, e44224. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Crous, P.W.; Redhead, S.A.; Reynolds, D.R.; Samson, R.A.; Seifert, K.A.; Taylor, J.W.; Wingfield, M.J.; Abaci, O.; Aime, C.; et al. The amsterdam declaration on fungal nomenclature. IMA Fungus 2011, 2, 105–112. [Google Scholar] [CrossRef]
- Fiedorova, K.; Radvansky, M.; Nemcova, E.; Grombirikova, H.; Bosak, J.; Cernochova, M.; Lexa, M.; Smajs, D.; Freiberger, T. The Impact of DNA Extraction Methods on Stool Bacterial and Fungal Microbiota Community Recovery. Front. Microbiol. 2019, 10, 821. [Google Scholar] [CrossRef]
- Angebault, C.; Ghozlane, A.; Volant, S.; Botterel, F.; d’Enfert, C.; Bougnoux, M.E. Combined bacterial and fungal intestinal microbiota analyses: Impact of storage conditions and DNA extraction protocols. PLoS ONE 2018, 13, e0201174. [Google Scholar] [CrossRef] [PubMed]
- Thielemann, N.; Herz, M.; Kurzai, O.; Martin, R. Analyzing the human gut mycobiome—A short guide for beginners. Comput. Struct. Biotechnol. J. 2022, 20, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Lucking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Gdanetz, K.; Benucci, G.M.N.; Vande Pol, N.; Bonito, G. CONSTAX: A tool for improved taxonomic resolution of environmental fungal ITS sequences. BMC Bioinform. 2017, 18, 538. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).