Fusarium mindanaoense sp. nov., a New Fusarium Wilt Pathogen of Cavendish Banana from the Philippines Belonging to the F. fujikuroi Species Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. Genomic DNA Extraction
2.3. Gene Prediction
2.4. Phylogenetic Analyses
2.5. Morphological Analyses
2.6. Pathogenicity Test
2.7. Detection of Secreted in Xylem Genes among the Whole-Genome Data
2.8. Identification of the Homologous SIX Genes of PD20-05 in the Foc TR4 Genome
3. Results
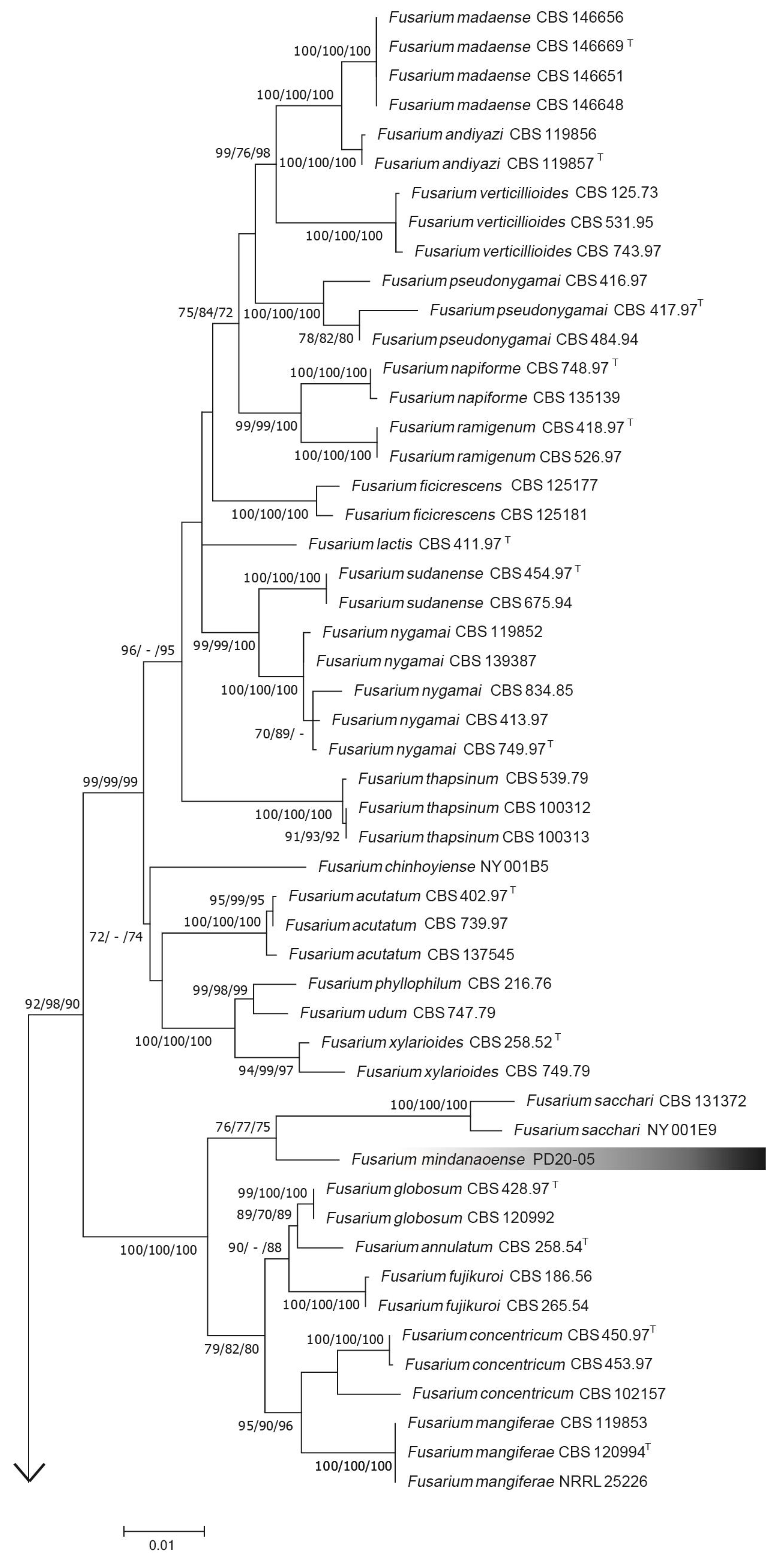
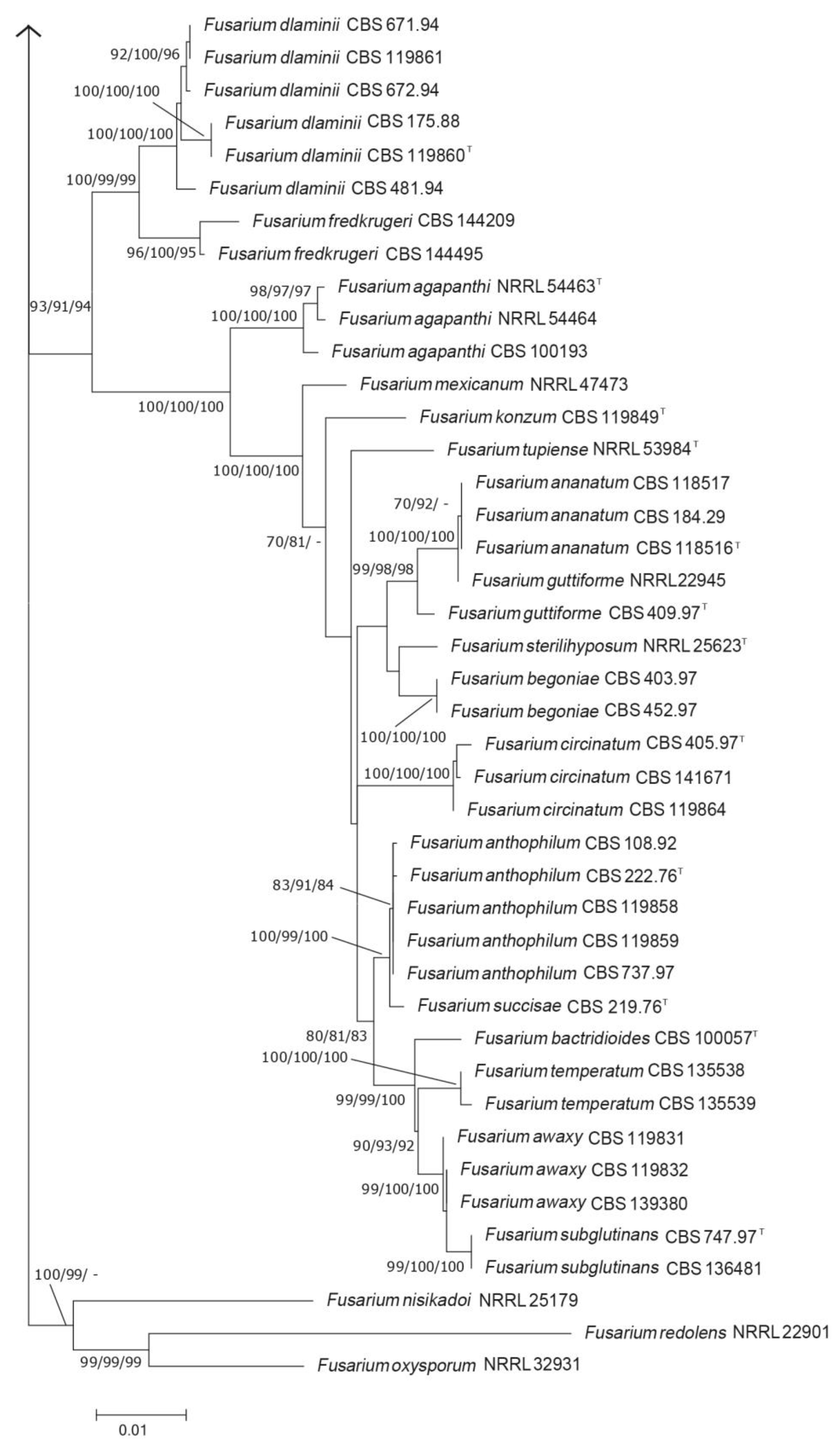
3.1. Phylogenetic Analysis
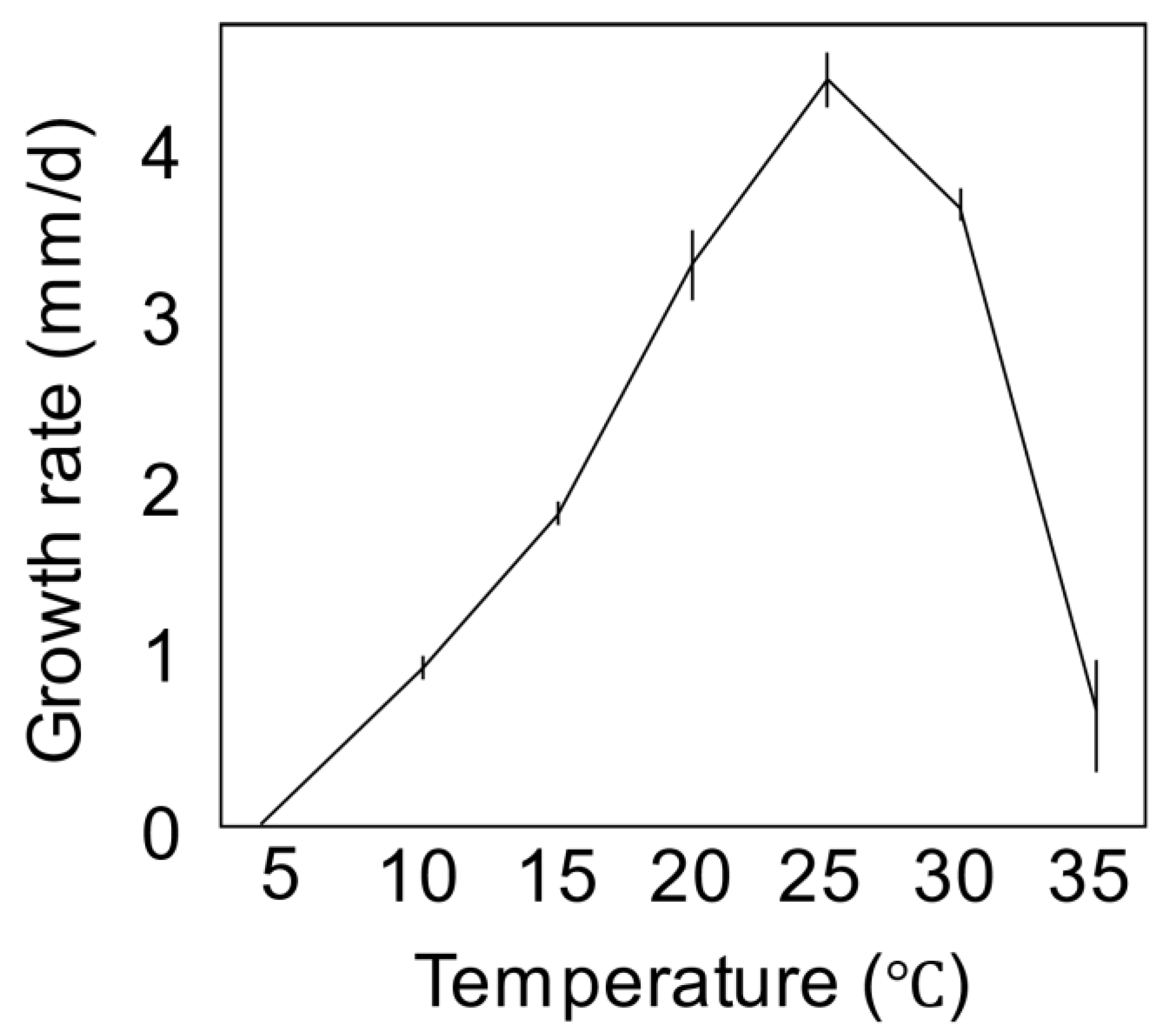
3.2. Taxonomy
- Fusarium mindanaoense Nozawa & Watanabe, sp. nov.
- Mycobank MB 848129; Figure 3.
- Etymology: the name refers to Mindanao, the region where the ex-type strain was obtained.
- Holotype: PD20-05S.
- Ex-holotype: PD20-05.

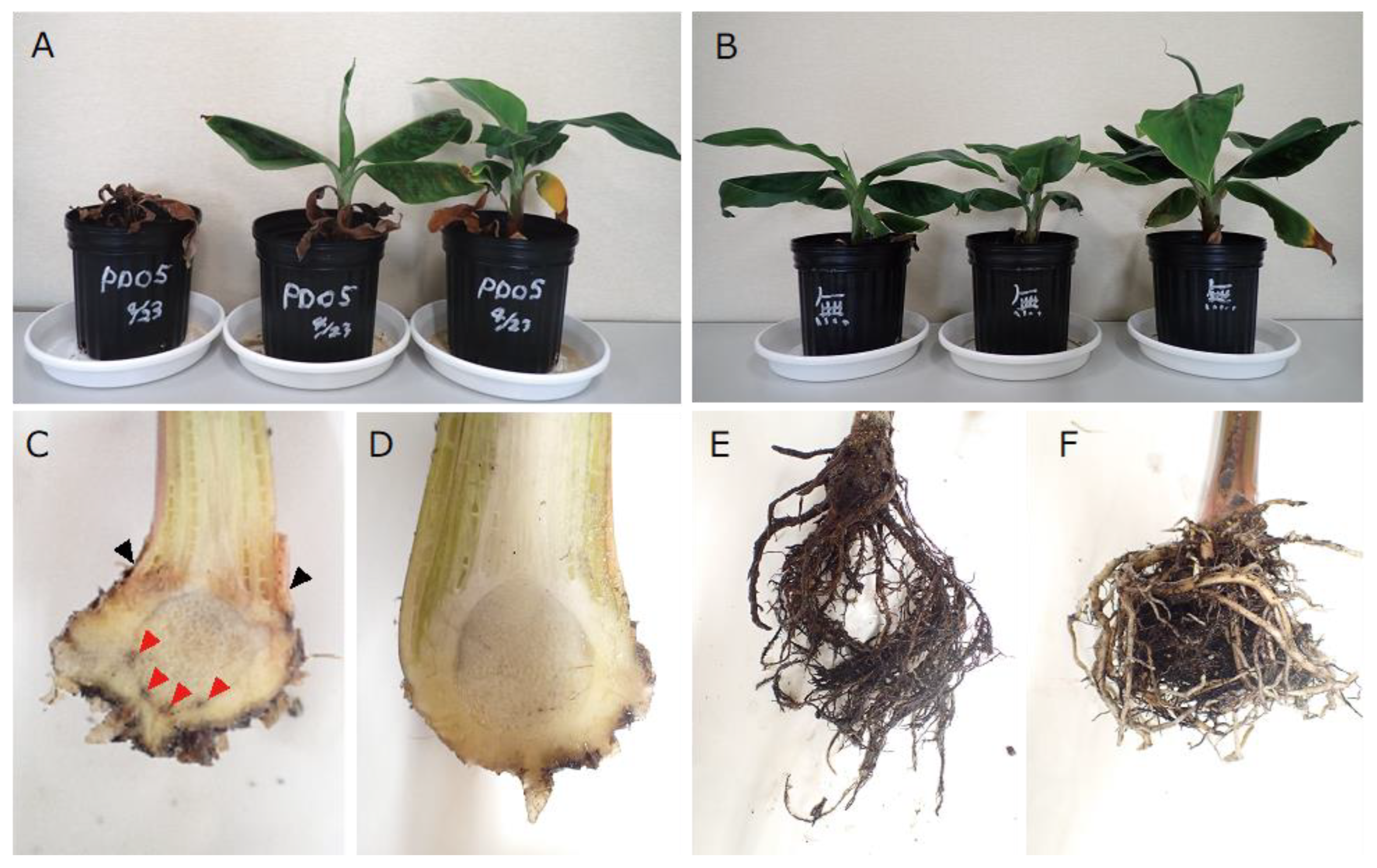
3.3. Pathogenicity Test
3.4. Detection of Secreted in Xylem Genes in Whole-Genome Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nayar, N.M. The bananas: Botany, origin, dispersal. Hortic. Res. 2010, 36, 118–164. [Google Scholar]
- Ploetz, R.C. Panama disease: Return of the first banana menace. Int. J. Pest. Manag. 1994, 40, 326–336. [Google Scholar] [CrossRef]
- Catambacan, D.G.; Cumagun, C.J. Weed-associated fungal endophytes as biocontrol agents of Fusarium oxysporum f. sp. cubense TR4. J. Fungus 2021, 7, 224. [Google Scholar] [CrossRef]
- Mostert, D.; Molina, A.B.; Daniells, J.; Fourie, G.; Hermanto, C.; Chao, C.P.; Fabregar, E.; Sinohin, V.G.; Masdek, N.; Thangavelu, R.; et al. The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE 2017, 12, e0181630. [Google Scholar] [CrossRef]
- Solpot, T.C.; Pangga, I.B.; Baconguis, R.D.; Cumagun, C.J. Occurrence of Fusarium oxysporum f. sp. cubense tropical race 4 and other genotypes in banana in south-Central Mindanao, Philippines. Philipp. Agric. Sci. 2016, 99, 370–378. [Google Scholar]
- Fraser-Smith, S.; Czislowski, E.; Meldrum, R.A.; Zander, M.; Balali, G.R.; Aitken, E.A.B. Sequence variation in the putative effector gene SIX8 facilitates molecular differentiation of Fusarium oxysporum f. sp. cubense. Plant Pathol. 2014, 63, 1044–1052. [Google Scholar] [CrossRef]
- Guo, L.; Han, L.; Yang, L.; Zeng, H.; Fan, D.; Zhu, Y.; Feng, Y.; Wang, G.; Peng, C.; Jiang, X.; et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS ONE 2014, 9, e95543. [Google Scholar] [CrossRef]
- Farah, N.; Belaffif, M.B.; Aryantha, I.N.P.; Esyanti, R.R. SIX6 shows a high divergence in Fusarium oxysporum f. sp. cubense TR4. Int. J. Agric. Biol. 2021, 25, 1331–1338. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, R.; Yoshimura, D.; Okuno, M.; Minakuchi, Y.; Kagoshima, H.; Fujiyama, A.; Kubokawa, K.; Kohara, Y.; Toyoda, A.; Itoh, T. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 2019, 10, 1702. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. Augustus: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Yilmaz, N.; Sandoval-Denis, M.; Lombard, L.; Visagie, C.M.; Wingfield, B.D.; Crous, P.W. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 2021, 46, 129–162. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- Hofstetter, V.; Miadlikowska, J.; Kauff, F.; Lutzoni, F. Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: A case study of the Lecanoromycetes (Ascomycota). Mol. Phylogenet. Evol. 2007, 44, 412–426. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N.; et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nirenberg, H.I.; O’Donnell, K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 1998, 90, 434–458. [Google Scholar] [CrossRef]
- Booth, C. Fusarium. Laboratory Guide to the Identification of the Major Species; Commonwealth Mycological Institute: Kew, UK, 1977; 58p. [Google Scholar]
- Fisher, N.L.; Burgess, W.; Toussoun, T.A.; Nelson, P.E. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 1982, 72, 151–153. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Leiva, A.M.; Rouard, M.; Lopez-Alvarez, D.; Cenci, A.; Breton, C.; Acuña, R.; Rojas, J.C.; Dita, M.; Cuellar, W.J. Draft genome sequence of Fusarium oxysporum f. sp. cubense tropical race 4 from Peru, obtained by nanopore and illumina hybrid assembly. Microbiol. Resoure Announc. 2022, 11, e00347-22. [Google Scholar] [CrossRef]
- Bugnicourt, P. Note on the mycoflora of rice seed in the territories of the South Pacific. Rev. Mycol. 1952, 17, 26–29. [Google Scholar]
- Britz, H.; Steenkamp, E.T.; Coutinho, T.A.; Wingfield, B.D.; Marasas, W.F.; Wingfield, M.J. Two new species of Fusarium section Liseola associated with mango malformation. Mycologia 2002, 94, 722–730. [Google Scholar] [CrossRef]
- Nirenberg, H. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. In Biologische Bundesanstalt für Land- und Forstwirtschaft; Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft, Berlin-Dahlem; Kommissionsverlag Paul Parey: Berlin, Germany, 1976; Volume 169, pp. 1–117. [Google Scholar]
- Van Dam, P.; Rep, M. The distribution of miniature impala elements and SIX genes in the Fusarium genus is suggestive of horizontal gene transfer. J. Mol. Evol. 2017, 85, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wu, B.; Peng, A.; Song, X.; Chen, X. The genome of banana leaf blight pathogen Fusarium sacchari str. FS66 harbors widespread gene transfer from Fusarium oxysporum. Front. Plant Sci. 2021, 12, 629859. [Google Scholar] [CrossRef]
- Hirata, T.; Kimishima, E.; Aoki, T.; Nirenberg, H.I.; O’Donnell, K. Morphological and molecular characterization of Fusarium verticillioides from rotten banana imported into Japan. Mycoscience 2001, 42, 155–166. [Google Scholar] [CrossRef]
- Maldonado-Bonilla, L.D.; Calderón-Oropeza, M.A.; Villarruel-Ordaz, J.L.; Sánchez-Espinosa, A.C. Identification of novel potential causal agents of Fusarium wilt of Musa sp. AAB in southern Mexico. J. Plant Pathol. Microbiol. 2019, 10, e479. [Google Scholar] [CrossRef]
- Czislowski, E.; Zeil-Rolfe, I.; Aitken, E.A.B. Effector profiles of endophytic Fusarium associated with asymptomatic banana (Musa sp.) Hosts. Int. J. Mol. Sci. 2021, 22, 2508. [Google Scholar] [CrossRef]
- Huang, S.P.; Wei, J.G.; Guo, T.X.; Li, Q.L.; Tang, L.H.; Mo, J.Y.; Wei, J.F.; Yang, X.B. First report of sheath rot caused by Fusarium proliferatum on pisang awak banana (Musa ABB) in China. J. Plant Pathol. 2019, 101, 1271–1272. [Google Scholar] [CrossRef]
- Maryani, N.; Sandoval-Denis, M.; Lombard, L.; Crous, P.W.; Kema, G.H.J. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Pers. Mol. Phylogeny Evol. Fungi 2019, 43, 48–69. [Google Scholar] [CrossRef]
- Molnár, O.; Bartók, T.; Szécsi, Á. Occurrence of Fusarium verticillioides and Fusarium musae on banana fruits marketed in Hungary. Acta Microbiol. Immunol. Hung. 2015, 62, 109–119. [Google Scholar] [CrossRef]
- Van Hove, F.; Waalwijk, C.; Logrieco, A.; Munaut, F.; Moretti, A. Gibberella musae (Fusarium musae) sp. nov., a recently discovered species from banana is sister to F. verticillioides. Mycologia 2011, 103, 570–585. [Google Scholar] [CrossRef]
- Thi, L.L.; Mertens, A.; Vu, D.T.; Vu, T.D.; Minh, P.L.A.; Duc, H.N.; De Backer, S.; Swennen, R.; Vandelook, F.; Panis, B.; et al. Diversity of Fusarium associated banana wilt in northern Viet Nam. MycoKeys 2022, 87, 53–76. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Pu, J.; Qi, Y.; Yu, Q.; Xie, Y.; Peng, J. Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp. cubense tropical race 4 in soil. PLoS ONE 2013, 8, e82841. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, N.; Salacinas, M.; Mendes, O.; Seidl, M.F.; Meijer, H.J.G.; Schoen, C.D.; Kema, G.H.J. A loop-mediated isothermal amplification (LAMP) assay based on unique markers derived from genotyping by sequencing data for rapid in planta diagnosis of Panama disease caused by tropical Race 4 in banana. Plant Pathol. 2019, 68, 1682–1693. [Google Scholar] [CrossRef]
- Thangavelu, R.; Edwinraj, E.; Gopi, M.; Pushpakanth, P.; Sharmila, K.; Prabaharan, M.; Loganathan, M.; Uma, S. Development of PCR-based race-specific markers for differentiation of Indian Fusarium oxysporum f. sp. cubense, the causal agent of fusarium wilt in banana. J. Fungi 2022, 8, 53. [Google Scholar] [CrossRef] [PubMed]


| Species | Culture Collections | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|
| tef1 | tub2 | cmdA | rpb2 | rpb1 | ||
| F. acutatum | CBS 402.97 T | MW402125 | MW402323 | MW402459 | MW402768 | MW402653 |
| CBS 739.97 | AF160276 | MW402348 | AF158329 | MN193883 | MW402696 | |
| CBS 137545 | MN533987 | MN534062 | MN534147 | MN534228 | MW402587 | |
| F. agapanthi | CBS 100193 | MW401959 | MW402160 | MW402363 | MW402727 | MW402491 |
| NRRL 54463 T | KU900630 | KU900635 | KU900611 | KU900625 | KU900620 | |
| NRRL 54464 | MN193856 | KU900637 | KU900613 | KU900627 | MW402718 | |
| F. ananatum | CBS 118516 T | LT996091 | MN534089 | MW402376 | LT996137 | MW402507 |
| CBS 118517 | MN533988 | MN534090 | MN534157 | MN534229 | MW402508 | |
| CBS 184.29 | MW402105 | MW402303 | MW402445 | MW402809 | MW402629 | |
| F. andiyazi | CBS 119856 | MN533989 | MN534081 | MN534174 | MN534286 | MW402523 |
| CBS 119857 T | MN193854 | LT996113 | MN534175 | LT996138 | MW402524 | |
| F. annulatum | CBS 258.54 T | MT010994 | MT011041 | MT010908 | MT010983 | MT010944 |
| F. anthophilum | CBS 108.92 | MW401965 | MW402166 | MW402368 | MW402783 | MW402498 |
| CBS 119858 | MN533990 | MN534091 | MN534158 | MN534232 | MW402525 | |
| CBS 119859 | MN533991 | MN534092 | MN534164 | MN534233 | MW402526 | |
| CBS 222.76 ET | MW402114 | MW402312 | MW402451 | MW402811 | MW402641 | |
| CBS 737.97 | MN533992 | MN534093 | MN534160 | MN534234 | MW402695 | |
| F. awaxy | CBS 119831 | MN534056 | MN534108 | MN534167 | MN534237 | MW402514 |
| CBS 119832 | MN534057 | MN534106 | MN534170 | MN534240 | MW402515 | |
| CBS 139380 | MN534058 | MN534107 | MN534172 | MN534238 | MW402597 | |
| F. bactridioides | CBS 100057 T | MN533993 | MN534112 | MN534173 | MN534235 | MW402490 |
| F. begoniae | CBS 403.97 | MN193858 | U61543 | MW402460 | MN193886 | MW402654 |
| CBS 452.97 T | MN533994 | MN534101 | MN534163 | MN534243 | MW402675 | |
| F. chinhoyiense | NY 001B5 | MN534051 | MN534083 | MN534197 | MN534263 | MW402725 |
| F. circinatum | CBS 405.97 T | MN533997 | MN534097 | MN534199 | MN534252 | MW402656 |
| CBS 119864 | MW401996 | MW402196 | MW402389 | MW402736 | MW402528 | |
| CBS 141671 | MW402083 | MW402282 | MW402427 | MW402807 | MW402610 | |
| F. concentricum | CBS 450.97 T | AF160282 | MW402334 | MW402467 | JF741086 | MW402674 |
| CBS 453.97 | MN533998 | MN534123 | MN534216 | MN534264 | MW402676 | |
| CBS 102157 | MW401963 | MW402164 | MW402367 | MW402728 | MW402496 | |
| F. dlaminii | CBS 175.88 | MN534002 | MN534138 | MN534150 | MN534256 | MW402623 |
| CBS 481.94 | MN534003 | MN534139 | MN534151 | MN534257 | MW402679 | |
| CBS 671.94 | MN534004 | MN534136 | MN534152 | MN534254 | MW402690 | |
| CBS 672.94 | MN534005 | MN534137 | MN534153 | MN534255 | MW402691 | |
| CBS 119860 T | MW401995 | MW402195 | MW402388 | KU171701 | KU171681 | |
| CBS 119861 | MN534001 | MN534135 | MN534149 | MN534253 | MW402527 | |
| F. ficicrescens | CBS 125177 | MN534006 | MN534071 | MN534176 | MN534281 | MW402545 |
| CBS 125181 | MN534007 | MN534072 | MN534177 | MN534282 | MW402548 | |
| F. fredkrugeri | CBS 144209 T | LT996097 | LT996118 | LT996181 | LT996147 | LT996199 |
| CBS 144495 | LT996096 | LT996117 | LT996180 | LT996146 | LT996198 | |
| F. fujikuroi | CBS 186.56 | MW402108 | MW402306 | MW402447 | MW402765 | MW402632 |
| CBS 265.54 | MN534011 | MN534132 | MN534222 | MN534268 | MW402650 | |
| F. globosum | CBS 428.97 T | KF466417 | MN534124 | MN534218 | KF466406 | MW402668 |
| CBS 120992 | MW401998 | MW402198 | MW402390 | MW402788 | MW402529 | |
| F. guttiforme | CBS 409.97 T | MT010999 | MT011048 | MT010901 | MT010967 | MT010938 |
| NRRL 22945 | AF160297 | U34420 | AF158350 | JX171618 | JX171505 | |
| F. konzum | CBS 119849 T | LT996098 | MN534095 | LT996182 | MW402733 | MW402519 |
| F. lactis | CBS 411.97 ET | MN193862 | MN534077 | MN534178 | MN534275 | MW402659 |
| F. madaense | CBS 146648 | MW402095 | MW402294 | MW402436 | MW402761 | MW402616 |
| CBS 146651 | MW402096 | MW402295 | MW402437 | MW402762 | MW402617 | |
| CBS 146656 | MW402097 | MW402296 | MW402438 | MW402763 | MW402618 | |
| CBS 146669 T | MW402098 | MW402297 | MW402439 | MW402764 | MW402619 | |
| F. mangiferae | CBS 119853 | MN534016 | MN534140 | MN534225 | MN534270 | MW402522 |
| CBS 120994 T | MN534017 | MN534128 | MN534224 | MN534271 | MW402530 | |
| NRRL 25226 | AF160281 | U61561 | AF158334 | HM068353 | MW402712 | |
| F. mexicanum | NRRL 47473 | GU737416 | GU737308 | GU737389 | LR792615 | LR792579 |
| F. mindanaoense (this study) | PD20-05 | LC720609 | LC720611 | LC720610 | LC720608 | LC720612 |
| F. napiforme | CBS 748.97 T | MN193863 | MN534085 | MN534192 | MN534291 | MW402701 |
| CBS 135139 | MN534019 | MN534084 | MN534183 | MN534290 | MW402572 | |
| F. nygamai | CBS 413.97 | MW402127 | MW402325 | MW402462 | MW402815 | MW402660 |
| CBS 749.97 T | MW402151 | MW402352 | MW402479 | EF470114 | MW402703 | |
| CBS 834.85 | MW402154 | MW402355 | MW402482 | MW402821 | MW402707 | |
| CBS 119852 | MW401992 | MW402192 | MW402386 | MW402734 | MW402521 | |
| CBS 139387 | MW402073 | MW402272 | MW402419 | MW402753 | MW402601 | |
| F. phyllophilum | CBS 216.76 T | MN193864 | KF466443 | KF466333 | KF466410 | MW402637 |
| F. pseudonygamai | CBS 416.97 | MN534030 | MN534064 | MN534194 | MN534283 | MW402663 |
| CBS 417.97 T | AF160263 | MN534066 | AF158316 | MN534285 | MW402664 | |
| CBS 484.94 | MN534031 | MN534065 | MN534195 | MN534284 | MW402681 | |
| F. ramigenum | CBS 418.97 T | KF466423 | MN534145 | MN534187 | KF466412 | MW402665 |
| CBS 526.97 | MN534032 | MN534086 | MN534188 | MN534292 | MW402682 | |
| F. sacchari | CBS 131372 | MN534033 | MN534134 | MN534226 | MN534293 | MW402560 |
| NY 001E9 | MN534034 | MN534133 | MN534227 | MN534294 | MW402726 | |
| F. sterilihyposum | NRRL 25623 T | MN193869 | AF160316 | AF158353 | MN193897 | MW402713 |
| F. subglutinans | CBS 747.97 NT | MW402150 | MW402351 | MW402478 | MW402773 | MW402700 |
| CBS 136481 | MW402059 | MW402258 | MW402413 | MW402748 | MW402585 | |
| F. succisae | CBS 219.76 ET | AF160291 | U34419 | AF158344 | MW402766 | MW402639 |
| F. sudanense | CBS 454.97 T | MN534037 | MN534073 | MN534179 | MN534278 | MW402677 |
| CBS 675.94 | MN534038 | MN534074 | MN534182 | MN534279 | MW402693 | |
| F. temperatum | CBS 135538 | MN534039 | MN534111 | MN534168 | MN534239 | MW402575 |
| CBS 135539 | MN534040 | MN534110 | MN534169 | MN534242 | MW402576 | |
| F. thapsinum | CBS 539.79 | MW402140 | MW402340 | MW402472 | MW402818 | MW402686 |
| CBS 100312 | MW401961 | MW402162 | MW402365 | MW402780 | MW402494 | |
| F. thapsinum | CBS 100313 | MW401962 | MW402163 | MW402366 | MW402781 | MW402495 |
| F. tupiense | NRRL 53984 T | GU737404 | GU737296 | GU737377 | LR792619 | LR792583 |
| F. udum | CBS 747.79 | MN193872 | MN534141 | MN534154 | MN534258 | MW402699 |
| F. verticillioides | CBS 125.73 | MW402012 | MW402212 | MW402392 | MW402791 | MW402543 |
| CBS 531.95 | MW402136 | MW402336 | MW402468 | MW402771 | MW402683 | |
| CBS 734.97 | MW402146 | MW402346 | AF158315 | EF470122 | MW402694 | |
| F. xylarioides | CBS 258.52 T | MN193874 | AY707118 | MW402455 | HM068355 | MW402646 |
| CBS 749.79 | MN534049 | MN534143 | AF158326 | MN534259 | MW402702 | |
| Species | Size (µm) | Septate | Shape | Substrate/ Media | References |
|---|---|---|---|---|---|
| Fusarium mindanaoense | 37.7–52.6 × 3.5–5.0 (av. ± sd. 45.9 ± 3.8 × 4.2 ± 0.35; 3-septate) 50.4–66.4 × 3.1–4.6 (av. ± sd. 57.6 ± 3.4 × 3.7 ± 0.39; 4-septate) 55.5–67.8 × 3.2–4.1 (av. ± sd. 62.1 ± 4.3 × 4.1 ± 0.7; 5-septate) | 3-5 | Slightly curved | CLA | This study |
| F. annulatum | 13–58 × 1.9–3.3 | 3-6 | Menidiform or annular | Not mentioned | Bugnicourt [30] |
| F. concentricum | 53.5–61.4 × 3.7–4 (avg. 57.4 × 3.7) | 3-5 | Slightly curved | SNA | Nirenberg and O’Donnell [23] |
| F. mangiferae | 43.1–61.4 × 3 1.9–3.4 (avg. 51.8 × 2.3) | 3-5 | Slightly curved | CLA | Britz et al. [31] |
| F. sacchari | 35.5–49.5 × 3.3–4.1 | 1-5 | Slightly curved | SNA | Nirenberg [32] |
| Hit_Defintion | Score | e-Value | Query_from | Query_to | Hit_from | Hit_to | Identity |
|---|---|---|---|---|---|---|---|
| KAF5645217.1 secreted in xylem Fusarium sp. NRRL 25303 | 504.597 | 0 | 1 | 246 | 1 | 246 | 241 |
| KAG4288609.1 secreted in xylem 6 Fusarium proliferatum | 503.056 | 0 | 1 | 247 | 1 | 247 | 240 |
| KAG4277728.1 secreted in xylem 6 Fusarium proliferatum | 501.13 | 0 | 1 | 247 | 1 | 247 | 240 |
| KAG4252980.1 secreted in xylem 6 Fusarium proliferatum | 499.204 | 0 | 9 | 247 | 1 | 239 | 239 |
| RBA12867.1 secreted in xylem 6 Fusarium proliferatum | 498.049 | 0 | 1 | 247 | 1 | 247 | 239 |
| KAF5709672.1 secreted in xylem Fusarium globosum | 498.049 | 0 | 9 | 246 | 1 | 238 | 238 |
| KAF4501124.1 secreted in xylem 6 Fusarium agapanthi | 484.567 | 3.32 × 10−178 | 1 | 246 | 1 | 246 | 230 |
| KAF5689079.1 secreted in xylem Fusarium denticulatum | 476.093 | 6.97 × 10−175 | 1 | 246 | 1 | 246 | 224 |
| KAF5626692.1 secreted in xylem 6 Fusarium tjaetaba | 475.707 | 1.07 × 10−174 | 1 | 246 | 1 | 246 | 224 |
| XP_037203386.1 secreted in xylem 6 Fusarium tjaetaba | 475.707 | 1.07 × 10−174 | 1 | 246 | 1 | 246 | 224 |
| KAF5565621.1 secreted in xylem 6 Fusarium napiforme | 473.781 | 4.55 × 10−174 | 1 | 246 | 1 | 246 | 223 |
| KAF5589364.1 secreted in xylem 6 Fusarium pseudocircinatum | 470.315 | 1.08 × 10−172 | 1 | 246 | 1 | 246 | 223 |
| KAF5661873.1 secreted in xylem 6 Fusarium circinatum | 466.463 | 3.68 × 10−171 | 9 | 246 | 1 | 238 | 221 |
| KAF5538596.1 secreted in xylem 6 Fusarium phyllophilum | 444.121 | 1.90 × 10−162 | 9 | 246 | 1 | 238 | 210 |
| KAF5719947.1 secreted in xylem 6 Fusarium mundagurra | 423.32 | 4.06 × 10−154 | 1 | 246 | 1 | 242 | 200 |
| KAF5588511.1 secreted in xylem 6 Fusarium pseudoanthophilum | 403.675 | 2.64 × 10−146 | 1 | 246 | 1 | 242 | 203 |
| KAF5973041.1 Secreted in xylem 6 Fusarium coicis | 348.977 | 3.92 × 10−125 | 1 | 246 | 1 | 208 | 180 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nozawa, S.; Seto, Y.; Takata, Y.; Narreto, L.A.; Valle, R.R.; Okui, K.; Taida, S.; Alvindia, D.G.; Reyes, R.G.; Watanabe, K. Fusarium mindanaoense sp. nov., a New Fusarium Wilt Pathogen of Cavendish Banana from the Philippines Belonging to the F. fujikuroi Species Complex. J. Fungi 2023, 9, 443. https://doi.org/10.3390/jof9040443
Nozawa S, Seto Y, Takata Y, Narreto LA, Valle RR, Okui K, Taida S, Alvindia DG, Reyes RG, Watanabe K. Fusarium mindanaoense sp. nov., a New Fusarium Wilt Pathogen of Cavendish Banana from the Philippines Belonging to the F. fujikuroi Species Complex. Journal of Fungi. 2023; 9(4):443. https://doi.org/10.3390/jof9040443
Chicago/Turabian StyleNozawa, Shunsuke, Yosuke Seto, Yoshiki Takata, Lalaine Albano Narreto, Reynaldo R. Valle, Keiju Okui, Shigeya Taida, Dionisio G. Alvindia, Renato G. Reyes, and Kyoko Watanabe. 2023. "Fusarium mindanaoense sp. nov., a New Fusarium Wilt Pathogen of Cavendish Banana from the Philippines Belonging to the F. fujikuroi Species Complex" Journal of Fungi 9, no. 4: 443. https://doi.org/10.3390/jof9040443
APA StyleNozawa, S., Seto, Y., Takata, Y., Narreto, L. A., Valle, R. R., Okui, K., Taida, S., Alvindia, D. G., Reyes, R. G., & Watanabe, K. (2023). Fusarium mindanaoense sp. nov., a New Fusarium Wilt Pathogen of Cavendish Banana from the Philippines Belonging to the F. fujikuroi Species Complex. Journal of Fungi, 9(4), 443. https://doi.org/10.3390/jof9040443







