Phenylalanine Ammonia-Lyase: A Key Gene for Color Discrimination of Edible Mushroom Flammulina velutipes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strain Culture and Genomic DNA Isolation
2.2. Genome Sequencing and Identification of Variations
2.3. Primer Design and Amplification Refractory Mutation System PCRs
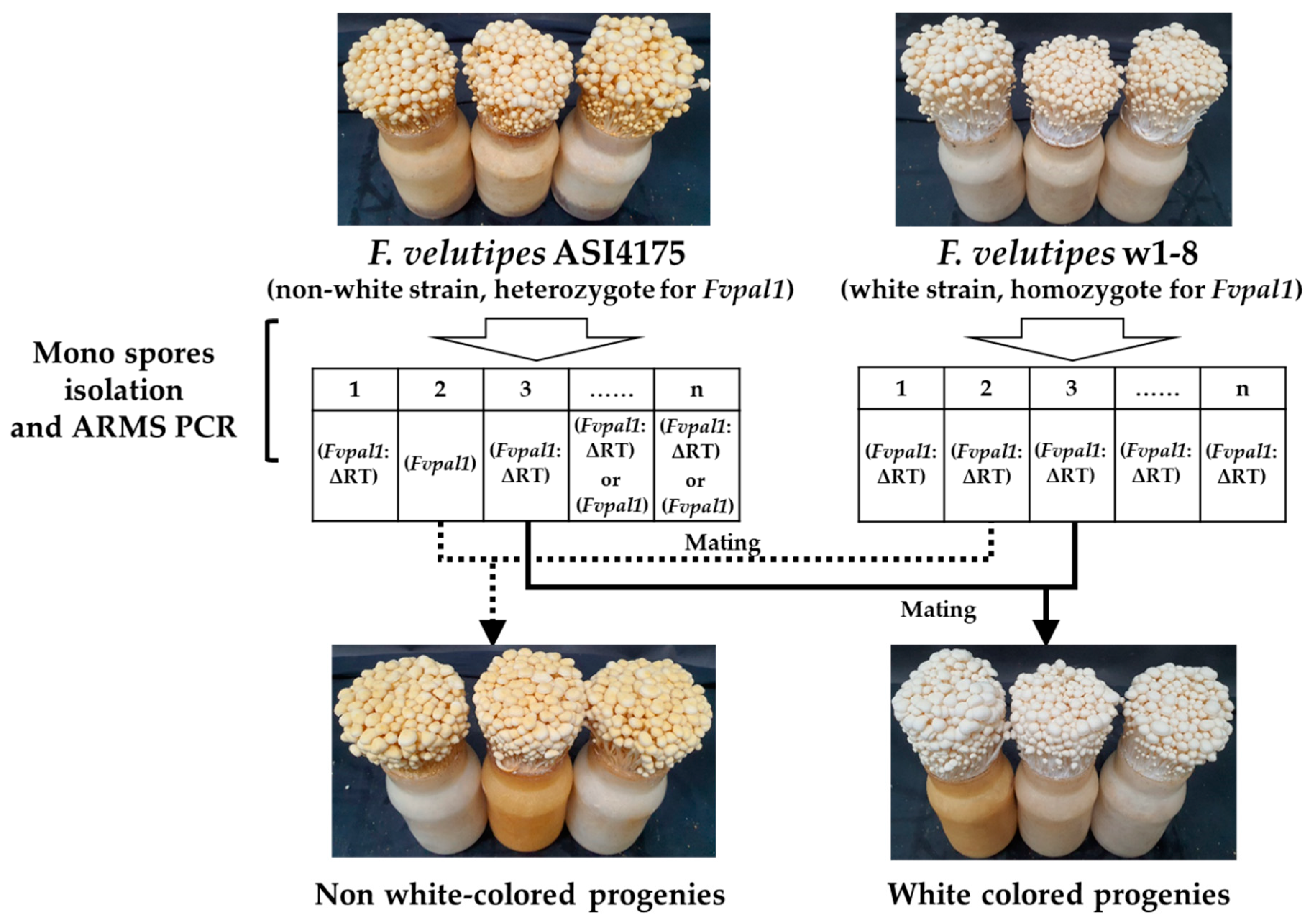
2.4. Mating of F. velutipes Strains
3. Results and Discussion
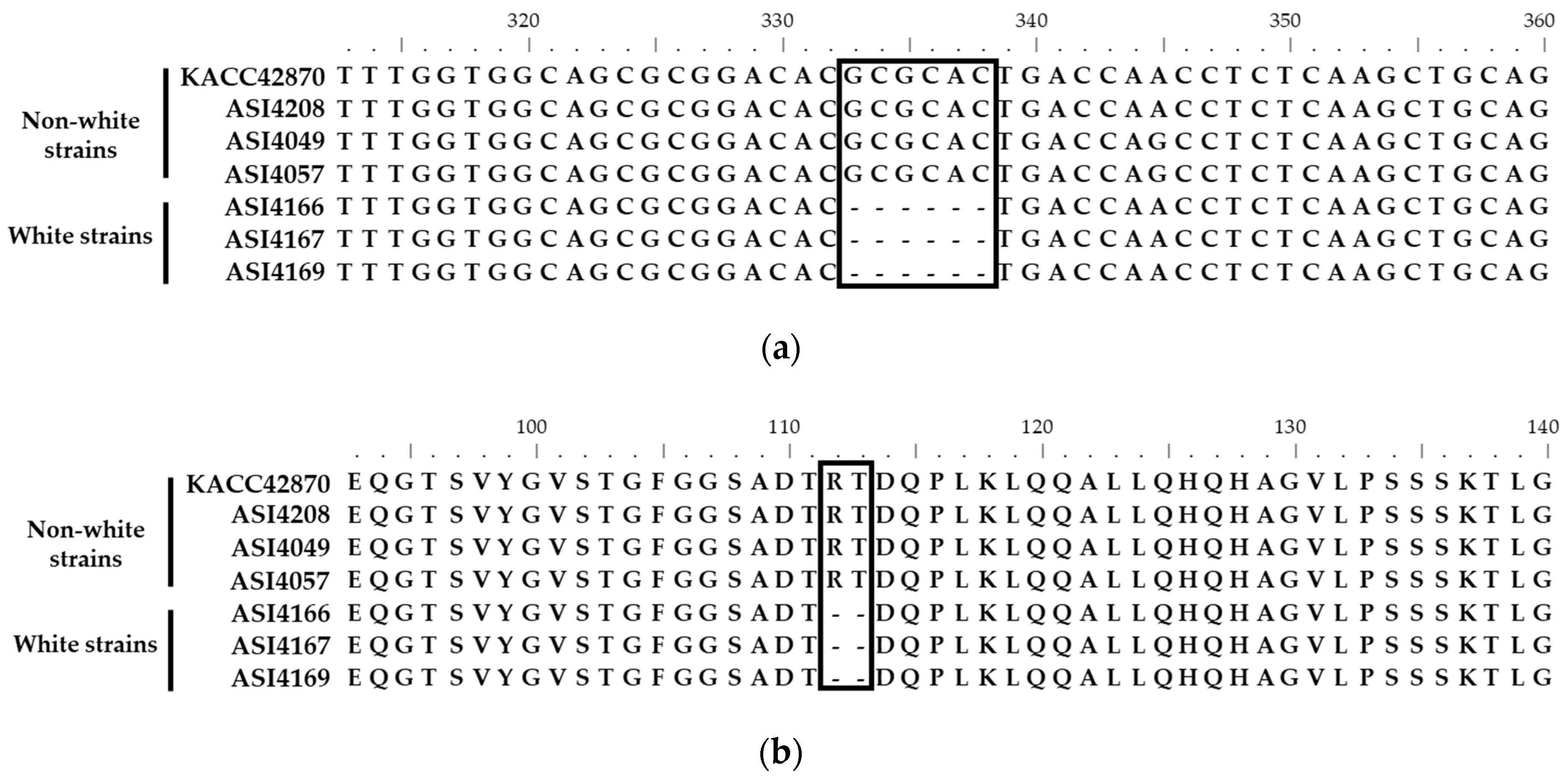
3.1. Identification of Fruiting Body Color-Specific Mutation of F. velutipes
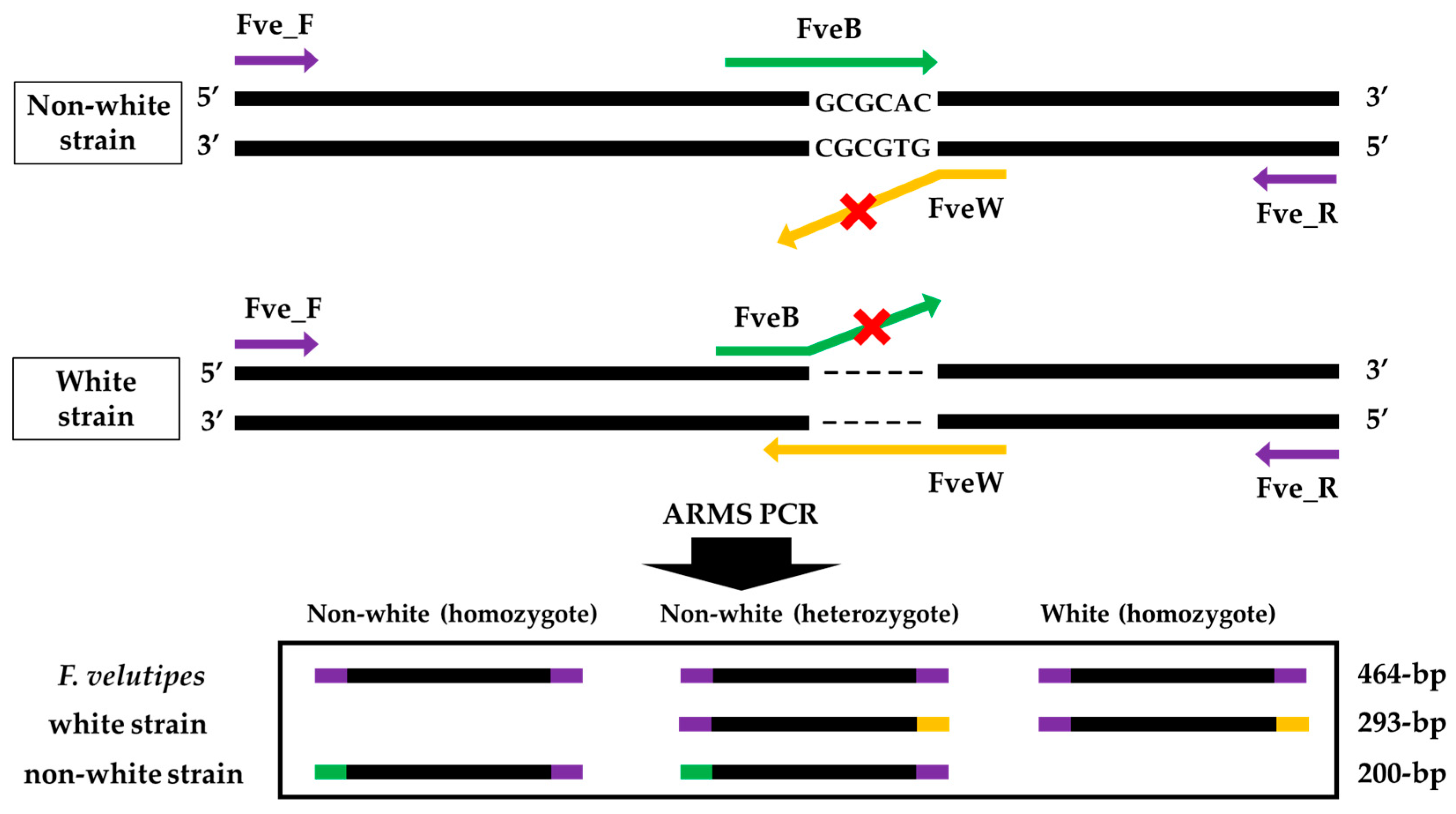
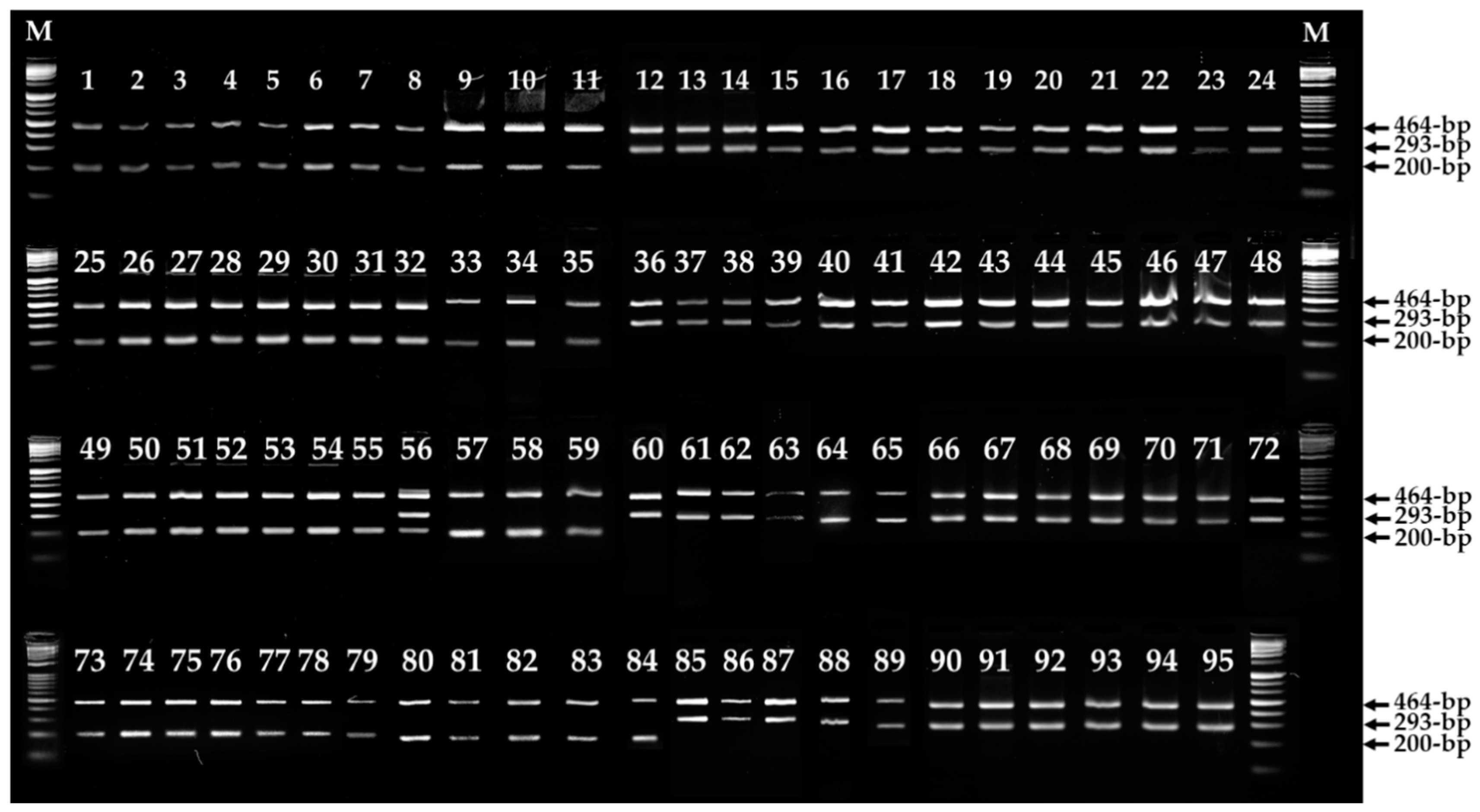
3.2. Primers Design and ARMS PCR for Fruiting Body Color Discrimination of F. velutipes
3.3. Mutation in the Fvpal1 Gene Affect Fruiting Body Color of F. velutipes
3.4. Structural Characteristics of the PAL of F. velutipes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, Y.J.; Baek, J.H.; Lee, S.; Kim, C.; Rhee, H.; Kim, H.; Seo, J.S.; Park, H.R.; Yoon, D.E.; Nam, J.Y.; et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS ONE 2014, 9, e93560. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Woo, S.I.; Oh, M.; Oh, Y.L.; Shin, P.G.; Jnag, K.Y.; Kong, W.S.; Lee, C.S. Characteristics of ‘Baekjung’, a variety adaptable to high temperature in Flammulina velutipes. J. Mushrooms 2015, 13, 203–206. [Google Scholar] [CrossRef]
- Kong, W.S.; Jang, K.Y.; Lee, C.Y.; Koo, J.; Shin, P.G.; Jhune, C.S.; Oh, Y.; Yoo, Y.B.; Suh, J.S. Breeding progress and characterization of a Korean white variety ‘Baek-A’ in Flammulina velutipes. J. Mushrooms 2013, 11, 159–163. [Google Scholar] [CrossRef]
- Kitamoto, Y.; Nakamata, M.; Masuda, P. Production of novel white Flammulina velutipes by breeding. In Geneticsand Breeding of Edible Mushrooms, 1st ed.; Chang, S.T., Buswell, J.A., Miles, P.G., Eds.; Routledge: London, UK, 1993; pp. 65–86. [Google Scholar] [CrossRef]
- Woo, S.I.; Kong, W.S.; Jang, K.Y. Characteristics of ’Baekseung’, a new cultivar Flammulina velutipes. J. Mushrooms 2017, 15, 25–30. [Google Scholar] [CrossRef]
- Kim, M.J.; Lee, K.W.; Chang, W.B.; Jeon, J.O.; Kim, I.J. Characteristics and breeding of ‘Yeoreumhyang2ho’: A new blackish-brown variety of Flammulina velutipes that is adaptable to high temperature. J. Mushrooms 2018, 15, 192–197. [Google Scholar] [CrossRef]
- Kim, M.J.; Lee, K.W.; Chang, W.B.; Jeon, J.O.; Kim, I.J. Characteristics and breeding of ‘Yeoreumhyang1ho’: A new light brown variety of Flammulina velutipes adaptable to high temperature. J. Mushrooms 2018, 16, 287–292. [Google Scholar] [CrossRef]
- Kim, M.J.; Lee, K.W.; Chang, W.B.; Jeon, J.O.; Kim, I.J. Characteristics and breeding of ‘Geumhyang2ho’, a new brown and labor-saving variety of Flammulina velutipes. J. Mushrooms 2018, 16, 293–298. [Google Scholar] [CrossRef]
- Im, J.H.; Jang, K.Y.; Oh, Y.L.; Oh, M.; Raman, J.; Kong, W.S. Breeding on a new cultivar of golden Flammulina velutipes ‘Auram’. J. Mushrooms 2019, 17, 218–223. [Google Scholar] [CrossRef]
- Kim, M.J.; Chang, H.B.; Choi, J.S.; Lee, K.W.; Joo, G.N.; Kim, Y.G. Characteristics and breeding of a new brown variety ‘Geumhyang’ with short cultivation period in Flammulina velutipes. J. Mushrooms 2015, 13, 92–96. [Google Scholar] [CrossRef]
- Kim, M.J.; Chang, H.B.; Choi, J.S.; Lee, K.W.; Joo, G.N.; Kim, Y.G. Characteristics and breeding of a new brown variety ‘Heukhyang’ with good taste in Flammulina velutipes. J. Mushrooms 2015, 13, 103–107. [Google Scholar] [CrossRef]
- Jhune, C.S.; Yun, H.S.; Leem, H.T.; Kong, W.S.; Lee, K.H.; Sung, G.H.; Cho, J.H. Changes of saccharide content in fruitbody composition of Flammulina velutipes during storage. J. Mushrooms 2011, 9, 123–131. [Google Scholar] [CrossRef]
- Jhune, C.S.; Yun, H.S.; Leem, H.T.; Gong, W.S.; Sung, G.H.; Cho, J.H.; Park, K.M. The change of amino acid content in fruit-body of winter mushroom according to the storage period and strains. J. Mushrooms 2012, 10, 224–235. [Google Scholar] [CrossRef]
- Kim, J.T.; Kim, M.J.; Jhune, C.S.; Shin, P.G.; Oh, Y.L.; Yoo, Y.B.; Kong, W.S. Comparison of amino acid and free amino acid contents between cap and stipe in Flammulina velutipes and Pleurotus ostreatus. J. Mushrooms 2014, 12, 341–349. [Google Scholar] [CrossRef]
- Kim, K.J.; Jin, S.W.; Choi, B.S.; Kim, J.K.; Koh, Y.W.; Ban, S.E.; Seo, K.S. Evaluation of the nutrition properties of Flammulina velutipes. J. Mushrooms 2016, 14, 44–50. [Google Scholar] [CrossRef]
- An, G.H.; Han, J.G.; Cho, J.H. Comparisons of biological activities and amino acid contents of edible mushrooms extracted using different solvents. J. Mushrooms 2020, 18, 53–62. [Google Scholar] [CrossRef]
- An, G.H.; Han, J.G.; Kim, O.T.; Cho, J.H. Changes of biological activity and nutritional content by processing methods of Flammulina velutipes, Grifola frondosa, and Sparassis crispa. J. Mushrooms 2020, 18, 403–409. [Google Scholar] [CrossRef]
- Nerya, O.; Ben-Arie, R.; Luzzatto, T.; Musa, R.; Khativ, S.; Vaya, J. Prevention of Agaricus bisporus postharvest browning with tyrosinase inhibitors. Postharvest Biol. Technol. 2006, 39, 272–277. [Google Scholar] [CrossRef]
- Jolivet, S.; Arpin, N.; Wichers, H.J.; Pellon, G. Agaricus bisporus browning: A review. Mycol. Res. 1988, 102, 1459–1483. [Google Scholar] [CrossRef]
- Claus, H.; Decker, H. Bacterial tyrosinases. Sys. Appl. Microbiol. 2006, 29, 3–14. [Google Scholar] [CrossRef]
- Lee, D.; Jang, E.H.; Lee, M.; Kim, S.W.; Lee, Y.; Lee, K.T.; Bahn, Y.S. Unraveling melanin biosynthesis and signaling networks in Cryptococcus neoformans. mBio 2019, 10, e02267-19. [Google Scholar] [CrossRef]
- Conesa, A.; Punt, P.J.; van den Hondel, C.A.M.J.J. Fungal peroxidases: Molecular aspects and applications. J. Biotechnol. 2002, 93, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 897–971. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Jeong, Y.U.; Kong, W.S. Genome sequencing and carbohydrate-active enzyme (CAZyme) repertoire of the white rot fungus Flammulina elastica. Int. J. Mol. Sci. 2018, 19, 2379. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Kong, W.S.; Park, Y.J. Genome sequencing and genome-wide identification of carbohydrate-active enzymes (CAZymes) in the white rot fungus Flammulina fennae. Microbiol. Biotechnol. Lett. 2018, 46, 300–312. [Google Scholar] [CrossRef]
- Park, Y.J.; Kong, W.S. Genome-wide comparison of carbohydrate-active enzymes (CAZymes) repertoire of Flammulina ononidis. Mycobiology 2018, 46, 349–360. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, C.S.; Kong, W.S. Genomic insights into the fungal lignocellulolytic machinery of Flammulina rossica. Microorganisms 2019, 7, 421. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.W.; Im, J.H.; Kong, W.S.; Park, Y.J. Comparative analysis of carbohydrate active enzymes in the Flammulina velutipes var. lupinicola genome. Microorganisms 2021, 9, 20. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute. Available online: http://broadinstitute.github.io/picard/ (accessed on 11 August 2022).
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Palmer, J.M.; Stajich, J.E. Funannotate: Eukaryotic Genome Annotation Pipeline. Available online: http://funannotate.readthedocs.io (accessed on 11 August 2022).
- Little, S. Amplification-refractory mutation system (ARMS) analysis of point mutations. Curr. Protoc. Hum. Genet. 2001, 7, 9–18. [Google Scholar] [CrossRef]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and plant phenylalanine ammonia-lyase. Mycobiology 2011, 39, 257–265. [Google Scholar] [CrossRef]
- Kong, J.Q. Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RCS Adv. 2015, 5, 62587–62603. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; Loaiza-Velarde, J.; Bonfanti, A.; Saltveit, M.E. Early wound- and ethylene-induced changes in phenylpropanoid metabolism in harvested lettuce. J. Am. Soc. Hortic. Sci. 1997, 122, 399–404. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; Gil-Muñoz, M.I.; Castañer, M.; Artés, F.; Saltveit, M.E. Effect of selected browning inhibitors on phenolic metabolism in stem tissue of harvested lettuce. J. Agric. Food Chem. 1997, 45, 583–589. [Google Scholar] [CrossRef]
- Ke, D.; Saltveit, M.E. Wound-induced ethylene production, phenolic metabolism and susceptibility to russet spotting in iceberg lettuce. Physiol. Plant. 1989, 76, 412–418. [Google Scholar] [CrossRef]
- Sameshima, N.; Nishimura, M.; Murakami, K.; Kogo, Y.; Shimamura, Y.; Sakuta, M.; Murata, M. Cloning of phenylalanine ammonia-lyase and its role in enzymatic browning of mung bean sprout during cold storage. Food Sci. Technol. Res. 2016, 22, 255–260. [Google Scholar] [CrossRef]
- Barroga, C.F.; Laurena, A.C.; Mendoza, E.M.T. Polyphenols in mung bean (Vigna radiata (L.) Wilczek): Determination and removal. J. Agric. Food Chem. 1985, 33, 1006–1009. [Google Scholar] [CrossRef]
- Nishimura, M.; Sameshima, N.; Joshita, K.; Murata, M. Regulation of enzymatic browning of mung bean sprout by heat-shock treatment. Food Sci. Technol. Res. 2012, 18, 412–417. [Google Scholar] [CrossRef]
- Power, D.M.; Towers, G.H.; Neish, A.C. Biosynthesis of phenolic acids by certain wood-destroying Basidiomycetes. Can. J. Biochem. 1965, 43, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Subba Rao, P.V.; Towers, G.H. Degradation of phenylalanine and tyrosine by Basidiomycetes. Life Sci. 1967, 6, 2629–2633. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Nabe, K.; Izuo, N.; Nakamichi, K.; Chibata, I. Production of l-phenylalanine from trans-cinnamic acid with Rhodotorula glutinis containing l-phenylalanine ammonia lyase activity. Appl. Environ. Microbiol. 1981, 42, 773–778. [Google Scholar] [CrossRef]
- Moore, K.; Subba Rao, P.V.; Towers, G.H. Degradation of phenylalanine and tyrosine by Sporobolomyces roseus. Biochem. J. 1968, 106, 507–514. [Google Scholar] [CrossRef]
- Leal, G.A.; Gomes, L.H.; Albuquerque, P.S.; Tavares, F.C.; Figueira, A. Searching for Moniliophthora perniciosa pathogenicity genes. Fungal Biol. 2010, 114, 842–854. [Google Scholar] [CrossRef]
- Yoon, H.; You, Y.H.; Kim, Y.E.; Kim, Y.J.; Kong, W.S.; Kim, J.G. Cloning and mRNA expression analysis of the gene encoding phenylalanine ammonia-lyase of the ectomycorrhizal fungus Tricholoma matsutake. J. Microbiol. Biotechnol. 2013, 23, 1055–1059. [Google Scholar] [CrossRef]
- Yun, Y.H.; Koo, J.S.; Kim, S.H.; Kong, W.S. Cloning and expression analysis of phenylalanine ammonia-lyase gene in the mycelium and fruit body of the edible mushroom Flammulina velutipes. Mycobiology 2015, 43, 327–332. [Google Scholar] [CrossRef]
- Oufedjikh, H.; Mahrouz, M.; Amiot, M.J.; Lacroix, M. Effect of γ-irradiation on phenolic compounds and phenylalanine ammonia-lyase activity during storage in relation to peel injury from peel of Citrus clementina Hort. Ex. Tanaka. J. Agric. Food Chem. 2000, 48, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Poppe, L. Methylidene-imidazolone: A novel electrophile for substrate activation. Curr. Opin. Chem. Biol. 2001, 5, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Heberling, M.M.; Wu, B.; Bartsch, S.; Janssen, D.B. Priming ammonia lyases and aminomutases for industrial and therapeutic applications. Curr. Opin. Chem. Biol. 2013, 17, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.D.; Qiu, J.Q.; Fan, X.W.; Jia, S.R.; Tan, Z.L. Biotechnological production and applications of microbial phenylalanine ammonia lyase: A recent review. Crit. Rev. Biotechnol. 2014, 34, 258–268. [Google Scholar] [CrossRef]
- Rother, D.; Poppe, L.; Morlock, G.; Viergutz, S.; Retey, J. An active site homology model of phenylalanine ammonia-lyase from Petroselinum crispum. Eur. J. Biochem. 2002, 269, 3065–3075. [Google Scholar] [CrossRef]
- Schuster, B.; Retey, J. The mechanism of action of phenylalanine ammonia-lyase: The role of prosthetic dehydroalanine. Proc. Natl. Acad. Sci. USA. 1995, 92, 8433–8437. [Google Scholar] [CrossRef]
- Abell, C.W.; Shen, R.S. Phenylalanine ammonia-lyase from the yeast Rhodotorula glutinis. Methods Enzymol. 1987, 142, 242–253. [Google Scholar] [CrossRef]
- Goldson-Barnaby, A.; Scaman, C.H. Purification and characterization of phenylalanine ammonia lyase from Trichosporon cutaneum. Enzym. Res. 2013, 2013, 670702. [Google Scholar] [CrossRef]
- Vannelli, T.; Xue, Z.; Breinig, S.; Qi, W.W.; Sariaslani, F.S. Functional expression in Escherichia coli of the tyrosine inducible tyrosine ammonia-lyase enzyme from yeast Trichosporon cutaneum for production of p-hydroxycinnamic acid. Enzym. Microb. Technol. 2007, 41, 413–422. [Google Scholar] [CrossRef]
- Ameyama, M.; Shinagawa, E.; Matsushita, K.; Adachi, O. Growth stimulation of microorganisms by pyrroloquinoline quinone. Agri. Biol. Chem. 1984, 48, 2909–2911. [Google Scholar] [CrossRef]
- Xue, Z.; McCluskey, M.; Cantera, K.; Ben-Bassat, A.; Sariaslani, F.S.; Huang, L. Improved production of p-hydroxycinnamic acid from tyrosine using a novel thermostable phenylalanine/tyrosine ammonia lyase enzyme. Enzyme Microb. Technol. 2007, 42, 58–64. [Google Scholar] [CrossRef]

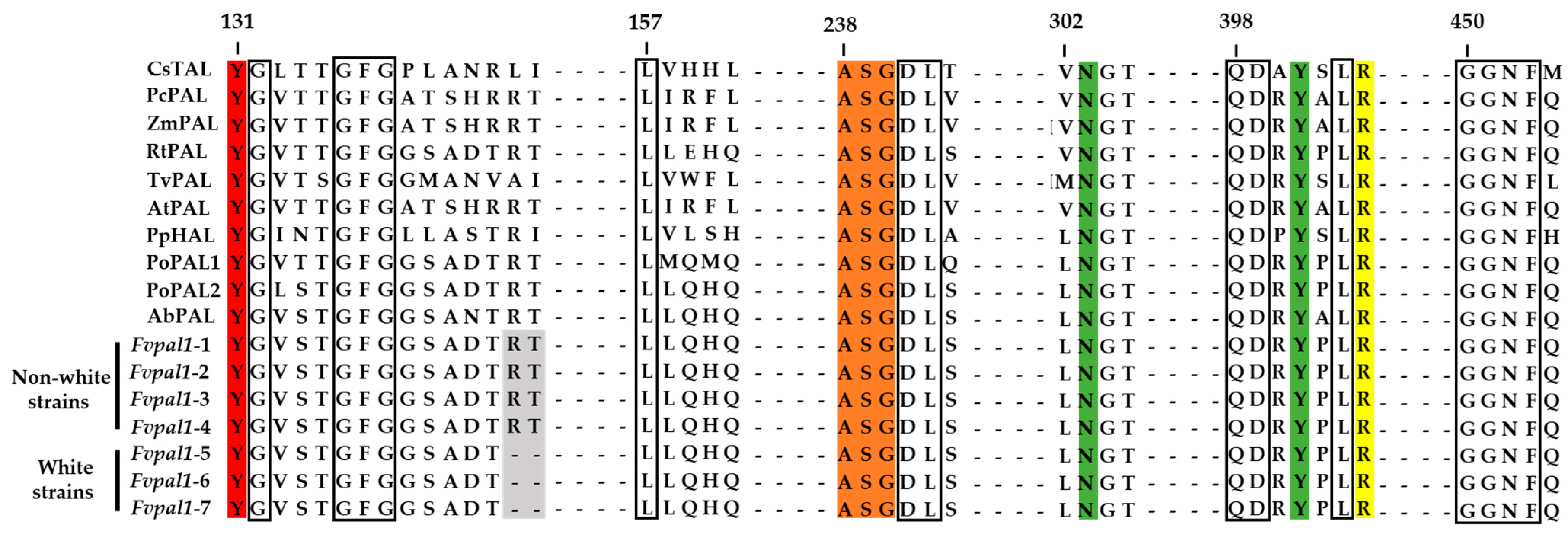
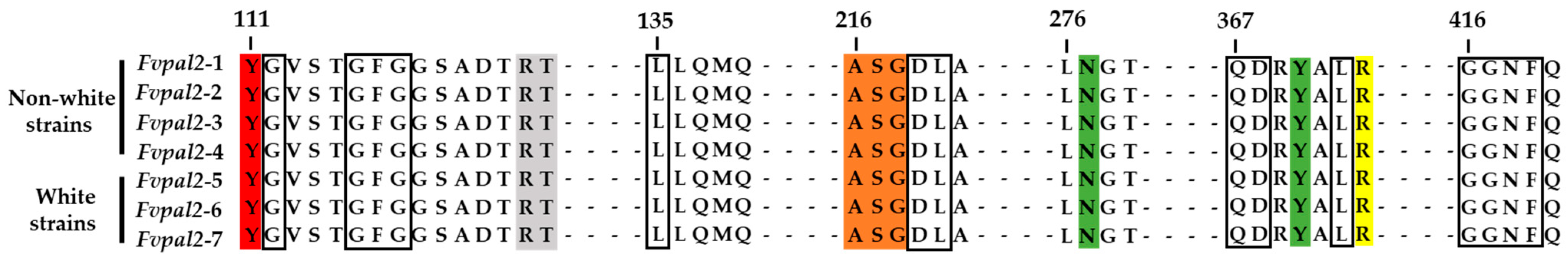
| No. | Strains | Color | No. | Strains | Color | No. | Strains | Color |
|---|---|---|---|---|---|---|---|---|
| 1 | ASI4019 | non-white | 41 | T011 | white | 81 | B21 | non-white |
| 2 | ASI4019-20 | non-white | 42 | honam | white | 82 | B8 | non-white |
| 3 | ASI4019-1820 | non-white | 43 | daeheung | white | 83 | B26 | non-white |
| 4 | ASI4028 | non-white | 44 | W6-8 | white | 84 | B39 | non-white |
| 5 | ASI4049 | non-white | 45 | W6-18 | white | 85 | baegi | white |
| 6 | ASI4019-2003 | non-white | 46 | W6-19 | white | 86 | baekseung | white |
| 7 | ASI4057 | non-white | 47 | W6-14 | white | 87 | baekjung | white |
| 8 | ASI4064 | non-white | 48 | W1-9 | white | 88 | 7937 | white |
| 9 | ASI4067 | non-white | 49 | hwanggeum | non-white | 89 | 8492 | white |
| 10 | ASI4069 | non-white | 50 | B151 | non-white | 90 | naju | white |
| 11 | ASI4072 | non-white | 51 | B129 | non-white | 91 | ASI4100 | white |
| 12 | ASI4074 | white | 52 | B112 | non-white | 92 | ASI4102 | white |
| 13 | ASI4166 | white | 53 | B13 | non-white | 93 | hampyeong | white |
| 14 | ASI4167 | white | 54 | yeoreumhyang2ho | non-white | 94 | W8-17 | white |
| 15 | ASI4168 | white | 55 | B16 | non-white | 95 | ASI4168 | white |
| 16 | ASI4169 | white | 56 | 4175 | non-white | |||
| 17 | ASI4178 | white | 57 | B17 | non-white | |||
| 18 | ASI4029 | white | 58 | B18 | non-white | |||
| 19 | ASI4210 | white | 59 | B62 | non-white | |||
| 20 | ASI4216 | white | 60 | W5-2 | white | |||
| 21 | ASI4217 | white | 61 | W1-18 | white | |||
| 22 | ASI4226 | white | 62 | W6-13 | white | |||
| 23 | ASI4228 | white | 63 | W8-18 | white | |||
| 24 | ASI4230 | white | 64 | W1-23 | white | |||
| 25 | ASI4083 | non-white | 65 | W1-8 | white | |||
| 26 | ASI4090 | non-white | 66 | W3-24 | white | |||
| 27 | ASI4103 | non-white | 67 | W4-16 | white | |||
| 28 | ASI4111 | non-white | 68 | W5-3 | white | |||
| 29 | ASI4146 | non-white | 69 | W5-16 | white | |||
| 30 | ASI4148 | non-white | 70 | W6-13 | white | |||
| 31 | ASI4149 | non-white | 71 | W6-16 | white | |||
| 32 | ASI4163 | non-white | 72 | W8-1 | white | |||
| 33 | ASI4218 | non-white | 73 | B70 | non-white | |||
| 34 | ASI4219 | non-white | 74 | B74 | non-white | |||
| 35 | ASI4232 | non-white | 75 | B87 | non-white | |||
| 36 | ASI4231 | white | 76 | B66 | non-white | |||
| 37 | ASI0003 | white | 77 | B127 | non-white | |||
| 38 | ASI0019 | white | 78 | B162 | non-white | |||
| 39 | jeonnam | white | 79 | B63 | non-white | |||
| 40 | cheongdo | white | 80 | B121 | non-white |
| Primer | Sequences (5′–3′) | Target | Amplicon Size (Base Pair) |
|---|---|---|---|
| Fve_F | TCTCCACTTACCTTCTCCTA | Flammulina velutipes strains | 464 |
| Fve_R | TATGGTAAGTACACGGTCAG | ||
| Fve_F | TCTCCACTTACCTTCTCCTA | Flammulina velutipes white strains | 293 |
| FveW | TTGAGAGGTTGGTCAGTGTC | ||
| Fve_R | TATGGTAAGTACACGGTCAG | Flammulina velutipes non-white strains | 200 |
| FveB | TTCCTAGCGGACACGCGCAC |
| Strains | Fruiting Body Color | Sequencing Reads | Mapping Rate (%) | References or Accession Number | |
|---|---|---|---|---|---|
| Total Reads | Trimmed Reads (%) | ||||
| F. velutipes KACC42870 | non-white | Reference strain | [1] | ||
| F. velutipes KACC43778 | non-white | Reference strain | [1] | ||
| F. velutipes ASI4028 | non-white | 7,510,278 | 6,355,902 (84.63) | 77.93 | NN-1534-000001 |
| F. velutipes ASI4049 | non-white | 11,535,948 | 9,447,436 (81.9) | 80.13 | NN-1535-000001 |
| F. velutipes ASI4057 | non-white | 8,134,556 | 6,949,358 (85.43) | 77.58 | NN-1537-000001 |
| F. velutipes ASI4166 | white | 29,145,728 | 26,852,658 (92.13) | 73.15 | NN-0798-000001 |
| F. velutipes ASI4167 | white | 13,665,794 | 11,211,378 (82.04) | 75.89 | NN-1545-000001 |
| F. velutipes ASI4169 | white | 17,619,236 | 14,367,908 (81.55) | 78.61 | NN-1546-000001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Im, J.-H.; Yu, H.-W.; Park, C.-H.; Kim, J.-W.; Shin, J.-H.; Jang, K.-Y.; Park, Y.-J. Phenylalanine Ammonia-Lyase: A Key Gene for Color Discrimination of Edible Mushroom Flammulina velutipes. J. Fungi 2023, 9, 339. https://doi.org/10.3390/jof9030339
Im J-H, Yu H-W, Park C-H, Kim J-W, Shin J-H, Jang K-Y, Park Y-J. Phenylalanine Ammonia-Lyase: A Key Gene for Color Discrimination of Edible Mushroom Flammulina velutipes. Journal of Fungi. 2023; 9(3):339. https://doi.org/10.3390/jof9030339
Chicago/Turabian StyleIm, Ji-Hoon, Hye-Won Yu, Che-Hwon Park, Jin-Woo Kim, Ju-Hyeon Shin, Kab-Yeul Jang, and Young-Jin Park. 2023. "Phenylalanine Ammonia-Lyase: A Key Gene for Color Discrimination of Edible Mushroom Flammulina velutipes" Journal of Fungi 9, no. 3: 339. https://doi.org/10.3390/jof9030339
APA StyleIm, J.-H., Yu, H.-W., Park, C.-H., Kim, J.-W., Shin, J.-H., Jang, K.-Y., & Park, Y.-J. (2023). Phenylalanine Ammonia-Lyase: A Key Gene for Color Discrimination of Edible Mushroom Flammulina velutipes. Journal of Fungi, 9(3), 339. https://doi.org/10.3390/jof9030339





