Small Signals Lead to Big Changes: The Potential of Peptide-Induced Resistance in Plants
Abstract
1. The Plant Immune System
2. Phytocytokines in Basal Immunity
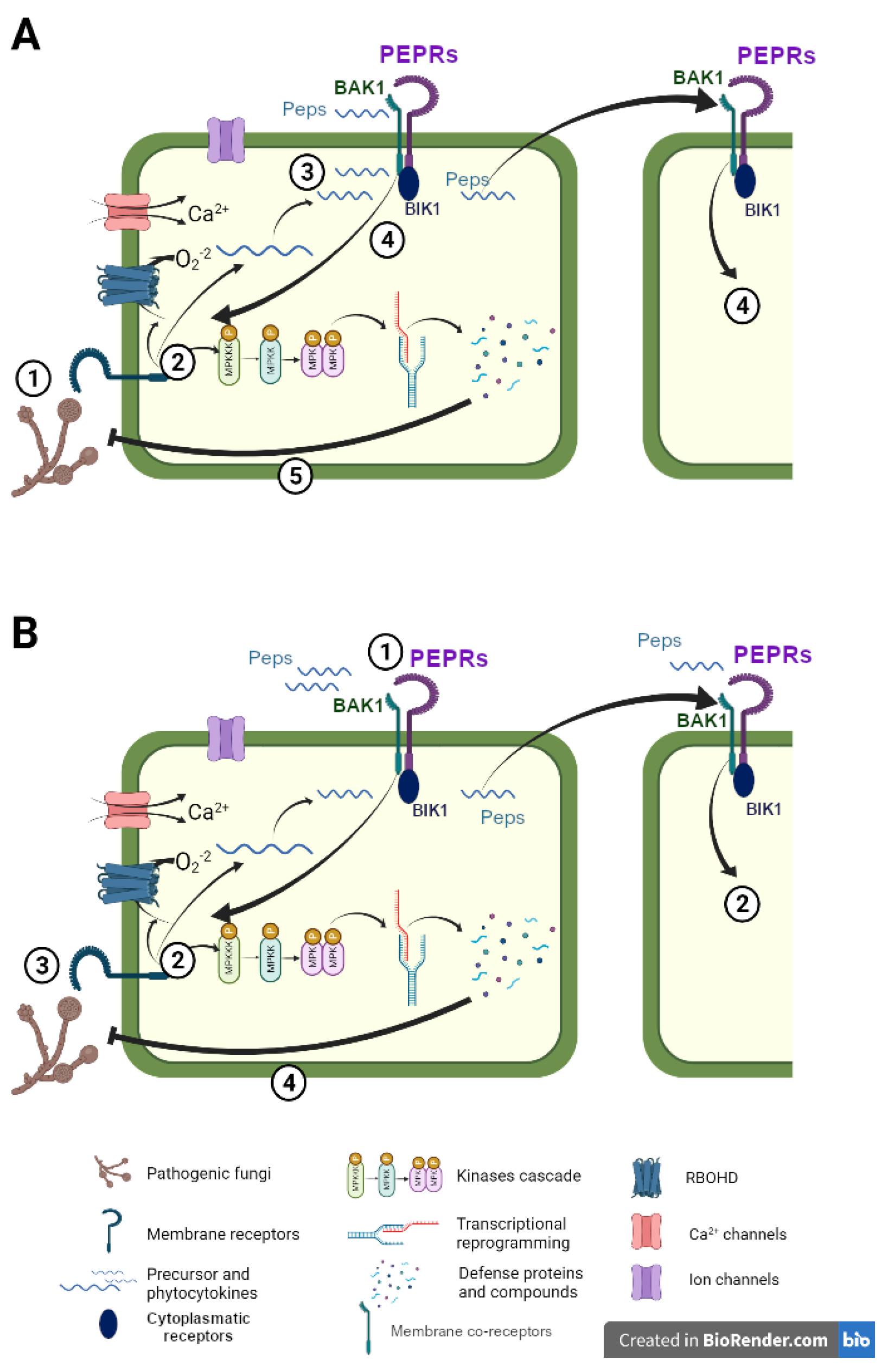
2.1. Peptides’ Perception and Signal Transduction
2.2. Peptides’ Perception and Signal Transduction
2.2.1. Increment of Cytosolic Ca2+
2.2.2. Effect on Ion Channels and Extracellular pH
2.2.3. Production of Reactive Oxygen Species (ROS) and Activation of Mitogen-Activated Protein Kinases (MAPK)
2.2.4. Expression of Defense-Related Genes and Protease Inhibitors
2.2.5. Hormonal following Phytocytokine Perception
2.2.6. Other Basal Inducible Defense Responses Triggered by Phtocytokines
2.3. Role of Phytocytokines in the Defense Response of Peptide-Induced Resistancegainst Pests and Pathogens
| Plan Species of Origin | Peptide/Precursor | Recipient Plant/Organism | Effect | References |
|---|---|---|---|---|
| Arabidopsis | PROPEP1 | Arabidopsis | Resistance to Pythium irregulare and Pseudomonas syringae | [11] |
| Arabidopsis | PrePIP1 | Arabidopsis | Resistance to foc 699 | [14] |
| Arabidopsis | SCOOP | Arabidopsis | Resistance to Alternaria brassicicola | [16] |
| Susceptibility against E. amylovora | [21] | |||
| Arabidopsis | RALF23 | Arabidopsis | Susceptibility to Pto DC3000 COR and P. cucumerina | [23] |
| Arabidopsis | IDL6 | Arabidopsis | Susceptibility to P. syringae Pst DC3000 | [24] |
| Arabidopsis | GRI | Arabidopsis | Susceptibility to P. syringae Pst DC3000 | [25,26] |
| Arabidopsis | CEP4 | Arabidopsis | Resistance to P. syringae Pto | [27] |
| Tomato | ProSystemin | Tomato | Resistance to herbivore | [80] |
| Resistance to aphids | [80] | |||
| Resistance to B. cinerea and A. alternata | [80] | |||
| Reduced susceptibility to Cucumber mosaic virus | [81] | |||
| Tomato | ProSystemin | Arabidopsis | Resistance to B. cinerea | [86] |
| Tomato | PSK | Arabidopsis | Susceptibility to Fusarium oxysporum | [34] |
| Arabidopsis | PSK | Tomato | Botrytis cinerea | [35] |
| Maize | Zip1 | Ustilago maydis | Resistance against Ustilago maydis | [37] |
| Tobacco | HypSys | Tobacco | Resistance to Helicovera armigera | [82] |
3. Phytocytokines/Peptides in Plant-Induced Resistance and Priming
3.1. Peptide-Induced Resistance against Pests and Pathogens
3.2. Cross-Species Perception and Peptide-Induced Resistance
4. Cooperative Functioning of Peptides in Innate Immunity and Induced Resistance
5. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, Y.; Zhou, J.; Shan, L.; Meng, X. Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 2018, 131, jcs209353. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Pruitt, R.; Nürnberger, T. Sensing Danger: Key to Activating Plant Immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Heil, M. Damaged-self recognition in plant herbivore defence. Trends Plant Sci. 2009, 14, 356–363. [Google Scholar] [CrossRef]
- Albert, M. Peptides as triggers of plant defence. J. Exp. Bot. 2013, 64, 5269–5279. [Google Scholar] [CrossRef]
- Tanaka, K.; Heil, M. Damage-Associated Molecular Patterns (DAMPs) in plant innate immunity: Applying the danger model and evolutionary perspectives. Annu. Rev. Phytopathol. 2021, 59, 53–75. [Google Scholar] [CrossRef]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Walker, R.K.; Berkowitz, G.A. Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+-dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 2013, 163, 1459–1471. [Google Scholar] [CrossRef]
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 1991, 253, 895–898. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Dafoe, N.J.; Schmelz, E.A. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 2011, 155, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Barona, G.; Ryan, C.A.; Pearce, G. GmPep914, an eight-amino acid peptide isolated from soybean leaves, activates defense-related genes. Plant Physiol. 2011, 156, 932–942. [Google Scholar] [CrossRef]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turrà, D.; Di Pietro, A.; Zhang, W. The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7. PLoS Pathog. 2014, 10, e1004331. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Lee, C.Y.; Cheng, K.T.; Chang, W.H.; Huang, R.N.; Nam, H.G.; Chen, Y.R. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 2014, 26, 4135–4148. [Google Scholar] [CrossRef]
- Gully, K.; Pelletier, S.; Guillou, M.-C.; Ferrand, M.; Aligon, S.; Pokotylo, I.; Perrin, A.; Vergne, E.; Fagard, M.; Ruelland, E.; et al. The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 1349–1365. [Google Scholar] [CrossRef]
- Huang, C.Y.; Araujo, K.; Sánchez, J.N.; Kund, G.; Trumble, J.; Roper, C.; Godfrey, K.E.; Jin, H. A stable antimicrobial peptide with dual functions of treating and preventing citrus Huanglongbing. Proc. Natl. Acad. Sci. USA 2021, 118, e2019628118. [Google Scholar] [CrossRef]
- Bartels, S.; Boller, T. Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef]
- Rhodes, J.; Yang, H.; Moussu, S.; Boutrot, F.; Santiago, J.; Zipfel, C. Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nat. Commun. 2021, 12, 705. [Google Scholar] [CrossRef]
- Hou, S.; Liu, D.; Huang, S.; Luo, D.; Liu, Z.; Xiang, Q.; Wang, P.; Mu, R.; Han, Z.; Chen, S. The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes. Nat. Commun. 2021, 12, 5494. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Kim, S.-K.; Lee, N.; Ahn, C.-Y.; Ryu, C.-M. d-Lactic acid secreted by Chlorella fusca primes pattern-triggered immunity against Pseudomonas syringae in Arabidopsis. Plant J. 2020, 102, 761–778. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, S.; Wu, Q.; Lin, M.; Acharya, B.R.; Wu, D.; Zhang, W. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. Plant J. 2017, 89, 250–263. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosche, M.; Kollist, H.; Kangasjarvi, J. Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc. Natl. Acad. Sci. USA 2009, 106, 5412–5417. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhöner, B.; Lamminmäki, A.; Staes, A.; et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef]
- Rzemieniewski, J.; Leicher, H.; Lee, H.; Broyart, C.; Nayem, S.; Wiese, C.; Maroschek, J.; Camgoez, Z.; Lalun, V.; Djordjevic, M.; et al. Phytocytokine signaling integrates cell surface immunity and nitrogen limitation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Lin, J. Respuestas de defensa sistémica a larga distancia mediadas por Systemin. N. Phytol. 2020, 226, 1573–1582. [Google Scholar] [CrossRef]
- Molisso, D.; Coppola, M.; Aprile, A.M.; Avitabile, C.; Natale, R.; Romanelli, A.; Chiaiese, P.; Rao, R. Colonization of Solanum melongena and Vitis vinifera Plants by Botrytis cinerea Is Strongly Reduced by the Exogenous Application of Tomato Systemin. J. Fungi 2021, 7, 15. [Google Scholar] [CrossRef]
- Constabel, C.P.; Yip, L.; Ryan, C.A. Prosystemin from potato, black nightshade, and bell pepper: Primary structure and biological activity of predicted systemin polypeptides. Plant Mol. Biol. 1998, 36, 55–62. [Google Scholar] [CrossRef]
- Pearce, G.; Ryan, C.A. Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense signalling glycopeptide hormones coded in a single precursor gene. J. Biol. Chem. 2003, 278, 30044–30050. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Koramutla, M.K.; Negi, M.; Pearce, G.; Ryan, C.A. Hydroxyproline-rich glycopeptide signals in potato elicit signalling associated with defense against insects and pathogens. Plant Sci. 2013, 207, 88–97. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A. Production of multiple plant hormones from a single polyprotein precursor. Nature 2001, 411, 817–820. [Google Scholar] [CrossRef]
- Shen, Y.; Diener, A.C. Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet 2013, 9, e1003525. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Z.; Lei, C.; Zheng, C.; Wang, J.; Shao, S.; Li, X.; Xia, X.; Cai, X.; Zhou, J.; et al. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 2018, 30, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, S.; van der Linde, K.; Lahrmann, U.; Acar, B.; Kaschani, F.; Colby, T.; Kaiser, M.; Ding, Y.; Schmelz, E.; Huffaker, A.; et al. An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 2018, 4, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Lundquist PMichael Udvardi, M.; Wolf-Rüdiger Scheible, W. Small and Mighty: Peptide hormones in plant biology. Plant Cell 2018, 30, tpc.118.tt0718. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Sakagami, Y. Peptide hormones in plants. Annu. Rev. Plant Biol. 2006, 57, 649–674. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Huffaker, A. Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 2011, 14, 351–357. [Google Scholar] [CrossRef]
- Beloshistov, R.E.; Dreizler, K.; Galiullina, R.A.; Alexander, I.; Tuzhikov, A.I.; Serebryakova, M.V.; Reichardt, S.; Shaw, J.; Taliansky, M.E.; Pfannstiel, J.; et al. Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. N. Phytol. 2017, 218, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Hander, T.; Fernández-Fernández, Á.D.; Kumpf, R.P.; Willems, P.; Schatowitz, H.; Rombaut, D.; Staes, A.; Nolf, J.; Pottie, R.; Yao, P.; et al. Damage on plants activates Ca2+-dependent meta caspases for release of immunomodulatory peptides. Science 2019, 363, eaar7486. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y. Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol. 2014, 65, 385–413. [Google Scholar] [CrossRef] [PubMed]
- Stührwohldt, N.; Schaller, A. Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 2019, 21, 49–63. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Sakagami, Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 1996, 93, 7623–7627. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Mou, Z. Perception of Damaged Self in Plants. Plant Physiol. 2020, 182, 1545–1565. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Pearce, G.; Ryan, C.A. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 2006, 103, 10104–10109. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, Y.; Yang, F.; Zhang, Y.; Chen, S.; Xie, Q.; Tian, X.; Zhou, J.M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 6205–6210. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Yang, Y.; Bao, Q.; Li, Y.; Wang, H.; Hou, S. EWR1 as a SCOOP peptide activates MIK2-dependent immunity in Arabidopsis. J. Plant Interact. 2022, 17, 562–568. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamashita-Yamada, M.; Hirase, T.; Fujiwara, T.; Tsuda, K.; Hiruma, K.; Saijo, Y. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 2016, 35, 46–61. [Google Scholar] [CrossRef]
- Wang, L.; Einig, E.; Almeida-Trapp, M.; Albert, M.; Fliegmann, J.; Mithöfer, A.; Kalbacher, H.; Felix, G. The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants 2018, 4, 152–156. [Google Scholar] [CrossRef]
- Xu, S.; Liao, C.J.; Jaiswal, N.; Lee, S.; Yun, D.J.; Lee, S.Y.; Garvey, M.; Kaplan, I.; Mengiste, T. Tomato PEPR1 ORTHOLOG RECEPTOR-LIKE KINASE1 regulates responses to systemin, necrotrophic fungi, and insect herbivory. Plant Cell 2018, 30, 2214–2229. [Google Scholar] [CrossRef]
- Olsson, V.; Joos, L.; Zhu, S.; Gevaert, K.; Butenko, M.A.; De Smet, I. Look Closely, the Beautiful May Be Small: Precursor-Derived Peptides in Plants. Annu. Rev. Plant Biol. 2019, 70, 153–186. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense Priming: An Adaptive Part of Induced Resistance. Anuu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Moyen, C.; Hammond-Kosack, K.E.; Jones, J.; Knight, M.R.; Johannes, E. Systemin triggers an increase in cytoplasmic calcium in tomato mesophyll cells: Ca2+ mobilization from intra- and extracellular compartments. Plant Cell Environ. 1998, 21, 1101–1111. [Google Scholar] [CrossRef]
- Ma, Y.; Walker, R.K.; Zhao, Y.; Berkowitz, G.A. Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl Acad. Sci. USA 2012, 109, 19852–19857. [Google Scholar] [CrossRef]
- Shen, W.; Liu, J.; Li, J.F. Type-II Metacaspases Mediate the Processing of Plant Elicitor Peptides in Arabidopsis. Mol. Plant 2019, 12, 1524–1533. [Google Scholar] [CrossRef]
- Jeworutzki, E.; Roelfsema, M.R.; Anschütz, U.; Krol, E.; Elzenga, J.T.; Felix, G.; Boller, T.; Hedrich, R.; Becker, D. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J. 2010, 62, 367–378. [Google Scholar] [CrossRef]
- Schaller, A. Oligopeptide signalling and the action of systemin. Plant Mol. Biol. 1999, 40, 763–769. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Mittler, R. Rapid systemic signaling during abiotic and biotic stresses: Is the ROS wave master of all trades? Plant J. 2020, 102, 887–896. [Google Scholar] [CrossRef]
- Testerink, C.; Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 2011, 62, 2349–2361. [Google Scholar] [CrossRef]
- Stratmann, J.W.; Ryan, C.A. Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors. Proc. Natl. Acad. Sci. USA 1997, 94, 11085–11089. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Fernández, J.; Sánchez-Bel, P.; Gamir, J.; Pastor, V.; Sanmartín, N.; Cerezo, M.; Andrés-Moreno, S.; Flors, V. Tomato Systemin induces resistance against Plectosphaerella cucumerina in Arabidopsis through the induction of phenolic compounds and priming of tryptophan derivatives. Plant Sci. 2022, 321, 111321. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef]
- Coppola, M.; Di Lelio, I.; Romanelli, A.; Gualtieri, L.; Molisso, D.; Ruocco, M.; Avitabile, C.; Natale, R.; Cascone, P.; Guerrieri, E.; et al. Tomato Plants Treated with Systemin Peptide Show Enhanced Levels of Direct and Indirect Defense Associated with Increased Expression of Defense-Related Genes. Plants 2019, 8, 395. [Google Scholar] [CrossRef]
- Green, T.R.; Ryan, C.A. Wound-Induced Proteinase Inhibitor in Plant Leaves: A Possible Defense Mechanism against Insects. Science 1972, 175, 776–777. [Google Scholar] [CrossRef]
- Sun, J.Q.; Jiang, H.L.; Li, C.Y. Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef]
- Chen, J.; Yu, F.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodríguez, P.L.; Liu, X.; Li, D.; et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E5519–E5527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Z.; Wu, D.; Yu, F. RALF-FERONIA signaling: Linking plant immune response with cell growth. Plant Commun. 2020, 1, 100084. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Voigt, C.A. Callose biosynthesis in Arabidopsis with a focus on pathogen response: What we have learned within the last decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- Ficarra, F.A.; Grandellis, C.; Garavaglia, B.S.; Gottig, N.; Ottado, J. Bacterial and plant natriuretic peptides improve plant defence responses against pathogens. Mol. Plant Pathol. 2018, 19, 801–811. [Google Scholar] [CrossRef]
- Corrado, G.; Sasso, R.; Pasquariello, M.; Iodice, L.; Carretta, A.; Cascone, P.; Ariati, L.; Digilio, M.C.; Guerrieri, E.; Rao, R. Systemin regulates both systemic and volatile signaling in tomato plants. J. Chem. Ecol. 2007, 33, 669–681. [Google Scholar] [CrossRef]
- Coppola, M.; Cascone, P.; Madonna, V.; Di Lelio, I.; Esposito, F.; Avitabile, C.; Romanelli, A.; Guerrieri, E.; Vitiello, A.; Pennacchio, F.; et al. Plant-to-plant communication triggered by systemin primes anti-herbivore resistance in tomato. Sci. Rep. 2017, 7, 15522. [Google Scholar] [CrossRef]
- Coppola, M.; Corrado, G.; Coppola, V.; Cascone, P.; Martinelli, R.; Digilio, M.C.; Pennacchio, F.; Rao, R. Prosystemin Overexpression in Tomato Enhances Resistance to Different Biotic Stresses by Activating Genes of Multiple Signaling Pathways. Plant Mol. Biol. Rep. 2015, 33, 1270–1285. [Google Scholar] [CrossRef]
- Bubici, G.; Carluccio, A.V.; Stavolone, L.; Cillo, F. Prosystemin overexpression induces transcriptional modifications of defense-related and receptor-like kinase genes and reduces the susceptibility to Cucumber mosaic virus and its satellite RNAs in transgenic tomato plants. PLoS ONE 2017, 12, e0171902. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Lu, Y. Overexpression of tobacco hydroxyproline-rich glycopeptide systemin precursor A gene in transgenic tobacco enhances resistance against Helicoverpa armigera larvae. Plant Sci. 2006, 171, 286–292. [Google Scholar] [CrossRef]
- Orozco-Cardenas, M.; McGurl, B.; Ryan, C.A. Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 1993, 90, 8273–8276. [Google Scholar] [CrossRef] [PubMed]
- Mosher, S.; Seybold, H.; Rodriguez, P.; Stahl, M.; Davies, K.A.; Dayaratne, S.; Morillo, S.A.; Wierzba, M.; Favery, B.; Keller, H.; et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 2013, 73, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, D.; Tsuda, K.; Katagiri, F. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J. 2012, 71, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, P.; Zhao, J.; Jiang, H.; Wang, H.; Zhu, Y.; Botella, M.A.; Šamaj, J.; Li, C.; Lin, J. Expression of tomato prosystemin gene in Arabidopsis reveals systemic translocation of its mRNA and confers necrotrophic fungal resistance. N. Phytol. 2017, 27, 799–812. [Google Scholar] [CrossRef]
- De Kessel, J.; Conrath, U.; Flors, V.; Luna, E.; Mageroy, M.H.; Mauch-Mani, B.; Pastor, V.; Pozo, M.J.; Pieterse, C.M.J.; Ton, J.; et al. The Induced Resistance Lexicon: Do’s and Don’ts. Trends Plant Sci. 2021, 26, 685–691. [Google Scholar] [CrossRef]
- Pastor, V.; Luna, E.; Ton, J.; Cerezo, M.; García-Agustín, P.; Flors, V. Fine Tuning of Reactive Oxygen Species Homeostasis Regulates Primed Immune Responses in Arabidopsis. MPMI 2013, 26, 1334–1344. [Google Scholar] [CrossRef]
- Thevenet, D.; Pastor, V.; Baccelli, I.; Balmer, A.; Vallat, A.; Neier, R.; Glauser, G.; Mauch-Mani, B. The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. N. Phytol. 2017, 213, 552–559. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Walters, D.R.; Ratsep, J.; Havis, N.D. Controlling crop diseases using induced resistance: Challenges for the future. J. Exp. Bot. 2013, 64, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.M.J.J.; Pozo, M.J.; Ton, J.; van Dam, N.M.; Conrath, U. Recognizing plant defense priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef]
- van Hulten, M.; Pelser, M.; van Loon, L.C.; Pieterse, C.M.; Ton, J. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 5602–5607. [Google Scholar] [CrossRef]
- Pastor, V.; Balmer, A.; Gamir, J.; Flors, V.; Mauch-Mani, B. Preparing to fight back: Generation and storage of priming compounds. Front. Plant Sci. 2014, 5, 295. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Fernández, J.; Gamir, J.; Pastor, V.; Sanchez-Bel, P.; Sanmartín, N.; Cerezo, M.; Flors, V. Arabidopsis Plants Sense Non-self Peptides to Promote Resistance Against Plectosphaerella cucumerina. Front. Plant Sci. 2020, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Scheer, J.M.; Pearce, G.; Ryan, C.A. Generation of systemin signaling in tobacco by transformation with the tomato systemin receptor kinase gene. Proc. Natl. Acad. Sci. USA 2003, 100, 10114–10117. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Higgins, R.; Luo, Y.; Piper, L.; Nazir, A.; Bajwa, V.S.; Clouse, S.D.; Thompson, P.R.; Stratmann, J.W. The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Mol. Biol. 2009, 70, 603–616. [Google Scholar] [CrossRef]
- Rocco, M.; Corrado, G.; Arena, S.; D’Ambrosio, C.; Tortiglione, C.; Sellaroli, S.; Marra, M.; Rao, R.; Scaloni, A.l. The expression of tomato prosystemin gene in tobacco plants highly affects host proteomic repertoire. J. Proteom. 2008, 71, 176–185. [Google Scholar] [CrossRef]
- Marmiroli, N.; Maestri, E. Plant peptides in defense and signaling. Peptides 2014, 56, 30–44. [Google Scholar] [CrossRef]
- Narváez-Vásquez, J.; Orozco-Cárdenas, M.L.; Ryan, C.A. Systemic wound signaling in tomato leaves is cooperatively regulated by systemin and hydroxyproline-rich glycopeptide signals. Plant Mol. Biol. 2007, 65, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Klauser, D.; Flury, P.; Boller, T.; Bartels, S. Several MAMPs, including chitin fragments, enhance AtPep-triggered oxidative burst independently of wounding. Plant Signal Behav. 2013, 8, e25346. [Google Scholar] [CrossRef] [PubMed]
- Pastor, V.; Cervero, R.; Gamir, J. The simultaneous perception of self- and non-self-danger signals potentiates plant innate immunity responses. Planta 2022, 256, 10. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yasuda, S.; Hyodo, K.; Tani, R.; Hojo, Y.; Fujiwara, Y.; Hiruma, K.; Ishizaki, T.; Fujita, Y.; Saijo, Y.; et al. Integration of danger peptide signals with herbivore-associated molecular pattern signaling amplifies anti-herbivore defense responses in rice. Plant J. 2018, 94, 626–637. [Google Scholar] [CrossRef]
| Phytocytokine | Species of Origin | Signal Transduction | Induced Defense Responses and Signaling | References |
|---|---|---|---|---|
| Peps | Arabidopsis | PEPR1 and PEPR2 | media alkalinization, H2O2 PDF1.2 and PROPEPs expression | [11] |
| BAK1 | Ca2+, ET, callose | [9] | ||
| BIK1/PBL1 | Ca2+, H2O2 NO MPK3 and WRKY33 expression | [18] | ||
| ZmPep1 | Maize | JA, ET, defense gene expression, defense metabolites accumulation | [12] | |
| ZmPep3 | Maize | JA, ET, defense gene expression, volatiles emission, phytoalexin | [19] | |
| PIP1 | Arabidopsis | RLK7 | ROS, FRK1, WRKY30, WRKY33, WRKY53, MYB51 and PR1 expression | [14] |
| partially BAK1- dependent | MAPK, Callose, Stomatal closure | |||
| SCOOP12 | Arabidopsis | MIK2-BAK1/SERK4 | ROS, callose | [16] |
| Phosphatidic acid (PA) FRK1 expression | [20] | |||
| SCOOPs | BIK1/PBL1 | Ca2+, ROS, MAPK Ethylene, defense gene expression | [21] | |
| PNP-A | Arabidopsis | PNP-R2 | antagonizes SA responses, stomatal closure | [22] |
| RALF23 | Arabidopsis | FER-BAK1 | Ca2+, Media alkalinization | [23] |
| Antagonizes PAMP-induced ROS | ||||
| IDL6 | Arabidopsis | HAE and HSL2 | Poligalacturonase gene ADPG2 | [24] |
| GRI | Arabidopsis | PRK5 | ROS-dependent Cell death, hormones | [25,26] |
| CEP4 | Arabidopsis | CEPR1/2 and RLK7 | Ca2+, MAPK Ethylene, FRK1 expression | [27] |
| Systemin | Tomato | SYR1 | Opening of ion channels, Ca2+, MAPKs JA, defense genes | [10] |
| SYR2 | CDPKs, ROS Protease inhibitors | [28] | ||
| PORK1 | CAT and APX activity Volatiles emission | [29] | ||
| PotSys1 and 2 | Potato | SYR1 and SYR2 | Proteinase inhibitors | [30] |
| PepSys | Pepper | |||
| NishSys | Nightshade | |||
| HypSys1, 2 and 3 | Tomato | Media alkalinization JA, PI-I, and PI-II | [31] | |
| Potato | H2O2 PIs, JA, defense-related genes, antioxidant defensive enzymes | [32] | ||
| TobHypSys 1 and 2 | tobacco | Media alkalinization, MAPK Proteinase inhibitors | [33] | |
| CAPE1 | Tomato | H2O2 SA, defense gene expression | [15] | |
| PSK | Arabidopsis | PSRKs | Ca2+ IAA and Auxin-dependent responses | [34] |
| Tomato | [35] | |||
| PSY1 | Arabidopsis | PSY1R | [34] | |
| SubPep | Soybean | Media alkalinization Chitinase1b, CYP93A1, chalcone synthase and PDR12 gene expression | [36] | |
| Pep914 | Soybean | Media alkalinization CYP93A1, Chib1-1, and chalcone synthase gene expression | [13] | |
| Pep890 | ||||
| Zip1 | Maize | SA, SA, and JA marker genes, defense-related genes | [37] | |
| SAMP | Citrus | Defense gene expression | [17] |
| Plant Species of Origin | Peptide | Recipient Plant | Effect | References |
|---|---|---|---|---|
| Arabidopsis | Pep3 | Arabidopsis | Resistance to Pst DC 3000 | [9] |
| Arabidopsis | PIP1 | Arabidopsis | Resistance to Pst DC 3000 | [14] |
| Arabidopsis | SCOOP12 | Arabidopsis | Resistance to Pst DC 3000 | [16] |
| Maize | ZmPep1 | Maize | Resistance to Cochliobolis heterostrophus and C. graminicola | [12] |
| Maize | ZmPep3 | Maize | Resistance to Spodoptera exigua | [19] |
| Tomato | CAPE1 | Tomato | Resistance to Spodoptera litura | [15] |
| Resistance to Pst DC 3000 | ||||
| Tomato | PSK | Tomato | Resistance to B. cinerea | [35] |
| Tomato | Systemin | Tomato | Resistance to Spodoptera litoralis | [67] |
| Resistance to B. cinerea | ||||
| Arabidopsis | PNP-A | Arabidopsis | Susceptibility to P. syringae | [22] |
| Maize | Zip1 | Maize | Susceptibility to B. cinerea | [37] |
| Tomato | Systemin | Arabidopsis | Resistance to P. cucumerina | [96] |
| Potato | PotSysII | |||
| Pepper | PepSys | |||
| Nightshade | Nishsys | |||
| Tomato | HypSys | |||
| Radish | AFP’s | |||
| Arabidopsis | Pep1 | Arabidopsis | Resistance to P. cucumerina | [96] |
| Tomato | Systemin | Eggplant | Resistance to B. cinerea | [29] |
| Vitis vinifera | ||||
| Citrus | SAMP | Citrus | Resistance to Candidatus liberibacter asiaticus | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pastor-Fernández, J.; Sánchez-Bel, P.; Flors, V.; Cerezo, M.; Pastor, V. Small Signals Lead to Big Changes: The Potential of Peptide-Induced Resistance in Plants. J. Fungi 2023, 9, 265. https://doi.org/10.3390/jof9020265
Pastor-Fernández J, Sánchez-Bel P, Flors V, Cerezo M, Pastor V. Small Signals Lead to Big Changes: The Potential of Peptide-Induced Resistance in Plants. Journal of Fungi. 2023; 9(2):265. https://doi.org/10.3390/jof9020265
Chicago/Turabian StylePastor-Fernández, Julia, Paloma Sánchez-Bel, Víctor Flors, Miguel Cerezo, and Victoria Pastor. 2023. "Small Signals Lead to Big Changes: The Potential of Peptide-Induced Resistance in Plants" Journal of Fungi 9, no. 2: 265. https://doi.org/10.3390/jof9020265
APA StylePastor-Fernández, J., Sánchez-Bel, P., Flors, V., Cerezo, M., & Pastor, V. (2023). Small Signals Lead to Big Changes: The Potential of Peptide-Induced Resistance in Plants. Journal of Fungi, 9(2), 265. https://doi.org/10.3390/jof9020265







