Transcriptome Analysis of T. asperellum GDFS 1009 Revealed the Role of MUP1 Gene on the Methionine-Based Induction of Morphogenesis and Biological Control Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. T. asperellum GDFS 1009 Fermentation Culture
2.2. RNA Isolation, cDNA Library Preparation and Sequencing
2.3. Validation of Transcriptome Data by Reverse Transcription Quantitative PCR (RT-qPCR)
2.4. Construction of MUP1 Gene Knockout and Overexpression Strains
2.5. Growth and Morphology of the TAWT, OEMup1–4, OEMup1–5, ΔMup1–1 and ΔMup1–10
2.6. cAMP Measurements
2.7. RT-qPCR Analysis of Gene Expression Related to Sporulation and Mycoparasitism
2.8. Mycoparasitism Assays
2.9. Biocontrol Assays
2.10. Statistical Analysis
3. Results
3.1. Morphological Growth Differences of T. asperellum GDFS 1009 in Solid-Surface Culture and Liquid-Shaking Culture
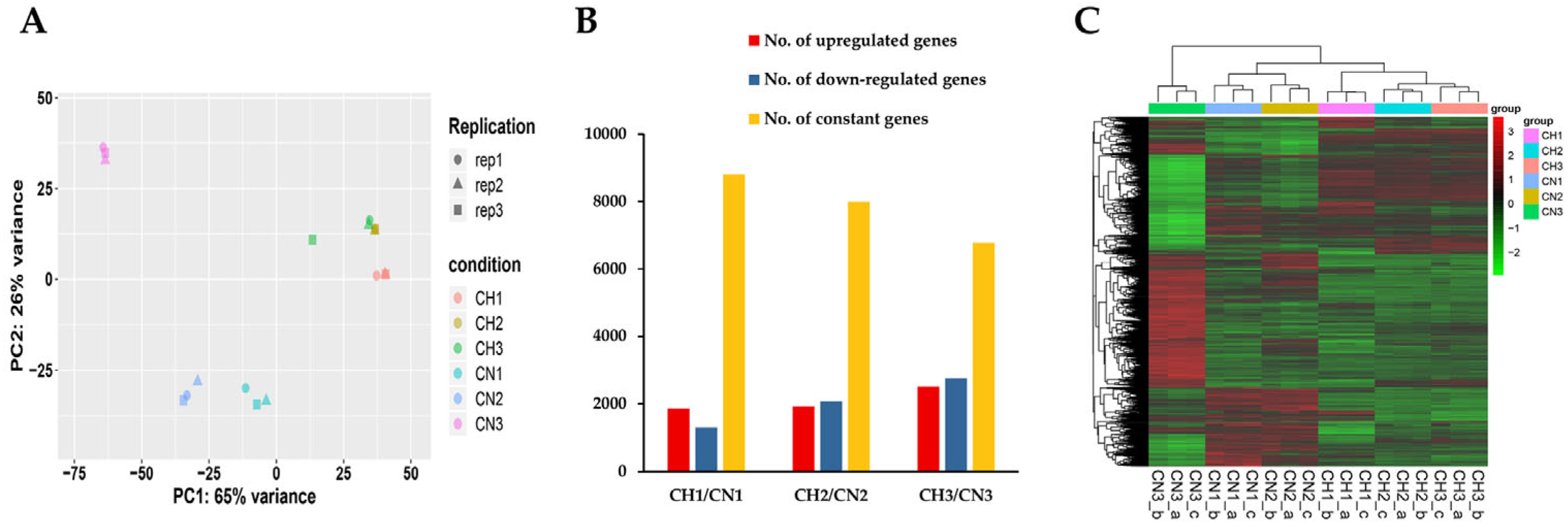
3.2. RNA Sequencing and De Novo Assembling of T. asperellum GDFS 1009 Transcriptome
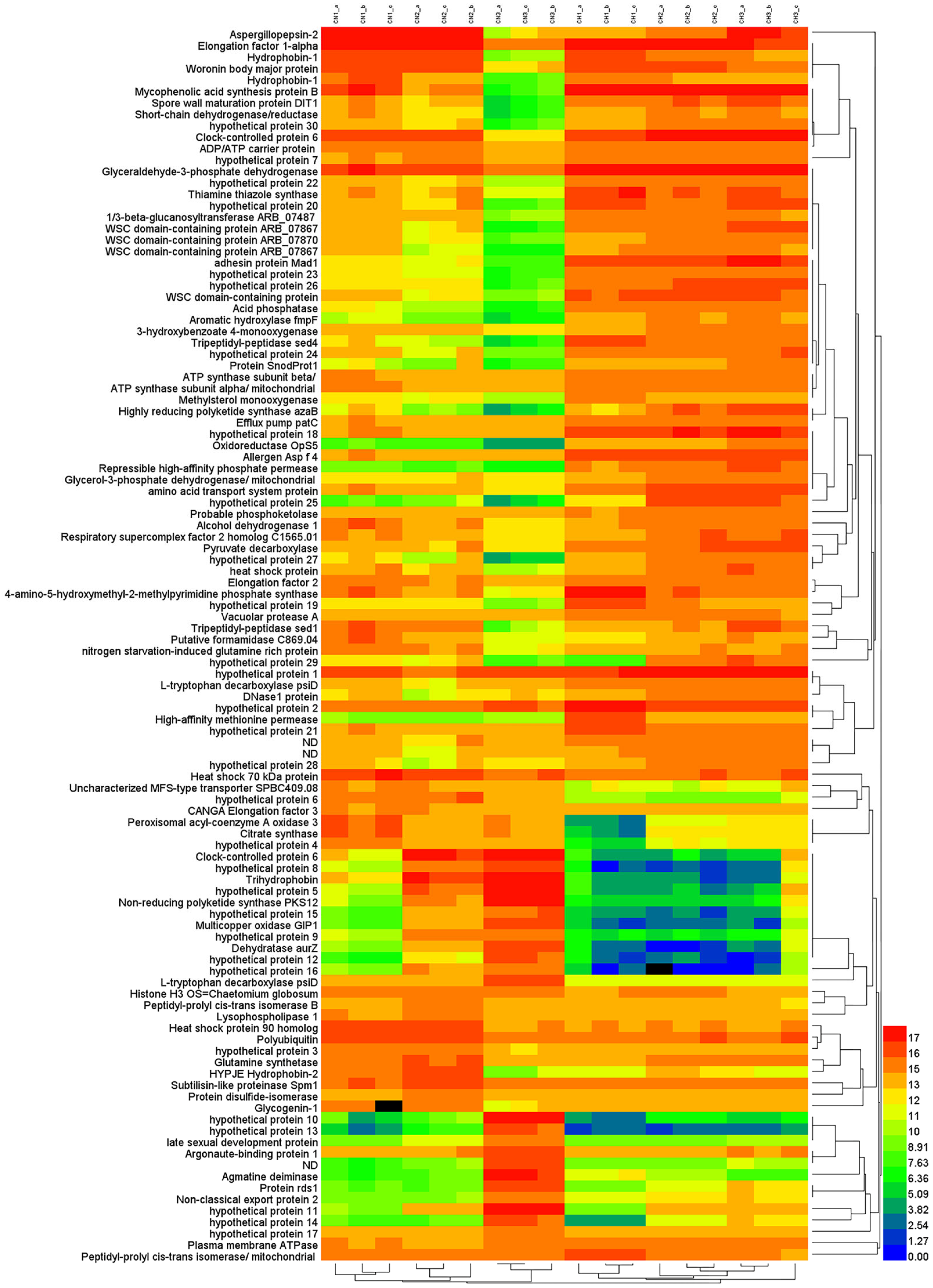
3.3. Differential Expression Genes of Liquid-Shaking Culture and Solid-Surface Culture
3.4. Gene Expression Analysis and qRT-PCR Validation
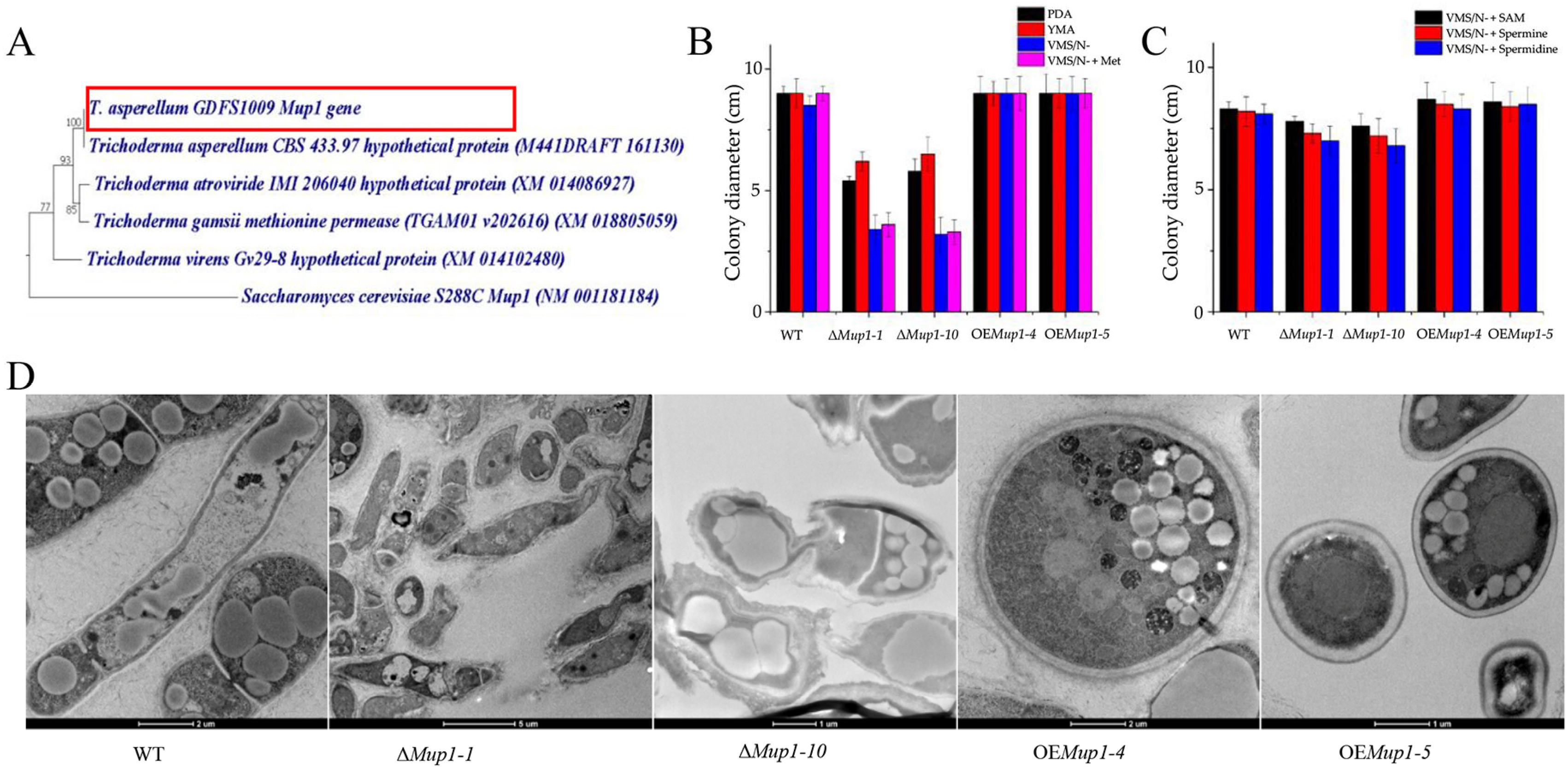
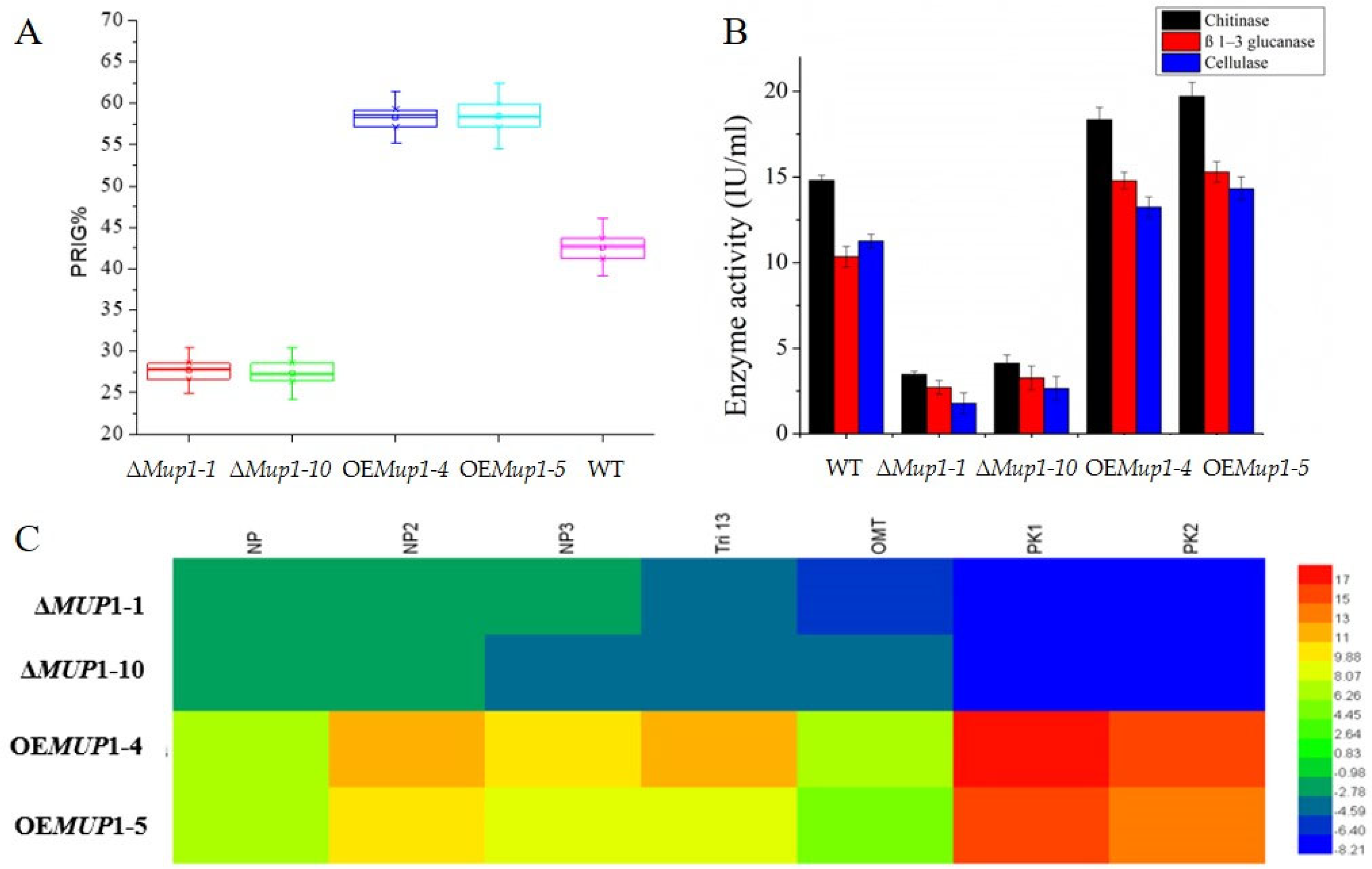
3.5. The High-Affinity Methionine Permease MUP1 Is Required for Morphogenesis
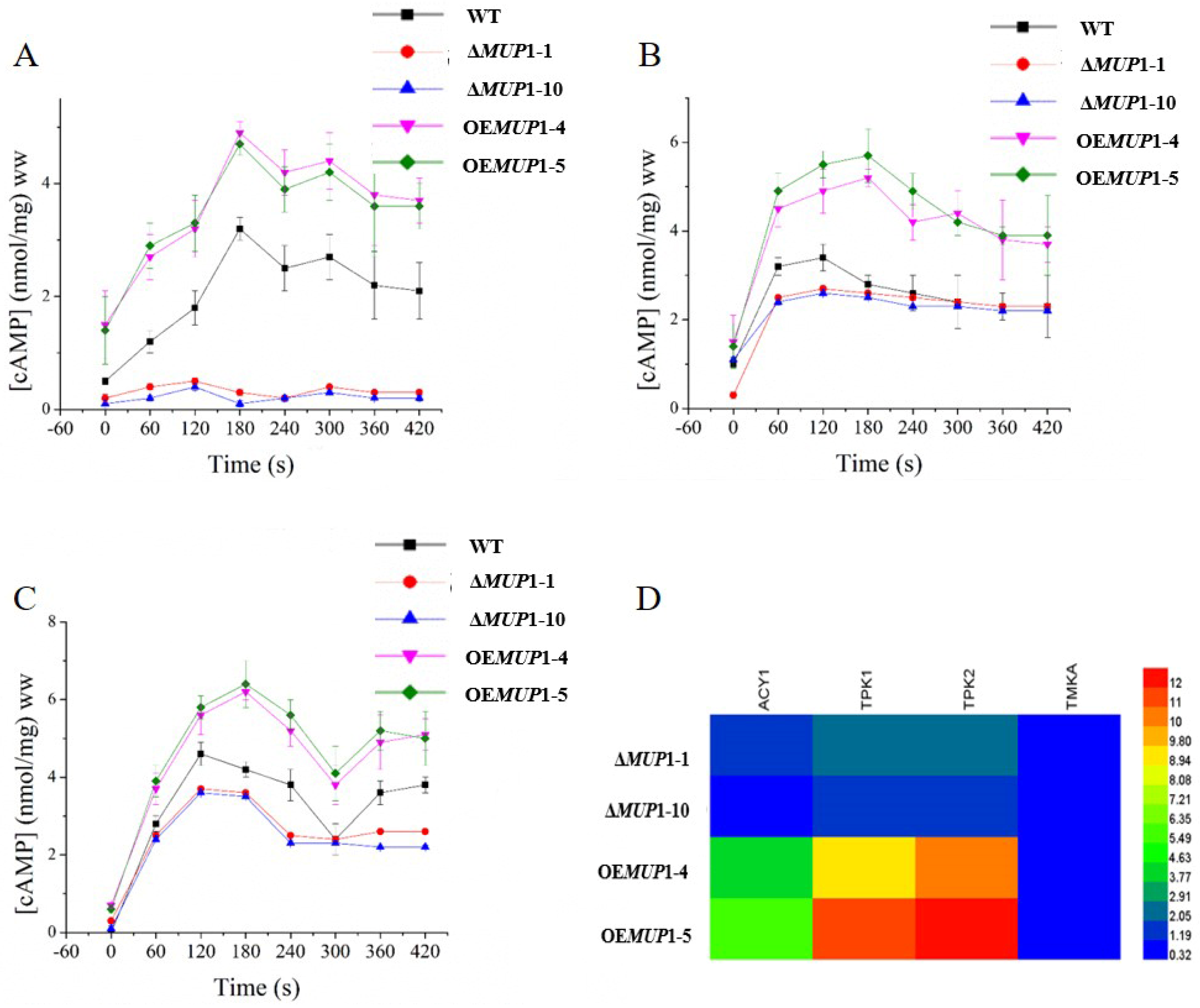
3.6. The Role of Mup1 in the Methionine-Induced PKA Signaling Pathway
3.7. Influence of T. asperellum MUP1 Gene on Mycoparasitism Activity
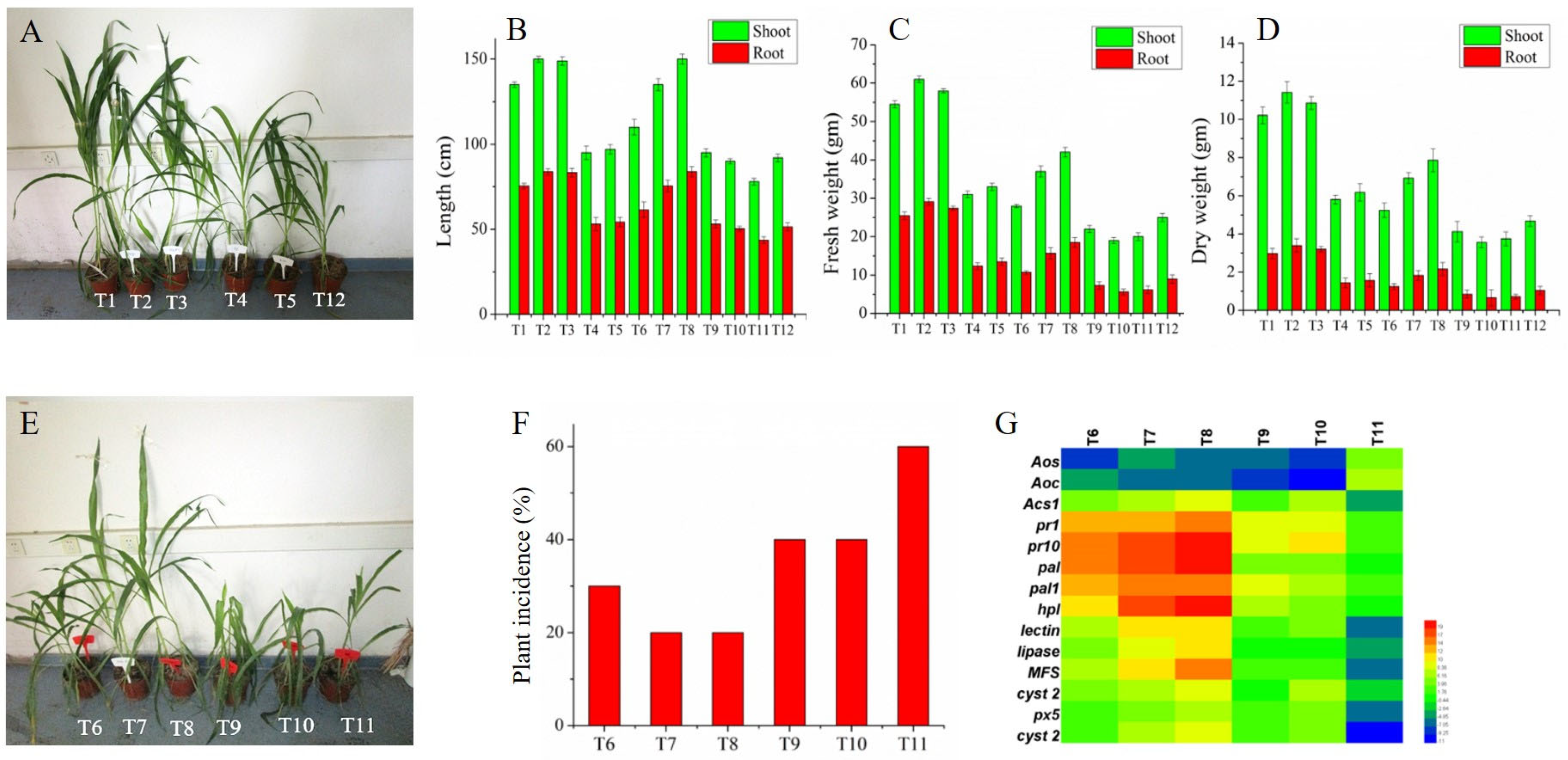
3.8. Influence of MUP1 Gene on the Maize Plant Growth and Biocontrol Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Esquivel-Naranjo, E.U.; Garcia-Esquivel, M.; Medina-Castellanos, E.; Correa-Perez, V.A.; Parra-Arriaga, J.L.; Landeros-Jaime, F.; Cervantes-Chavez, J.A.; Herrera-Estrella, A. A Trichoderma atroviride stress-activated MAPK pathway integrates stress and light signals. Mol. Microbiol. 2016, 100, 860–876. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Gao, R.; Zhao, Z.; Ding, M.; Jiang, S.; Yagtu, C.; Zhu, H.; Zhang, J.; Ebner, T.; Mayrhofer-Reinhartshuber, M.; et al. Evolutionary compromises in fungal fitness: Hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 2020, 14, 2610–2624. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, B.; Zhang, S.; Li, M.; Jiang, X. Transcriptome Dynamics Underlying Chlamydospore Formation in Trichoderma virens GV29-8. Front. Microbiol. 2021, 12, 654855. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.S.; Carlisle, P.L.; Kadosh, D. Coevolution of morphology and virulence in Candida species. Eukaryot. Cell 2011, 10, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Palige, K.; Linde, J.; Martin, R.; Bottcher, B.; Citiulo, F.; Sullivan, D.J.; Weber, J.; Staib, C.; Rupp, S.; Hube, B.; et al. Global transcriptome sequencing identifies chlamydospore specific markers in Candida albicans and Candida dubliniensis. PLoS ONE 2013, 8, e61940. [Google Scholar] [CrossRef]
- Francisco, C.S.; Ma, X.; Zwyssig, M.M.; McDonald, B.A.; Palma-Guerrero, J. Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Sci. Rep. 2019, 9, 9642. [Google Scholar] [CrossRef]
- Madrid, M.; Vázquez-Marín, B.; Franco, A.; Soto, T.; Vicente-Soler, J.; Gacto, M.; Cansado, J. Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast. Sci. Rep. 2016, 6, 37515. [Google Scholar] [CrossRef]
- Schrevens, S.; Van Zeebroeck, G.; Riedelberger, M.; Tournu, H.; Kuchler, K.; Van Dijck, P. Methionine is required for cAMP-PKA-mediated morphogenesis and virulence of Candida albicans. Mol. Microbiol. 2018, 108, 258–275. [Google Scholar] [CrossRef]
- Breton, A.; Surdin-Kerjan, Y. Sulfate uptake in Saccharomyces cerevisiae: Biochemical and genetic study. J. Bacteriol. 1977, 132, 224–232. [Google Scholar] [CrossRef]
- Gremel, G.; Dorrer, M.; Schmoll, M. Sulphur metabolism and cellulase gene expression are connected processes in the filamentous fungus Hypocrea jecorina (anamorph Trichoderma reesei). BMC Microbiol. 2008, 8, 174. [Google Scholar] [CrossRef]
- Gong, B.; Li, X.; VandenLangenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-L-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18, e13034. [Google Scholar] [CrossRef]
- Isnard, A.D.; Thomas, D.; Surdin-Kerjan, Y. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 1996, 262, 473–484. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Liao, W.; Yang, Y.; Li, Y.; Peng, M. Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental-stress-induced abscission. Sci. Rep. 2016, 6, 32006. [Google Scholar] [CrossRef]
- Fu, K.; Fan, L.; Li, Y.; Gao, S.; Chen, J. Tmac1, a transcription factor which regulated high affinity copper transport in Trichoderma reesei. Microbiol. Res. 2012, 167, 536–543. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Kenerley, C.M. Regulation of morphogenesis and biocontrol properties inTrichoderma virens by a VELVET Protein, Vel1. Appl. Environ. Microbiol. 2010, 76, 2345–2352. [Google Scholar] [CrossRef]
- Karuppiah, V.; Vallikkannu, M.; Li, T.; Chen, J. Simultaneous and sequential based co-fermentations of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841: A strategy to enhance the gene expression and metabolites to improve the bio-control and plant growth promoting activity. Microb. Cell Fact. 2019, 18, 185. [Google Scholar] [CrossRef]
- Karuppiah, V.; Sun, J.; Li, T.; Vallikkannu, M.; Chen, J. Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 Causes Differential Gene Expression and Improvement in the Wheat Growth and Biocontrol Activity. Front. Microbiol. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Maidan, M.M.; De Rop, L.; Serneels, J.; Exler, S.; Rupp, S.; Tournu, H.; Thevelein, J.M.; Van Dijck, P. The G protein-coupled receptor Gpr1 and the G alpha protein Gpa2 act through the cAMP-protein kinase a pathway to induce morphogenesis in candida albicans. Mol. Biol. Cell 2005, 16, 1971–1986. [Google Scholar] [CrossRef]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The “Secrets” of a Multitalented Biocontrol Agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.B.; Zhang, J.; Sun, M.H.; Li, S.D. Identification of genes related to chlamydospore formation in Clonostachys rosea 67-1. Microbiologyopen 2019, 8, e00624. [Google Scholar] [CrossRef] [PubMed]
- Kar, B.; Patel, P.; Ao, J.; Free, S.J. Neurospora crassa family GH72 glucanosyltransferases function to crosslink cell wall glycoprotein N-linked galactomannan to cell wall lichenin. Fungal Genet. Biol. 2019, 123, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, Y.; Jia, Z.; Ge, W.; Zhang, L.; Zhao, Q.; Song, S.; Huang, Y. Whole RNA-sequencing and gene expression analysis of Trichoderma harzianum Tr-92 under chlamydospore-producing condition. Genes Genom. 2019, 41, 689–699. [Google Scholar] [CrossRef]
- Reichard, U.; Léchenne, B.; Asif, A.R.; Streit, F.; Grouzmann, E.; Jousson, O.; Monod, M. Sedolisins, a new class of secreted proteases from Aspergillus fumigatus with endoprotease or tripeptidyl-peptidase activity at acidic pHs. Appl. Environ. Microbiol. 2006, 72, 1739–1748. [Google Scholar] [CrossRef]
- Coluccio, A.; Bogengruber, E.; Conrad, M.N.; Dresser, M.E.; Briza, P.; Neiman, A.M. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 1464–1475. [Google Scholar] [CrossRef]
- Wang, C.; St Leger, R.J. The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot. Cell 2007, 6, 808–816. [Google Scholar] [CrossRef]
- Rodrigues, M.L. The Multifunctional Fungal Ergosterol. mBio 2018, 9, e01755-18. [Google Scholar] [CrossRef]
- Walvekar, A.S.; Laxman, S. Methionine at the Heart of Anabolism and Signaling: Perspectives From Budding Yeast. Front. Microbiol. 2019, 10, 2624. [Google Scholar] [CrossRef]
- Slocum, R.D.; Kaur-Sawhney, R.; Galston, A.W. The physiology and biochemistry of polyamines in plants. Arch. Biochem. Biophys. 1984, 235, 283–303. [Google Scholar] [CrossRef]
- Poidevin, L.; Unal, D.; Belda-Palazón, B.; Ferrando, A. Polyamines as Quality Control Metabolites Operating at the Post-Transcriptional Level. Plants 2019, 8, 109. [Google Scholar] [CrossRef]
- Schumacher, J.; Kokkelink, L.; Huesmann, C.; Jimenez-Teja, D.; Collado, I.G.; Barakat, R.; Tudzynski, P.; Tudzynski, B. The cAMP-dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea. Mol. Plant Microbe Interact. 2008, 21, 1443–1459. [Google Scholar] [CrossRef]
- Malinich, E.A.; Wang, K.; Mukherjee, P.K.; Kolomiets, M.; Kenerley, C.M. Differential expression analysis ofTrichoderma virens RNA reveals a dynamic transcriptome during colonization of Zea mays roots. BMC Genom. 2019, 20, 280. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Kumar, D. Trichoderma: A beneficial antifungal agent and insights into its mechanism of biocontrol potential. Egypt. J. Biol. Pest Control 2020, 30, 133. [Google Scholar] [CrossRef]
| Incubation Time (Hour) | Conidiospore in Solid Surface Culture (Spores/mL) | Chlamydospores in Liquid Shaking Culture (Spores/mL) |
|---|---|---|
| 24 | - | - |
| 48 | - | - |
| 72 | (9.3 ± 0.47) × 105 | (5 ± 0.32) × 103 |
| 96 | (7.2 ± 0.52) × 107 | (9 ± 0.58) × 105 |
| 120 | - | (8.5 ± 0.41) × 108 |
| Gene Name | Real-Time PCR Data | Transcriptomics Data | Realtime PCR Data of Different Medium | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CH1vsCN1 | CH2vsCN2 | CH3vsCN3 | CH1vsCN1 | CH2vsCN2 | CH3vsCN3 | M1 | M2 | YM | |
| Thiamine thiazole synthase | 1.0045 ± 0.12 | 2.3830 ± 0.76 | 6.3995 ± 0.35 | 1.72 ± 0.21 | 2.10 ± 0.36 | 4.437 ± 0.51 | 0.231 ± 0.078 | 0.229 ± 0.012 | 0.73 ± 0.69 |
| Adhesin protein MAD1 | 11.01 ± 0.28 | 39.65 ± 0.53 * | 113.53 ± 0.92 * | 13.78 ± 0.48 | 23.055 ± 0.91 | 448.69 ± 0.27 * | 0.010 ± 0.002 | 0.033 ± 0.01 | 0.06 ± 0.002 |
| Acid phosphatases acp | 6.10 ± 0.72 | 9.11 ± 0.87 | 39.43 ± 1.5 * | 8.64 ± 0.95 | 6.15 ± 0.21 | 34.14 ± 1.93 | 0.066 ± 0.005 * | 0.058 ± 0.001 | 0.162 ± 0.05 |
| FAD binding domain-containing protein | 1.11 ± 0.03 | 2.77 ± 0.18 | 3.82 ± 0.095 | 1.31 ± 0.082 | 1.44 ± 0.02 | 2.621 ± 0.043 | 0.115 ± 0.0093 | 0.084 ± 0.002 | 0.1 ± 0.006 |
| glycosyltransferase | 321.00 ± 0.91 * | 559.94 ± 0.51 * | 419.21 ± 0.72 * | 266.34 ± 0.85 * | 575.74 ± 0.35 * | 409.46 ± 0.92 * | 42.12 ± 0.85 * | 68.67 ± 0.23 * | 142.36 ± 0.28 * |
| ABC1 domain-containing protein | 5.013 ± 0.07 | 11.037 ± 0.04 | 96.11 ± 0.02 * | 5.21 ± 065 | 9.64 ± 0.19 | 91.852 ± 0.36 * | 0.056 ± 0.02 | 0.004 | 0.0032 |
| Urea active transporter | 100.00 ± 0.47 * | 122.03 ± 0.76 * | 84.67 ± 0.37 * | 103.21 ± 0.82 * | 128.62 ± 0.56 * | 87.75 ± 0.49 | 0.033 ± 0.001 | 0.108 ± 0.003 * | 0.68 ± 0.006 |
| High-affinity methionine permease | 69.26 ± 0.45 * | 30.884 ± 0.67 * | 17.61 ± 0.71 | 69.51 ± 0.37 * | 34.76 ± 0.28 * | 17.36 ± 0.36 | 39.33 ± 0.42 * | 87.94 ± 0.93 * | 129.0 ± 0.89 * |
| Tripeptydyl peptidase-sedD | 0.22 ± 0.006 | 1.18 ± 0.003 | 78.27 ± 0.54 | 0.21 ± 0.003 | 1.002 ± 0.004 | 49.74 ± 0.3 | 0.4585 ± 0.001 | 0.054 ± 0.0001 | 1.05 ± 0.001 |
| Methylsterol monooxygenase | 9.007 ± 0.03 | 8.08 ± 0.1 | 38.92 ± 0.6 | 8.93 ± 0.1 | 5.71 ± 0.84 | 33.01 ± 0.92 | 0.0347 | 0.032 | 0.09 |
| Oligopeptide transporter 7 | 8.008 ± 0.67 | 8.82 ± 0.53 | 55.04 ± 0.98 | 8.41 ± 0.67 | 8.86 ± 0.04 | 47.92 ± 0.49 | 0.1471 ± 0.004 | 0.507 ± 0.008 | 7.87 ± 0.02 |
| Repressible high-affinity phosphate permease | 55.04 ± 0.84 | 213.90 ± 0.45 * | 717.12 ± 3.6 * | 52.69 ± 0.76 | 210.53 ± 1.45 * | 707.00 ± 1.23 * | 0.073 | 0.060 | 5.26 ± 0.006 |
| Spore wall maturation protein | −4.960 | −6.35 | 4.47 | −4.76 | −5.92 | 6.36 | ND | ND | ND |
| STRAINS | PDB | YMB | VMS/N− | VMS/N− +LMET | VMS/N− +DMET | VMS/N− +L&DMET |
|---|---|---|---|---|---|---|
| WT | 8.3 × 107 ± 0.4 | 9.1 × 108 ± 0.64 | 6.8 × 103 ± 0.61 | 4.5 × 106 ± 0.84 | 0.1 × 102 ± 0.34 | 0.01 × 102 ± 0.59 |
| ΔMup1–1 | 0 | 1.4 × 102 ± 0.78 | 0 | 0 | 0 | 0 |
| ΔMup1–10 | 0 | 1.8 × 102 ± 0.59 | 0 | 0 | 0 | 0 |
| OEMup1–4 | 9.2 × 107 ± 0.56 | 8.8 × 109 ± 0.85 | 3.4 × 104 ± 0.76 | 5.6 × 107 ± 0.75 | 0.2 × 102 ± 0.54 | 0.09 × 102 ± 0.78 |
| OEMup1–5 | 1.2 × 108 ± 0.46 | 7.9 × 109 ± 0.87 | 2.6 × 104 ± 0.68 | 4.96 × 107 ± 0.56 | 0.1 × 102 ± 0.42 | 0.07 × 102 ± 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karuppiah, V.; Zhang, C.; Liu, T.; Li, Y.; Chen, J. Transcriptome Analysis of T. asperellum GDFS 1009 Revealed the Role of MUP1 Gene on the Methionine-Based Induction of Morphogenesis and Biological Control Activity. J. Fungi 2023, 9, 215. https://doi.org/10.3390/jof9020215
Karuppiah V, Zhang C, Liu T, Li Y, Chen J. Transcriptome Analysis of T. asperellum GDFS 1009 Revealed the Role of MUP1 Gene on the Methionine-Based Induction of Morphogenesis and Biological Control Activity. Journal of Fungi. 2023; 9(2):215. https://doi.org/10.3390/jof9020215
Chicago/Turabian StyleKaruppiah, Valliappan, Cheng Zhang, Tong Liu, Yi Li, and Jie Chen. 2023. "Transcriptome Analysis of T. asperellum GDFS 1009 Revealed the Role of MUP1 Gene on the Methionine-Based Induction of Morphogenesis and Biological Control Activity" Journal of Fungi 9, no. 2: 215. https://doi.org/10.3390/jof9020215
APA StyleKaruppiah, V., Zhang, C., Liu, T., Li, Y., & Chen, J. (2023). Transcriptome Analysis of T. asperellum GDFS 1009 Revealed the Role of MUP1 Gene on the Methionine-Based Induction of Morphogenesis and Biological Control Activity. Journal of Fungi, 9(2), 215. https://doi.org/10.3390/jof9020215







