Abstract
Candida tropicalis is emerging as one of the most common Candida species causing opportunistic infections in Latin America. Outbreak events caused by C. tropicalis were reported, and antifungal resistant isolates are on the rise. In order to investigate population genomics and look into antifungal resistance, we applied a short tandem repeat (STR) genotyping scheme and antifungal susceptibility testing (AFST) to 230 clinical and environmental C. tropicalis isolates from Latin American countries. STR genotyping identified 164 genotypes, including 11 clusters comprised of three to seven isolates, indicating outbreak events. AFST identified one isolate as anidulafungin-resistant and harboring a FKS1 S659P substitution. Moreover, we identified 24 clinical and environmental isolates with intermediate susceptibility or resistance to one or more azoles. ERG11 sequencing revealed each of these isolates harboring a Y132F and/or Y257H/N substitution. All of these isolates, except one, were clustered together in two groups of closely related STR genotypes, with each group harboring distinct ERG11 substitutions. The ancestral C. tropicalis strain of these isolates likely acquired the azole resistance-associated substitutions and subsequently spread across vast distances within Brazil. Altogether, this STR genotyping scheme for C. tropicalis proved to be useful for identifying unrecognized outbreak events and better understanding population genomics, including the spread of antifungal-resistant isolates.
1. Introduction
Candida tropicalis is one of the most common causative species of candidemia, although epidemiology varies significantly in different parts of the world [1,2]. In parts of Latin American (LATAM) and Asian Pacific countries, C. tropicalis is the most common opportunistic Candida species, even surpassing Candida albicans and other non-Candida albicans candida (NCAC) species [3,4,5,6,7,8]. The yeast is considered the second most virulent after C. albicans and is reported as the most efficient biofilm producer compared to other pathogenic Candida species [9,10]. Besides C. tropicalis being part of the normal human microbiome, it is isolated from the environment, including from soil, plants, fresh water, marine ecosystems, and animals [7,11,12,13,14]. Globally, antifungal resistance is emerging, and C. tropicalis is no exception to this worrying trend [15,16,17,18,19]. Resistance to azoles, echinocandins, and amphotericin B were described [3,20,21,22,23]. Similar to other Candida species, upregulation of multidrug efflux pumps [22,24,25] and missense mutations in genes of the ergosterol pathway can lead to azole resistance, the latter primarily in the target ERG11 [23,26,27,28,29]. In addition to azole resistance, azole trailing is occasionally observed and is defined as persistent but reduced growth above minimal inhibitory concentrations (MICs) [11,30,31]. Echinocandin resistance in Candida species is frequently caused by substitutions in the FKS1 gene encoding 1,3-beta-D-glucan synthase, which is the target enzyme of echinocandins [32,33]. Resistance-associated substitutions are concentrated at so-called hot spot (HS) regions which echinocandins utilize as binding sites [15,34].
Most outbreaks caused by C. tropicalis were described in intensive care units (ICUs) [35,36]. Patients that are immunocompromised, receive broad-spectrum antibiotics, have indwelling catheters, are on hemodialysis, mechanical ventilation, and/or have had recent abdominal surgery are most notably at risk [10,37]. In order to investigate outbreaks and prevent further transmission, adequate genotyping is essential and could identify the source and transmission routes. Multiple genotyping schemes were developed for C. tropicalis, each with its own specific benefits and turn-offs. Multiple short tandem repeat (STR) genotyping schemes proved to have high resolution, short turnaround times, and high reproducibility. Moreover, STR genotyping assays were developed for C. tropicalis [38,39,40,41].
Previously, we developed STR genotyping for C. tropicalis and compared its outcomes with whole genome sequencing (WGS) single nucleotide polymorphism (SNP) analysis [42]. Here, we applied this STR genotyping scheme to a large collection of clinical and environmental Latin American (LATAM) isolates. Additionally, susceptibility testing and resistance-associated genes were investigated.
2. Materials and Methods
2.1. Isolates
All 230 C. tropicalis isolates, either clinical or environmental samples from LATAM countries, predominantly from Brazil, were collected between 2010 and 2022 (Table S1). Clinical isolates originated from blood cultures (taken from both peripheral blood and catheter), and environmental isolates were collected from beach sand (Ponta Negra Beach) in Natal, Brazil. Isolates were stored according to standard procedures at −80 °C. All isolates were identified by using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as previously described [43]. The ethics committee of the Federal University of São Paulo approved this study (Studies not enrolling human beings or animals; Approval code: 8260200319, 7 March 2019).
2.2. STR Genotyping and Data Analysis
All isolates were grown on Sabouraud dextrose agar (SDA) plates (Oxoid, Hampshire, United Kingdom) at 30 °C. A single colony was resuspended in 400 µL MagNA Pure bacteria lysis buffer and MagNA Lyser green beads (both Roche Diagnostics GmbH, Mannheim, Germany). The suspensions were mechanically lysed for 30 s at 6500 rpm using the MagNA Lyser (Roche Diagnostics). DNA was extracted and purified with the MagNA Pure 96 instrument and the MagNA Pure DNA and Viral NA Small Volume kit (Roche Diagnostics), following the manufacturer’s instructions and the Pathogen 200 SV protocol.
Multiplex PCR reactions amplifying six STR targets were performed on a thermocycler (Biometra, Westburg, Göttingen, Germany) using 1× FastStart Taq polymerase buffer without MgCl2, deoxynucleoside triphosphates (dNTPs) (0.2 mM), MgCl2 (3 mM), forward and reverse primers (1 to 5 µM), 1 U FastStart Taq polymerase (Roche Diagnostics), and isolated DNA. The thermal protocol for PCR amplification consisted of denaturation of 10 min at 95 °C followed by 30 cycles consisting of 30 sec denaturation at 95 °C, 30 s annealing at 60 °C, and 1 min of extension at 72 °C with a final incubation step for 10 min at 72 °C. PCR products were diluted 1:1000 in water, and 10 µL of the diluted product, in addition to 0.12 µL of the Orange 600 DNA size standard (NimaGen, Nijmegen, The Netherlands), was incubated for 1 min at 95 °C and analyzed on a 3500 XL genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Corresponding copy numbers were determined using GeneMapper software (Applied Biosystems). Stutter peaks below 50% of the intensity of the highest peak for an allele, minus-A peaks and bleed-through peaks, were discarded for all markers. Copy numbers were ensured to be rounded. Copy numbers were converted to a binary matrix: “1” if an isolate contained the allele and “0” if it did not. The relatedness between isolates was analyzed with BioNumerics v.7.6.1 software (Applied Maths NV, Sint-Martens-Latem, Belgium), utilizing the unweighted pair group method with arithmetic mean averages (UPGMA) and using the multistate categorical similarity coefficient as previously described [44].
2.3. Antifungal Susceptibility Testing (AFST) and Resistance-Associated Gene Investigation
In vitro AFSTs against fluconazole (MERK, Darmstadt, Germany), voriconazole (MERK, Darmstadt, Germany), amphotericin B (MERK, Darmstadt, Germany), and anidulafungin (MERK, Darmstadt, Germany) were performed via broth microdilution according to the Clinical and Laboratory Standard Institute (CLSI) M27-S4 (23). In short, colonies were diluted in RPMI medium, and final concentrations of 1 × 103–5 × 105 CFU/mL were obtained with a Genesys 20 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Microtiter plates were incubated at 35 °C and visually interpreted after 24 h. MIC values were read as the lowest antifungal concentration with a 50% growth reduction when compared to the growth control, except for amphotericin B with a 100% growth reduction. Interpretive categories were implemented according to the definition by CLSI [45]. For fluconazole, MIC breakpoints of ≤2, 4, and ≥8 mg/L were considered susceptible, susceptible-dose dependent, and resistant, respectively. For voriconazole, ≤0.125, 0.25–0.5, and ≥1 mg/L were indicated as susceptible, intermediate, and resistant, respectively. Anidulafungin breakpoints were ≤0.25, 0.5, and ≥1 mg/L for susceptible, intermediate, and resistant, respectively. Amphotericin B breakpoints were set as ≤1 mg/L for susceptible and ≥1 mg/L as resistant. Trailing was optically interpreted as persistent but with residual growth above the MIC value compared to the growth control [30,31]. ERG11 and FKS1 gene sequences were amplified as previously described [25,33]. Amplified DNA was purified using the D-pure purification protocol (Nimagen), sequenced on a 3500 XL genetic analyzer (Applied Biosystems), and analyzed using BioNumerics 7.6.1 (Applied Maths). Generated sequences of susceptible and resistant isolates were compared to each other with the use of the multiple alignment option with an open gap penalty of 100%. Lanosterol 14 α-demethylase (ERG11) protein homology modelling was conducted with SWISS-MODEL (https://swissmodel.expasy.org/interactive, accessed on 2 November 2022), by using the 5eqb.1A “Crystal structure of lanosterol 14 α demethylase (ERG11) with intact transmembrane domain bound to itraconazole” template [46]. The predicted lanosterol 14 α-demethylase (ERG11) protein model was visually inspected to ensure that all major P450 structural motifs (I-helix, FG loop, and the substrate access channel) were intact. The resulting PDB file was uploaded to PyMOL v2.0, and the amino acids Y132 and Y257 were subsequently marked red [47].
3. Results
3.1. Application of C. tropicalis STR Genotyping
A STR genotyping for C. tropicalis was applied to all 230 C. tropicalis isolates as previously identified by MALDI-TOF. Isolates were predominantly from São Paulo (n = 117, 50.9%), Curitiba (n = 27, 11.7%), and Rio de Janeiro (n = 25, 10.9%). Genotyping resulted in the identification of 164 different genotypes each consisting of one to seven isolates (Figure 1A). Overall genetic diversity was high, with the vast majority of isolates exhibiting copy number differences in more than three microsatellite markers. We found 32 small clusters of two isolates in addition to 11 large clusters with more than two isolates. Isolates from large clusters exclusively originated from São Paulo, Natal, Curitiba, Salvador, and Buenos Aires. Five of these large clusters spanned two different cities; 18 of 32 small clusters comprised two independent isolates taken from the same patient. All but one of the environmental isolates (ID: 1443/2017) from Ponta Negra Beach, Natal, Brazil (n = 12, 5.2%) exhibited highly similar genotypes (Gt 95 to 97) and were closely related to clinical isolates (Gt 93 and 94) with copy number differences in three alleles (Figure S1).
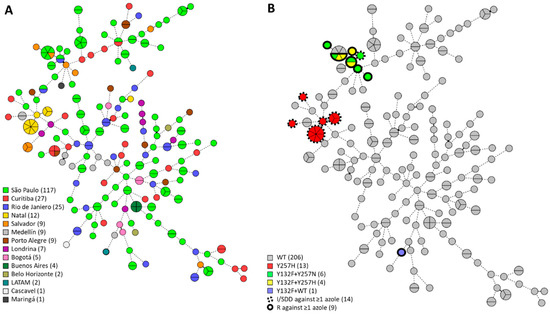
Figure 1.
Minimum-spanning tree of 230 C. tropicalis isolates. Branch lengths indicate the similarity between isolates with thick solid lines (variation in one allele), thin solid lines (variation in two alleles), and thin dotted lines (variation in three or more alleles). Azole-intermediate and azole-resistant isolates are demarcated in dotted black lines and solid black lines, respectively. On the left (A), the number of isolates per city are shown in the color key; on the right (B), isolates with ERG11 substitutions and reduced azole susceptibility are shown as in the color key. LATAM, Latin American.
3.2. AFST and Resistance-Associated Genes Investigation
AFST CLSI microbroth dilution was performed on all isolates using fluconazole, voriconazole, amphotericin B, and anidulafungin (Table S1). Elevated MICs against azoles and anidulafungin were found in 10.4% of all isolates, those being intermediate (6.1%; n = 15) or resistant (3.9%; n = 9) to at least one azole drug, and one isolate (0.4%) was found to be resistant to anidulafungin (Table 1). Trailing was observed in two isolates (IDs: 7225/2010 and 1535/2017) with reduced susceptibility to azoles (Table 2). ERG11 and FKS1 genes were sequenced to identify mutations conferring resistance. Y132F, Y257H, and Y257N substitutions in ERG11 were found exclusively in isolates with reduced susceptibility to at least one azole drug (Table 2). The azole-intermediate and -resistant isolates all originated from Brazil, except for one Colombian isolate (ID: 291/2018). These resistant Brazilian isolates were confined to two groups of related STR genotypes, despite isolates from both groups originating from diverse cities up to 2,600 km apart. One of these groups (n = 13, IDs: 7225/2010, 7416/2011, 1450/2017, 1452/2017, 1453/2017, 1463/2017, 1468/2017, 1471/2017, 1483/2017, 1486/2017, 1488/2017, 1491/2017, and 1498/2017) was solely intermediately resistant to azoles, harbored the ERG11 Y257H substitution, and included environmental isolates (n = 11). The other group comprised intermediate and resistant isolates (n = 10, IDs: 8095/2011, 9253/2013, 822/2015, 1534/2017, 1535/2017, 1743/2017, 681/2019, 683/2019, 1540/2020, and 1541/2020) and exhibited the ERG11 Y132F and Y257H/Y257N substitutions. The fluconazole-resistant Colombian isolate was not closely related to these two groups of intermediate and resistant isolates, and it harbored a heterozygous ERG11 Y132F substitution, whereas all previously mentioned substitutions were homozygous (Figure 1B, Table 2). Protein homology modelling for lanosterol 14 α-demethylase (ERG11) was conducted to visualize and inspect the resistance-associated substitutions (Figure S2). The ERG11 Y257H/N substitution was situated in the G-helix, which is at the rear of the protein at the putative substrate access channel. The echinocandin resistant isolate (ID: 1751/2017) exhibited a FKS1 S659P substitution located in HS-1 (Table 2).

Table 1.
MIC ranges, GM, MIC50, and MIC90 values found performing microbroth dilution for 24 h according to the CLSI M27-S4 standard. MIC, minimal inhibitory concentration; GM, geometric mean.

Table 2.
Overview of isolates resistant or intermediately resistant to azoles or echinocandins with corresponding ERG11 or FKS1 substitutions. MIC, minimal inhibitory concentration; FLC, fluconazole; VOR, voriconazole; AFG, anidulafungin.
4. Discussion
In this study, an STR genotyping scheme for C. tropicalis was applied to 230 clinical and environmental C. tropicalis isolates from Latin American countries. STR genotyping identified 164 different genotypes, indicating an overall high genetic diversity within LATAM countries, especially in Brazil. Limited data is available regarding C. tropicalis phylogenetic relationships within LATAM countries. One Brazilian study was in line with the present observation of an overall high genetic diversity, according to MLST genotyping [48]. In total, 11 large clusters consisting of more than two isolates were found. Using AFSTs of fluconazole, voriconazole, amphotericin B, and anidulafungin, 15 isolates were found to have intermediately resistant MICs to at least one azole drug, and nine isolates were resistant to at least one azole drug. Except for one isolate with a reduced susceptibility to azoles, all were confined to two groups of related STR genotypes. These isolates all harbored one or more of the azole resistance-associated substitutions ERG11 Y132F, Y257H, and Y257N. Additionally, one isolate was resistant to anidulafungin and exhibited an FKS1 S659P substitution in HS1.
4.1. Application of C. tropicalis STR Genotyping
Within the 164 different C. tropicalis genotypes, we found several clusters of varying sizes comprising two to seven isolates from different patients, suggesting outbreak events within multiple hospitals. The majority of these previously unrecognized outbreak events included isolates collected within a few months from each other, suggesting the possibility of persistent clonal spread, as was also found for C. parapsilosis in Brazilian hospital settings [8,49,50]. Multiple studies have identified C. tropicalis as the strongest biofilm producer when compared to other pathogenic Candida species, making this yeast highly capable of colonizing intravascular lines and devices [9,10,51,52]. Moreover, multiple studies identified healthcare personnel as the source of C. tropicalis outbreaks [35,53,54]. From 38 patients, two samples were taken on consecutive dates, and genotypes were identical. For nine patients, one sample was cultured from peripheral blood and one from catheter blood, both samples always exhibiting the same STR genotype. Taken together with the short timespan between these related infections within the same hospital wards, it is possible that nosocomial spread due to inadequate infection prevention and control measures was involved in these outbreaks.
Interestingly, five clusters represented isolates from two different cities up to 1.400 km apart from each other. These findings suggest the spread of multiple C. tropicalis genotypes across a vast distance, but WGS data analysis is necessary to confirm that hypothesis. The environmental isolates from Natal, Brazil were recovered from sand at the Ponta Negra beach, which is a popular recreational destination. Previously, C. tropicalis isolates were isolated from the same beach, and some were found to be resistant to azoles [13]. The environmental isolates in this study were related to clinical samples from Brazil and Colombia, suggesting the dissemination of C. tropicalis isolates due to human activities, as was previously also shown for C. tropicalis and C. auris [55,56]. Our findings highlight the importance of high-resolution genotyping to reveal outbreak events within hospitals and obtain a better understanding of population genomics.
4.2. AFST and Resistance Associated Genes Investigation
AFST identified 24 isolates (10.4%) that were intermediate or resistant to one or more azole drugs, in addition to one isolate (0.4%) being resistant to anidulafungin. Interestingly, all Brazilian isolates with a reduced susceptibility to azoles were confined to two separate groups of closely related STR genotypes despite originating from cities far apart. All isolates from the first group harbored the ERG11 Y257H substitution and were intermediately resistant to at least one azole drug. The absence of this mutation in other isolates suggests that spontaneous development of this mutation is unlikely and that this mutation may have originated from a common single ancestor C. tropicalis strain in Brazil. Widespread use of agricultural azoles is known to have driven the emergence of environmental resistance mechanisms in fungi, which could hypothetically be a contributing factor in this setting [57,58,59,60,61]. However, although 11 out of 23 of these isolates originated from the environment, a non-environmental source cannot be excluded, as these samples were acquired from a public beach. The ERG11 Y257H substitution was previously associated with reduced fluconazole susceptibility in C. tropicalis and C. albicans [62,63]. The MICs found in this study indicate the Y257H substitution lowers the susceptibility to azoles to an intermediate level. In C. albicans, the Y257H was thought to prevent fluconazole from entering the active site, leading to reduced susceptibility. The ERG11 Y257 in C. tropicalis and C. albicans correspond to the same position and are located in the G-helix; therefore, the mode of action in C. albicans is likely identical for C. tropicalis [28,42,64,65].
Resistant isolates in the other group of related STR genotypes all harbored an ERG11 Y132F substitution in combination with a Y257H/N substitution. Again, the unique presence of the Y132F mutation of this group as compared to the other C. tropicalis isolates in this study suggests that the mutation originated from an ancestor strain of this group, whereas the Y257H or Y257N substitutions were likely acquired more recently. The Y257H/N substitution might prevent fluconazole from entering the binding site because aromatic rings in asparagine and histidine are absent there but present in tyrosine. Aromatic rings of amino acids such as tyrosine form pi–pi binding interactions with other aromatic rings, as also present in fluconazole [66]. This potential interaction between the aromatic rings is postulated to facilitate the binding between azoles and ERG11, leading to reduced protein activity and, therewith, increased azole susceptibility [65,67]. Finally, one STR cluster of six isolates within this group contained three resistant and three susceptible isolates. In the latter three, substitutions conferring azole resistance were absent. These susceptible isolates all originated from one patient and were collected in 2021, whereas the resistant isolates were collected in 2019 and 2020. These three recent isolates might have lost the Y132F and Y257H/N substitutions in a patient or an environment without drug pressure, as azole resistance is also associated with a loss of fitness [16,68].
The present study has some limitations. Previously, STR and WGS SNP outcomes were highly concordant, indicating a high resolution for the STR typing scheme, but the exact resolution is unknown and might differ in the present isolate collection. Moreover, only the ERG11 gene was sequenced, without taking into account other resistance-associated genes like UPC2 and TAC1B, which could harbor mutations conferring resistance. In a follow-up study, WGS could be used to identify more mutations in resistance-associated genes and confirm the close relatedness of these isolates according to STR typing despite originating from diverse cities. Additionally, extensive environmental sampling and genotyping could trace the source of the clusters found in this study. The limited clinical data in the present study hampered the interpretation of genotypic clustering.
To summarize, we applied an STR genotyping scheme for C. tropicalis to clinical and environmental isolates from LATAM countries to identify clusters of previously unrecognized outbreak events. AFST identified 10% of isolates with reduced susceptibility to azoles. Except for one isolate, all were confined to two related groups of STR genotypes, and all isolates harbored resistance-associated substitutions in ERG11. Each group exhibited a different set of substitutions, suggesting an ancestral C. tropicalis strain in both groups that subsequently spread to diverse cities across vast distances. Only one isolate was anidulafungin-resistant and exhibited a FKS1 S659P substitution in HS1, reflecting the low prevalence of C. tropicalis echinocandin-resistance in LATAM countries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9020207/s1, Figure S1: Cluster analysis of 230 C. tropicalis isolates. Branch lengths indicate relatedness according to microsatellite alleles. The UPGMA dendrogram was generated with BioNumerics 7.6.1 (Applied Maths, Kortrijk, Belgium) using the multistate categorical similarity coefficient. Bootstrap values using 1000 replicates were above 95% or specified at the branch when lower. Gt, genotype; FLC, fluconazole; S, susceptible; SDD, susceptible-dose dependent; R, resistant; Figure S2: C. tropicalis lanosterol 14 α-demethylase (ERG11) homology model. This model was constructed with SWISS-MODEL and viewed from multiple angles. Front view (A), side view (B), and top view (C), with the Y132 and Y257 amino acids marked in red. Y132 is located near the core of the protein, whereas Y257 is located in the G-helix at the rear of the protein. Table S1: Overview of all C. tropicalis isolates, including AFST MICs according to microbroth dilutions performed to the CLSI M27-S4 standard and clinical details. Gt, genotype; FLC, fluconazole; VRC, voriconazole; AMB, amphotericin B; AFG, anidulafungin; ICU, intensive care unit.
Author Contributions
Conceptualization, J.F.M. and A.L.C.; methodology, B.S. and C.C.S.Z.B.; software, B.S.; validation, B.S., C.C.S.Z.B., and A.L.C.; formal analysis, B.S. and T.d.G.; investigation, B.S., C.C.S.Z.B., and A.L.C.; resources, A.L.C., J.N.d.A.J., I.B., N.B.F., G.M.C., and group members; data curation, A.L.C., E.F.J.M., and T.d.G.; writing—original draft preparation, B.S. and C.C.S.Z.B.; writing—review and editing, A.L.C., E.F.J.M., J.N.d.A.J., I.B., N.B.F., G.M.C., J.F.M., and T.d.G.; visualization, B.S. and T.d.G.; supervision, E.F.J.M. and T.d.G.; project administration, A.L.C. and J.F.M.; funding acquisition, J.F.M. and A.L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Canisius-Wilhelmina Hospital, grant number CWZ_001421. A.L.C. received support from the Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP, grant number 2017/02203-07.
Institutional Review Board Statement
The ethics committee of the Federal University of São Paulo approved this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable.
Acknowledgments
The members of the Latin American Group for investigating Candida tropicalis resistance are: Carla Sakuma de Oliveira, Hospital do Câncer-Uopeccan, Cascavel, Brazil; Flavio Queiroz Telles, Universidade Federal do Parana, Curitiba, Brazil; Vania Vicente, Universidade Federal do Parana, Curitiba, Brazil; Marcio Nucci, Universidade Federal do Rio de Janeiro, Brazil; Thais Guimaraes, Hospital do Servidor Público Estadual, São Paulo, Brazil; Tereza Cristina Teixeira Sukiennik, Segurança Assistencial, Controle de Infecção, Santa Casa de Misericórdia de Porto Alegre, Brasil; Maria Goreth, Barberino, Hospital São Rafael, Salvador, Brazil; Ana Verena Mendes, Hospital São Rafael, Salvador, Brazil.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zuza-Alves, D.L.; Silva-Rocha, W.P.; Chaves, G.M. An Update on Candida tropicalis Based on Basic and Clinical Approaches. Front. Microbiol. 2017, 8, 1927. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.Y.A.; Denning, D.W.; Warn, P. Candida tropicalis in human disease. Crit. Rev. Microbiol. 2010, 36, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Kothavade, R.J.; Kura, M.M.; Valand, A.G.; Panthaki, M.H. Candida tropicalis: Its prevalence, pathogenicity and increasing resistance to fluconazole. J. Med. Microbiol. 2010, 59, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Y.C.; Hsueh, P.R. Epidemiology of candidemia and antifungal susceptibility in invasive Candida species in the Asia-Pacific region. Future Microbiol. 2016, 11, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Y.; Hsu, L.Y.; Alejandria, M.M.; Chaiwarith, R.; Chinniah, T.; Chayakulkeeree, M.; Choudhury, S.; Chen, Y.H.; Shin, J.H.; Kiratisin, P.; et al. Antifungal susceptibility of invasive Candida bloodstream isolates from the Asia-Pacific region. Med. Mycol. 2016, 54, 471–477. [Google Scholar] [CrossRef]
- Da Matta, D.A.; Souza, A.C.R.; Colombo, A.L. Revisiting Species Distribution and Antifungal Susceptibility of Candida Bloodstream Isolates from Latin American Medical Centers. J. Fungi 2017, 3, 24. [Google Scholar] [CrossRef]
- Lima, R.; Ribeiro, F.C.; Colombo, A.L.; de Almeida Jr, J.N. The emerging threat antifungal-resistant Candida tropicalis in humans, animals, and environment. Front. Fungal Biol. 2022, 3, 957021. [Google Scholar] [CrossRef]
- Souza, A.C.; Fuchs, B.B.; Pinhati, H.M.; Siqueira, R.A.; Hagen, F.; Meis, J.F.; Mylonakis, E.; Colombo, A.L. Candida parapsilosis Resistance to Fluconazole: Molecular Mechanisms and In Vivo Impact in Infected Galleria mellonella Larvae. Antimicrob. Agents Chemother. 2015, 59, 6581–6587. [Google Scholar] [CrossRef]
- Pannanusorn, S.; Fernandez, V.; Romling, U. Prevalence of biofilm formation in clinical isolates of Candida species causing bloodstream infection. Mycoses 2013, 56, 264–272. [Google Scholar] [CrossRef]
- Negri, M.; Silva, S.; Henriques, M.; Oliveira, R. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur J. Clin. Microbiol. Infect. Dis. 2012, 31, 1399–1412. [Google Scholar] [CrossRef]
- Yang, Y.L.; Lin, C.C.; Chang, T.P.; Lauderdale, T.L.; Chen, H.T.; Lee, C.F.; Hsieh, C.W. Comparison of human and soil Candida tropicalis isolates with reduced susceptibility to fluconazole. PLoS ONE 2012, 7, e34609. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, C.E.; Oliveira-Pacheco, J.; Cinnéide, E.Ó.; Haase, M.A.B.; Hittinger, C.T.; Rogers, T.R.; Zaragoza, O.; Bond, U.; Butler, G. Population genomics of the pathogenic yeast Candida tropicalis identifies hybrid isolates in environmental samples. PLoS Pathog. 2021, 17, e1009138. [Google Scholar] [CrossRef] [PubMed]
- Zuza-Alves, D.L.; de Medeiros, S.S.; de Souza, L.B.; Silva-Rocha, W.P.; Francisco, E.C.; de Araújo, M.C.; Lima-Neto, R.G.; Neves, R.P.; de Azevedo Melo, A.S.; Chaves, G.M. Evaluation of Virulence Factors In vitro, Resistance to Osmotic Stress and Antifungal Susceptibility of Candida tropicalis Isolated from the Coastal Environment of Northeast Brazil. Front. Microbiol. 2016, 7, 1783. [Google Scholar] [CrossRef]
- Medeiros, A.O.; Kohler, L.M.; Hamdan, J.S.; Missagia, B.S.; Barbosa, F.A.; Rosa, C.A. Diversity and antifungal susceptibility of yeasts from tropical freshwater environments in Southeastern Brazil. Water Res. 2008, 42, 3921–3929. [Google Scholar] [CrossRef]
- Arendrup, M.C. Update on antifungal resistance in Aspergillus and Candida. Clin. Microbiol. Infect. 2014, 20, 42–48. [Google Scholar] [CrossRef]
- Berkow, E.L.; Lockhart, S.R. Fluconazole resistance in Candida species: A current perspective. Infect. Drug Resist. 2017, 10, 237–245. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Yang, F.; Lu, H.; Wu, H.; Fang, T.; Berman, J.; Jiang, Y.Y. Aneuploidy Underlies Tolerance and Cross-Tolerance to Drugs in Candida parapsilosis. Microbiol. Spectr. 2021, 9, e0050821. [Google Scholar] [CrossRef]
- Yang, Y.L.; Ho, Y.A.; Cheng, H.H.; Ho, M.; Lo, H.J. Susceptibilities of Candida species to amphotericin B and fluconazole: The emergence of fluconazole resistance in Candida tropicalis. Infect. Control Hosp. Epidemiol. 2004, 25, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Arango, A.C.; Rueda, C.; Roman, E.; Quintin, J.; Terron, M.C.; Luque, D.; Netea, M.G.; Pla, J.; Zaragoza, O. Cell Wall Changes in Amphotericin B-Resistant Strains from Candida tropicalis and Relationship with the Immune Responses Elicited by the Host. Antimicrob. Agents Chemother. 2016, 60, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, H.O.; Watanabe, A.; Kamei, K. Azole and echinocandin resistance mechanisms and genotyping of Candida tropicalis in Japan: Cross-boundary dissemination and animal-human transmission of C. tropicalis infection. Clin. Microbiol. Infect. 2022, 28, e5–e8. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standard Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Fourth Informational Supplement; CLSI Document M27-S4; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Jin, L.; Cao, Z.; Wang, Q.; Wang, Y.; Wang, X.; Chen, H.; Wang, H. MDR1 overexpression combined with ERG11 mutations induce high-level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect. Dis. 2018, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, P.; Larcher, G.; Berges, T.; Renier, G.; Chabasse, D.; Bouchara, J.P. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob. Agents Chemother. 2005, 49, 4608–4615. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Pereira, V.S.; Castelo-Branco, D.; Cordeiro, R.A.; Sidrim, J.J.C.; Brilhante, R.S.N.; Rocha, M.F.G. The yeast, the antifungal, and the wardrobe: A journey into antifungal resistance mechanisms of Candida tropicalis. Can. J. Microbiol. 2020, 66, 377–388. [Google Scholar] [CrossRef]
- Xisto, M.I.D.S.; Caramalho, R.D.; Rocha, D.A.; Ferreira-Pereira, A.; Sartori, B.; Barreto-Bergter, E.; Junqueira, M.L.; Lass-Flörl, C.; Lackner, M. Pan-azole-resistant Candida tropicalis carrying homozygous erg11 mutations at position K143R: A new emerging superbug? J. Antimicrob. Chemother. 2017, 72, 988–992. [Google Scholar] [CrossRef]
- Tan, J.; Zhang, J.; Chen, W.; Sun, Y.; Wan, Z.; Li, R.; Liu, W. The A395T mutation in ERG11 gene confers fluconazole resistance in Candida tropicalis causing candidemia. Mycopathologia 2015, 179, 213–218. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 2020, 55, 105799. [Google Scholar] [CrossRef]
- Chen, P.Y.; Chuang, Y.C.; Wu, U.I.; Sun, H.Y.; Wang, J.T.; Sheng, W.H.; Chen, Y.C.; Chang, S.C. Mechanisms of Azole Resistance and Trailing in Candida tropicalis Bloodstream Isolates. J. Fungi 2021, 7, 612. [Google Scholar] [CrossRef]
- Astvad, K.M.T.; Sanglard, D.; Delarze, E.; Hare, R.K.; Arendrup, M.C. Implications of the EUCAST Trailing Phenomenon in Candida tropicalis for the In Vivo Susceptibility in Invertebrate and Murine Models. Antimicrob. Agents Chemother. 2018, 62, e01624-18. [Google Scholar] [CrossRef]
- Jensen, R.H.; Johansen, H.K.; Arendrup, M.C. Stepwise development of a homozygous S80P substitution in Fks1p, conferring echinocandin resistance in Candida tropicalis. Antimicrob. Agents Chemother. 2013, 57, 614–617. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Mokaddas, E.; Meis, J.F.; Joseph, L.; Abdullah, A.; Vayalil, S. Development of Echinocandin Resistance in Candida tropicalis following Short-Term Exposure to Caspofungin for Empiric Therapy. Antimicrob. Agents Chemother. 2018, 62, e01926-17. [Google Scholar] [CrossRef] [PubMed]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Barac, A.; Cevik, M.; Colovic, N.; Lekovic, D.; Stevanovic, G.; Micic, J.; Rubino, S. Investigation of a healthcare-associated Candida tropicalis candidiasis cluster in a haematology unit and a systematic review of nosocomial outbreaks. Mycoses 2020, 63, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Becker, K.; Fegeler, W.; Gugnani, H.C.; Kapoor, L.; Randhawa, V.S.; Mehta, G. An outbreak of candidemia due to Candida tropicalis in a neonatal intensive care unit. Mycoses 2003, 46, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Mellinghoff, S.C.; Hartmann, P.; Cornely, F.B.; Knauth, L.; Kohler, F.; Kohler, P.; Krause, C.; Kronenberg, C.; Kranz, S.; Menon, V.; et al. Analyzing candidemia guideline adherence identifies opportunities for antifungal stewardship. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1563–1571. [Google Scholar] [CrossRef]
- de Groot, T.; Puts, Y.; Berrio, I.; Chowdhary, A.; Meis, J.F. Development of Candida auris Short Tandem Repeat Typing and Its Application to a Global Collection of Isolates. mBio 2020, 11, e02971-19. [Google Scholar] [CrossRef]
- van Haren, M.H.I.; de Groot, T.; Spruijtenburg, B.; Jain, K.; Chowdhary, A.; Meis, J.F. Development of a Multiplex PCR Short Tandem Repeat Typing Scheme for Candida krusei. J. Clin. Microbiol. 2022, 60, e0203221. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, H.J.; Che, J.; Li, W.G.; Bian, F.N.; Yu, S.B.; Zhang, L.J.; Lu, J. Multilocus microsatellite markers for molecular typing of Candida tropicalis isolates. BMC Microbiol. 2014, 14, 245. [Google Scholar] [CrossRef]
- Fan, X.; Xiao, M.; Liu, P.; Chen, S.; Kong, F.; Wang, H.; Zhang, L.; Hou, X.; Xu, Y.C. Novel Polymorphic Multilocus Microsatellite Markers to Distinguish Candida tropicalis Isolates. PLoS ONE 2016, 11, e0166156. [Google Scholar] [CrossRef]
- Spruijtenburg, B.; van Haren, M.H.I.; Chowdhary, A.; Meis, J.F.; de Groot, T. Development and Application of a Short Tandem Repeat Multiplex Typing Assay for Candida tropicalis. Microbiol. Spectr. 2023, 11, e0461822. [Google Scholar] [CrossRef] [PubMed]
- De Carolis, E.; Vella, A.; Vaccaro, L.; Torelli, R.; Posteraro, P.; Ricciardi, W.; Sanguinetti, M.; Posteraro, B. Development and validation of an in-house database for matrix-assisted laser desorption ionization-time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J. Clin. Microbiol. 2014, 52, 1453–1458. [Google Scholar] [CrossRef] [PubMed]
- de Valk, H.A.; Meis, J.F.; Curfs, I.M.; Muehlethaler, K.; Mouton, J.W.; Klaassen, C.H. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 2005, 43, 4112–4120. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Kiefer, F.; Arnold, K.; Kunzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. Available online: https://pymol.org/2/ (accessed on 10 October 2022).
- Magri, M.M.; Gomes-Gouvea, M.S.; de Freitas, V.L.; Motta, A.L.; Moretti, M.L.; Shikanai-Yasuda, M.A. Multilocus sequence typing of Candida tropicalis shows the presence of different clonal clusters and fluconazole susceptibility profiles in sequential isolates from candidemia patients in Sao Paulo, Brazil. J. Clin. Microbiol. 2013, 51, 268–277. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; de Almeida, J.N., Jr.; Sejas, O.N.E.; Del Negro, G.M.B.; Carvalho, G.; Gimenes, V.M.F.; de Souza, M.E.B.; Arastehfar, A.; Camargo, C.H.; Motta, A.L.; et al. Environmental Clonal Spread of Azole-Resistant Candida parapsilosis with Erg11-Y132F Mutation Causing a Large Candidemia Outbreak in a Brazilian Cancer Referral Center. J. Fungi 2021, 7, 259. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; Del Negro, G.M.B.; Ribeiro, L.B.; da Silva, M.; Carvalho, G.O.M.H.; Camargo, C.H.; de Almeida, J.N., Jr.; Motta, A.L.; Siciliano, R.F.; Sejas, O.N.E.; et al. A Brazilian Inter-Hospital Candidemia Outbreak Caused by Fluconazole-Resistant Candida parapsilosis in the COVID-19 Era. J. Fungi 2022, 8, 100. [Google Scholar] [CrossRef]
- Negri, M.; Silva, S.; Henriques, M.; Azeredo, J.; Svidzinski, T.; Oliveira, R. Candida tropicalis biofilms: Artificial urine, urinary catheters and flow model. Med. Mycol. 2011, 49, 739–747. [Google Scholar] [CrossRef]
- Negri, M.; Martins, M.; Henriques, M.; Svidzinski, T.I.; Azeredo, J.; Oliveira, R. Examination of potential virulence factors of Candida tropicalis clinical isolates from hospitalized patients. Mycopathologia 2010, 169, 175–182. [Google Scholar] [CrossRef]
- Maganti, H.; Yamamura, D.; Xu, J. Prevalent nosocomial clusters among causative agents for candidemia in Hamilton, Canada. Med. Mycol. 2011, 49, 530–538. [Google Scholar] [CrossRef][Green Version]
- Asmundsdottir, L.R.; Erlendsdottir, H.; Haraldsson, G.; Guo, H.; Xu, J.; Gottfredsson, M. Molecular epidemiology of candidemia: Evidence of clusters of smoldering nosocomial infections. Clin. Infect. Dis. 2008, 47, e17–e24. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Singh, P.; Wang, Y.; Yadav, A.; Pawar, K.; Singh, A.; Padmavati, G.; Xu, J.; Chowdhary, A. Environmental Isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India. mBio 2021, 12, e03181-20. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Rogerson, A.; Schatz, S.; Laubach, H.; Tallman, A.; Fell, J. Prevalence of yeasts in beach sand at three bathing beaches in South Florida. Water Res. 2007, 41, 1915–1920. [Google Scholar] [CrossRef]
- Castelo-Branco, D.; Lockhart, S.R.; Chen, Y.C.; Santos, D.A.; Hagen, F.; Hawkins, N.J.; Lavergne, R.; Meis, J.F.; Le Pape, P.; Rocha, M.F.G.; et al. Collateral consequences of agricultural fungicides on pathogenic yeasts: A One Health perspective to tackle azole resistance. Mycoses 2022, 65, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Brauer, V.S.; Rezende, C.P.; Pessoni, A.M.; De Paula, R.G.; Rangappa, K.S.; Nayaka, S.C.; Gupta, V.K.; Almeida, F. Antifungal Agents in Agriculture: Friends and Foes of Public Health. Biomolecules 2019, 9, 521. [Google Scholar] [CrossRef]
- Arastehfar, A.; Gabaldon, T.; Garcia-Rubio, R.; Jenks, J.D.; Hoenigl, M.; Salzer, H.J.F.; Ilkit, M.; Lass-Flörl, C.; Perslin, D.S. Drug-Resistant Fungi: An Emerging Challenge Threatening Our Limited Antifungal Armamentarium. Antibiotics 2020, 9, 877. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Meis, J.F. Emergence of azole-resistant aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013, 9, e1003633. [Google Scholar] [CrossRef]
- Yadav, A.; Jain, K.; Wang, Y.; Pawar, K.; Kaur, H.; Sharma, K.K.; Tripathy, V.; Singh, A.; Xu, J.; Chowdhary, A. Candida auris on Apples: Diversity and Clinical Significance. mBio 2022, 13, e0051822. [Google Scholar] [CrossRef]
- Fan, X.; Xiao, M.; Zhang, D.; Huang, J.J.; Wang, H.; Hou, X.; Zhang, L.; Kong, F.; Chen, S.C.A.; Tong, Z.H.; et al. Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin. Microbiol. Infect. 2019, 25, 885–891. [Google Scholar] [CrossRef]
- Keighley, C.; Gall, M.; van Hal, S.J.; Halliday, C.L.; Chai, L.Y.A.; Chew, K.L.; Biswas, C.; Slavin, M.A.; Meyer, W.; Sintchenko, V.; et al. Whole Genome Sequencing Shows Genetic Diversity, as Well as Clonal Complex and Gene Polymorphisms Associated with Fluconazole Non-Susceptible Isolates of Candida tropicalis. J. Fungi 2022, 8, 896. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Y.; Li, C. Association of T916C (Y257H) mutation in Candida albicans ERG11 with fluconazole resistance. Mycoses 2013, 56, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.J.; Liu, J.Y.; Ni, P.H.; Wang, S.; Shi, C.; Wei, B.; Ni, Y.X.; Ge, H.L. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013, 13, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Salonen, L.M.; Ellermann, M.; Diederich, F. Aromatic rings in chemical and biological recognition: Energetics and structures. Angew. Chem. Int. Ed. Engl. 2011, 50, 4808–4842. [Google Scholar] [CrossRef]
- Perez-Cantero, A.; Lopez-Fernandez, L.; Guarro, J.; Capilla, J. Azole resistance mechanisms in Aspergillus: Update and recent advances. Int. J. Antimicrob. Agents 2020, 55, 105807. [Google Scholar] [CrossRef] [PubMed]
- Sasse, C.; Dunkel, N.; Schafer, T.; Schneider, S.; Dierolf, F.; Ohlsen, K.; Morschhäuser, J. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 2012, 86, 539–556. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).