Abstract
Metabolites produced by filamentous fungi are used extensively in the food and drug industries. With the development of the morphological engineering of filamentous fungi, numerous biotechnologies have been applied to alter the morphology of fungal mycelia and enhance the yields and productivity of target metabolites during submerged fermentation. Disruption of chitin biosynthesis can modify the cell growth and mycelial morphology of filamentous fungi and regulate the biosynthesis of metabolites during submerged fermentation. In this review, we present a comprehensive coverage of the categories and structures of the enzyme chitin synthase, chitin biosynthetic pathways, and the association between chitin biosynthesis and cell growth and metabolism in filamentous fungi. Through this review, we hope to increase awareness of the metabolic engineering of filamentous fungal morphology, provide insights into the molecular mechanisms of morphological control via chitin biosynthesis, and describe strategies for the application of morphological engineering to enhance the production of target metabolites in filamentous fungi during submerged fermentation.
1. The Relationship between Fungal Morphology and Production Performance
Microbial secondary metabolites (MSMs), such as antibiotics, organic acids, hormones, and pigments, typically synthesized during the late growth phase of the producing microorganisms, are generally inessential for their growth [1]. In the year 2000, the market for antimicrobial secondary metabolites was 55 billion dollars [2], and that for antibiotics had increased to 66 billion dollars by 2007 [3]. In addition, Evaluate Pharma predicted that the biopharmaceutical (including hormones, enzymes, and vaccines) market would grow from 202 billion dollars in 2016 to 326 billion dollars in 2022. MSMs, used extensively in the food and pharmaceutical industries, make valuable contributions to health, nutrition, and the social economy. Among the microbial producers of these secondary metabolites, filamentous fungi, distinguished by a mycelium composed of septate hyphae or branching filaments, are the most frequently employed metabolically versatile cell factories in the biotechnology industry, utilized as sources of a diverse range of compounds, including industrial enzymes, penicillin, and citric acid [4,5]. In a previous review of the morphology of industrial fungi, Quintanilla et al. described three main advantages of the widespread application of filamentous fungi: (1) exceptional secretion of proteins or enzymes [6], (2) specialized post-transcriptional modification machinery facilitating glycosylation and correct protein folding [7], and (3) a diverse range of safe species, approved by the regulatory authorities and categorized as generally recognized as safe (GRAS) [4]. Compared with conventional solid-state fermentation, submerged fermentation has notable advantages, including simple parameter control processing, large volume processing, reduced fermentation time, low labor intensity, and ready scale-up, and this type of cultivation is becoming the primary industrial biotechnology used in the production of target fungal metabolites [8,9].
However, there are certain disadvantages associated with the submerged fermentation cultivation of filamentous fungi, such as the high viscosity of the fermentation medium determined by fungal morphology, considerably affecting the transfer of mass, heat, and momentum, and consequently, the yield and productivity of target products within bioreactors [5,10,11]. During submerged cultivation in bioreactors, filamentous fungi grow in one of three morphological forms, namely, dispersed mycelia, clumped aggregates, or mycelial pellets (including rough and smooth pellets) [12,13,14,15]. In addition, during submerged fermentation of filamentous fungi in conical flasks, an irregular mycelial block-type morphology is observed, as seen among species in the genus Monascus. Fungal morphology can be markedly and intricately determined by environmental conditions and inherent molecular or genetic biology. Indeed, the occurrence of different morphological forms of fungi when cultivated under submerged fermentation conditions, is one of the several engineering issues that require appropriate resolution. In this regard, Veiter et al. outlined the interlinkages between productivity, performance, and morphology among a range of different fungal species, particularly highlighting the relationship between fungal pellet morphology and metabolite productivity [16]. Figure 1 represents the mechanisms underlying the correlation between environmental conditions or genes, and fungal morphology and metabolite productivity during submerged fermentation. The generation of macroscopic clumps and spherical pellets is associated with a number of undesirable effects, including the limitation of diffusive mass transfer and reduced nutrient and oxygen levels, particularly in the centers of mycelial pellets and clumps. Furthermore, a dispersed mycelial morphology is often associated with an increase in the viscosity of the fermentation medium, thereby reducing mixing efficiency, hampering stirring, and limiting oxygen transport due to the strong non-Newtonian rheological properties of the fermentation medium [17]. Consequently, it is highly desirable to determine the optimal morphology of filamentous fungi during the submerged fermentation process, to enhance the yields of target products in the food and pharmaceutical industries.
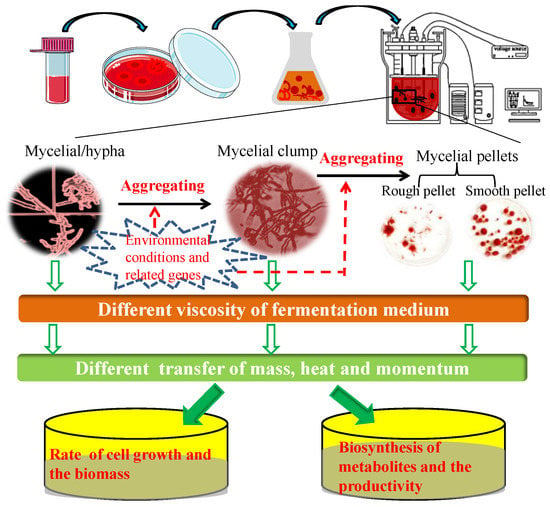
Figure 1.
Mechanisms underlying the correlations among environmental conditions or genes, fungal morphology, and the production of metabolites during submerged fermentation.
Often, it is not clear which morphological form is the most suitable for the efficient production of a given metabolite or enzyme. Dense mycelial pellets, within an appropriate range of diameters, are a favorable morphology for the biosynthesis of citric acid by Aspergillus niger [13,18,19], whereas, in Aspergillus oryzae, mycelium comprising numerous hyphal tips is more conducive to the production of enzymes, such as α-amylase and lipase [20,21,22]. Furthermore, the dispersed mycelium of Penicillium chrysogenum is generally regarded as a prerequisite to ensure the high productivity of penicillin in submerged fermentation [23]. Typically, however, the relationship between fungal morphology and production performance is complex, and different morphological forms have certain associated advantages. In 2001, Mclntyre et al. described the analytical tools employed in the study of hyphal morphology, the physiological aspects of morphological development, and molecular aspects of morphological control of Aspergillus [10]; they were the first to propose the term metabolic engineering of the morphology of filamentous fungi (MEMFF), a concept that covers the relationships between cell growth, phenotypes, and target products [10]. Since then, numerous regulatory genes, including the global regulator LaeA [24,25], transcription factors BrlA and WetA [26,27], CreA [28], and the Zn(II)2Cys6 family [29], have been identified and applied in MEMFF to enhance the production of target metabolites.
Chitin, a major structural component of the fungal cell wall, synthesized via the activity of the enzyme chitin synthase (chs), has become a high-profile target for investigating the effect on morphology, yield, and productivity in submerged fermentation. In this review, we summarize the classification of chs enzymes, the roles played by chs in the cell development and mycelial morphology of filamentous fungi, and the application of MEMFF. We provide a basis for understanding the inter-relationships between fungal differentiation, morphology, and productivity regulated by chs genes.
2. Chitin and Chitin Biosynthesis in Filamentous Fungi
2.1. Structure and Function of Chitin
After cellulose, chitin is the second most abundant natural polysaccharide, occurring widely in the exoskeletons of insects, crustaceans, and mollusks; it is also an important structural polysaccharide in fungal cell walls [30,31,32]. Notably, the chitin content in the fungal cell wall differs according to the morphological phase, accounting for only 1–2% of yeast cell wall dry weight [33,34], but reaching up to 10–20% of the cell wall dry weight of filamentous fungi (Aspergillus) [35]. Moreover, the content of chitin in the hyphal walls of Candida albicans is three times higher than that of other yeasts [36], whereas, in Paracoccidioides brasiliensis and Blastomyces dermatitidis, it is 25–30% higher than that in the yeast phase [37]. Chitin is a linear copolymer of N-acetyl-d-glucosamine (GlcNAc) and d-glucosamine units, linked by a β-(1–4) glycosidic bond, although predominantly comprising GlcNAc units [31]. Chitin chains of more than 100 and 190 GlcNAc monomers in length have been reported in cell walls and bud scars, respectively [33,38]. In addition, crystalline structural determinations have revealed that chitin can exist in three different forms, namely, α-, β-, and γ-chitin, representing antiparallel, parallel, and alternated arrangements of polymer chains, respectively [39,40]. In fungi, α-chitin is the major structural form [41], and γ-chitin is mainly found in the beetle family Lucanidae [42].
Chitosan is an important chitin derivative, generated by removing the acetyl group of chitin, either via treatment with concentrated alkali or the activity of chitin deacetylases. Chitin and its derivatives (chitosan and glucosamine series) have important applications in medicine and in the chemical industry, and as functional foods. Chitin and chitosan are considered advantageous biocompatible materials that can be used to augment or replace any tissue, organ, or function of the body [32,43]. Moreover, owing to their notable biological activities, including antibacterial, antifungal, antitumor, immunoregulatory, antioxidant, and anti-inflammatory properties, chitosan oligosaccharides have gained widespread application in the treatment and prevention of multiple life-threatening diseases and disorders, including cancer, heart disease, diabetes mellitus, and serious infections [44]. In addition, chitin and chitosan have a high absorptive capacity for wastewater pollutants, and thus have application potential in industrial wastewater treatment [45]. In filamentous fungi, chitin molecules form intrachain hydrogen bonds, facilitating assembly into fibrous microfibrils that form a basket-like scaffold surrounding cells. These fibrous microfibrils are characterized by considerable tensile strength, thereby maintaining cell wall integrity. As depicted in Figure 2, the cell wall comprises a twin-layer structure, the innermost layer of which is a relatively conserved structural skeletal layer (crosslinked chitin-glucan inner layer) comprising chitin and β-(1,3)-branched glucan, whereas the heterogeneous outer layer consists of other polysaccharides and glycoproteins [46,47]. The β-(1,3):β-(1,6)-branched glucan of the cell wall is bound to proteins or other polysaccharides, the composition of which may vary according to the fungal species, although it generally comprises highly mannosylated glycoproteins and mannoproteins. Chitin plays multiple roles in fungal species, including the maintenance of cell structural integrity, regulation of epithelial adhesion, the linkage between the cell wall and capsule, and antifungal resistance [46,47,48]. Accordingly, chitin is a key factor in maintaining normal cell growth and metabolism in filamentous fungi.
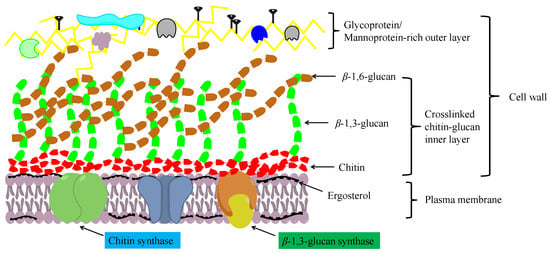
Figure 2.
A schematic diagram showing the structure of the fungal cell wall.
2.2. The Chitin Biosynthetic Pathway
In fungi, chitin is synthesized via a highly complex biosynthetic pathway involving a multifarious series of biochemical and physiological processes [49]. As substrates, glucose or one of its storage compounds (glycogen or trehalose) undergoes bioconversion to a polymer of the amino sugar GlcNAc via a series of enzymatic reactions divided into three sets of sub-reactions [49]. In the first set of sub-reactions (Figure 3), the biosynthesis of GlcNAc-1P proceeds via three steps: the substrate, fructose-6-phosphate (fructose-6P), generated from glycolysis (or the Embden–Meyerhof–Parnas (EMP) pathway), and trehalose are mobilized by hydrolysis to glucose catalyzed by trehalase [EC:3.2.1.28], and glycogen is converted to glucose-1-P by glycogen phosphorylase [EC:2.4.1.1]. During this stage, glucokinase [EC:2.7.1.2] or hexokinase [EC:2.7.1.1], and glutamine-fructose 6-phosphate transaminase (isomerizing) [EC:2.6.1.16] are the rate-limiting enzymes. In addition, glucose-6P can be used for the biosynthesis of β-(1,3) glucan via three reactions catalyzed by the enzymes phosphoglucomutase [EC:5.4.2.2], UTP-glucose-1-phosphate uridylyltransferase [EC:2.7.7.9], and 1,3-β-glucan synthase [EC:2.4.1.34]. In the second set of sub-reactions, GlcNAc-1P is catalyzed to generate the activated molecule amino sugar UDP-N-acetylglucosamine (UDP-GlcNAc) via the action of UDP-N-acetylglucosamine pyrophosphorylase [EC:2.7.7.23], an essential enzyme for chitin synthesis. In the final set of sub-reactions, the enzyme chs [EC:2.4.1.16] catalyzes a polymerization reaction to synthesize chitin using the activated UDP-GlcNAc as a sugar donor. The first two sets of sub-reactions occur within the cell cytoplasm, while the third reaction occurs in the chitosome, located within the plasma membrane of cells in the hyphal tips and cell cross-walls of filamentous fungi [50]. In the chitin biosynthetic pathway, glutamine-fructose 6-phosphate transaminase [EC:2.6.1.16], UDP-N-acetylglucosamine pyrophosphorylase [EC:2.7.7.23], and chs [EC:2.4.1.16] serve as the rate-limiting enzymes that dictate the rate at which chitin is synthesized, and are highly regulated in cells. Among these enzymes, chs catalyzes the final reaction, which is specifically and directly associated with the biosynthesis of chitin, and accordingly, is acknowledged to be the key enzyme in chitin biosynthesis. As described in the Introduction section, chs plays a vital role in cell development and the mycelial morphology of filamentous fungi, thereby having a prominent role in the application of MEMFF. Hence, we specifically focused on the chitin synthase in different fungal species and the involvement of chs.
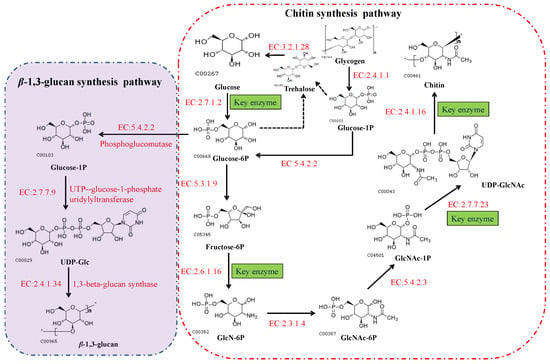
Figure 3.
The biosynthetic pathways of chitin and β-(1,3) glucan in fungi. These pathways can also be viewed at the Kyoto Encyclopedia of Genes and Genomes website (https://www.kegg.jp/pathway/map00520, accessed on 11 September 2021).
2.3. Classification of Chitin Synthase
Based on amino acid sequence homology, the chs enzyme family can be grouped into seven classes (I to VII), with different fungal species expressing varying numbers of chs genes [51]. In 2019, researchers summarized three chs genes (classes I–III) in Saccharomyces cerevisiae, four in Candida, and six to ten in filamentous fungi [52]. Among the seven classes, CHS III, V, VI, and VII are found exclusively in filamentous fungi [53]. In the same year, in their review of fungal chitin synthesis and degradation, Yang and Zhang described the members of the seven classes of chitin synthase in different fungi [47]. However, since then, details of chs genes and their classification have not been furthered. In this review, we further summarize and update the chs gene classes and number of chs genes in eukaryon, based on data obtained from the Kyoto Encyclopedia of Genes and Genomes (https://www.kegg.jp/kegg/genes.html) and National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) databases, with an emphasis on those expressed in filamentous fungi. As shown in Table 1, we found differences in the number of chs genes in Saccharomycotina, although these are generally grouped into three classes (CHS I–III). For instance, there are six (as opposed to the originally reported three), five, and four chs genes in S. cerevisiae S288c, Candida orthopsilosis Co 90-125, and Candida tropicalis MYA-3404, respectively, whereas eight chs genes have been identified in Sugiyamaella lignohabitans CBS 10342. Except for those in goldfish (seven chs genes), there are generally few chs genes in animals, including Eutheria, Amphibia, and Euteleostomi. Notably, the number and classes of chs genes in fungal genera are distinctly higher than those in the species of Saccharomycotina and animals. For example, among species of Pezizomycotina, such as Aspergillus fumigatus, Neurospora crassa, Cordyceps militaris, and Purpureocillium lilacinum, chs genes are generally grouped into seven classes, with seven to nine genes in each. Moreover, certain hypothetical proteins are identified as chs enzymes in Fusarium graminearum and Pestalotiopsis fici, thereby indicating the potential occurrence of up to 10 types of chs. Strains of filamentous fungi in the genus Monascus, an important industrialized fermentative microorganism, are noted for their production of MSMs, including Monascus pigments and monacolin K. In our laboratory, we have sequenced the whole genomes of M. purpureus LQ-6 (accession number of PRJNA503091) and its mutant strain M183 (accession number of JAACNI000000000) based on the combined application of single-molecule real-time DNA sequencing and next-generation sequencing. Accordingly, we identified eight genes encoding chs enzymes, the classification of which appears to be complex. In addition, nine genes encoding chs enzymes (including three hypothetical proteins) have been identified in the genome of M. purpureus HQ1 (accession number of VIFY00000000), mainly classified as CHS I, II, III, V, and VII. The larger number of chs genes in filamentous fungi compared to Saccharomycotina reflects the greater complexity of hyphal development and polarized growth, as well as a higher cell wall chitin content.

Table 1.
The members of the chitin synthase family in a section of diverse fungi. (Completely statistical data are shown in Table S1). chs, represents the gene of chitin synthase; CHS, represents the class of the members of chs family. The genes encoding hypothetical proteins, but mostly like chs, are marked in red.
With the increasing accumulation of genomic sequence data for fungi in recent years, the number of identified chs genes in different species has reached approximately 200. However, most of these genes have yet to be fully characterized. Generally, chs enzymes are grouped into two divisions, division I (containing CHS I–III) and division II (containing CHS IV–VII) [54]. Among the members of the chs enzyme family (CHS I–VII), 6–10 chs genes identified in different fungi species encode proteins with discernable structural differences. As shown in Figure 4, there are obvious differences in the tertiary structures that distinguish the different classes of chs proteins. All chs members have multiple transmembrane domains (TMD); however, CHS IV–VII enzymes typically contain a cytochrome b5-like heme/steroid-binding domain (Cyt-b5), which is not found in classes I to III. Furthermore, CHS V and CHS VI proteins both have an N-terminal myosin motor domain (MMD) and a C-terminal chitin synthase domain (CSD) [55]. Although the structures of CHS V and CHS VI proteins are highly similar and difficult to differentiate, the MMD of CHS V proteins contains conserved ATP-binding motifs (ABM, including p-Loop, Switch I, and Switch II) absent in class VI chitin synthases [56]. In addition, CHS I–III proteins are characterized by hydrophobic C-terminal and hydrophilic N-terminal regions containing a catalytic domain. In our laboratory, the chs protein-encoding gene Monascus_05162, detected in the M. purpureus LQ-6 genome, was identified as a CHS VI class enzyme based on the tertiary structure of the protein and conserved domain analysis [57].
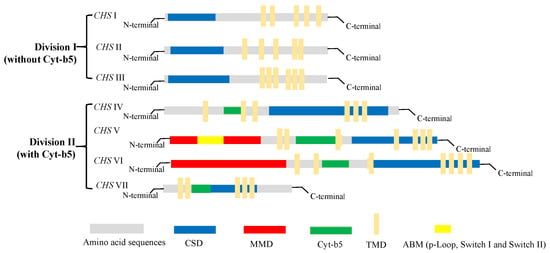
Figure 4.
The structure and classification of members of the chitin synthase family.
3. The Regulatory Effect of Chitin Biosynthesis on the Cell Growth and Morphology of Filamentous Fungi
In recent years, numerous studies have focused on the regulatory effects of chitin biosynthesis on the cell growth and morphology of filamentous fungi. Given its key role in chitin biosynthesis, chs has been preferentially selected to investigate its contribution to chitin biosynthesis and cell growth. A decade ago, Kong et al. reported that individual chs genes play diverse roles in the hyphal growth and conidiogenesis of the fungus Magnaporthe oryzae [58]. Members of the chs family have different functional effects with respect to the cell growth, stress tolerances, and cell wall integrity of Metarhizium acridum; of these, CHS III, V, and VII regulate the surface properties of conidia and hyphal bodies [52]. Kim et al. observed that CHS V and CHS VII gene knockout in Gibberella zeae was associated with the development of balloon-shaped hyphae and weak cell wall rigidity, with the mutants being unable to produce perithecia, thereby inducing disease symptoms in barley heads [59]. Larson et al. found that single or double deletion of CHS V and CHS VII genes in Fusarium verticillioides contributed to the poor growth of mutant strains, and reductions in the diameter and aerial mycelia of colonies cultivated on potato dextrose agar medium [54]. Moreover, when cultured in potato dextrose broth, ballooning of cell walls was observed in the hyphae of all three mutants. However, although the CHS V gene in S. cerevisiae is not essential for cell growth, it does play an important role in the mating of this yeast [60]. In addition, Amnuaykanjanasin and Epstein indicated that chs A (CHS V) is essential for conidial wall strength and contributes to the strength of hyphal tips [61]. Collectively, these findings indicate that CHS V and CHS VII genes are mainly required for normal hyphal growth, perithecia formation, and pathogenicity of filamentous fungi. Moreover, in our previous study, we observed that the deletion of the CHS VI gene in the filamentous fungus M. purpureus induced the development of balloon-tip-like structures in the hyphae, reduced hyphal branching efficiency, promoted hyphal elongation, and altered mycelial pellet formation during submerged fermentation [57]. Although CHS VI knockout reduced the maximum biomass obtained in submerged fermentation cultures by 19.63%, it did not alter the rate at which the colony diameter of M. purpureus grew [57]. In contrast, Cui et al. found that the radial growth of Botrytis cinerea colonies substantially reduced as a consequence of disrupting the CHS VI gene, and speculated that this gene is necessary for appropriate hyphal growth [55]. Similarly, Fajardo-Somera et al. reported that CHS VI primarily plays a role in hyphal extension and ascospore formation, based on a comparison of the functional importance of different CHS genes with respect to the cell growth and development of N. crassa [62]. Besides, it has been reported that inactivation of chs 1 (CHS I) in N. crassa resulted in extensive hyphal swelling and other hyphal abnormalities in liquid medium [63], but no significant changes after disruption of chs 2 (CHS II, a non-essential chitin synthase) [64]. In addition, the chs Z (class VI) gene of A. oryzae is specifically involved in hyphal extension and cell wall formation (filamentous cell morphogenesis) [65]. The findings of these studies indicate that CHS VI genes primarily play roles in hyphal elongation and determining mycelial morphology, although their contribution in determining the cell growth of filamentous fungi shows interspecific variation.
In this review, we highlight the fact that different chs genes have differential regulatory effects on the hyphal growth and morphology of filamentous fungi. For example, chs A (CHS I) and chs C (CHS III) genes are inessential for the normal cell growth of A. fumigatus and are not implicated in the regulation of colony diameter or growth rate [66]. However, deletion of chs B (class III in Aspergillus nidulans, class II in A. fumigatus) causes severe growth defects, thereby indicating that this gene plays an essential role in determining hyphal tip growth [54,67]. In a recent study on Verticillium dahlia, Qin et al. examined the effects of knocking out eight chs genes (chs 1–8) on cell growth and virulence, and accordingly established the respective differential requirements for these eight genes [68]. Similarly, Hiroyuki Horiuchi cloned six chs genes from A. nidulans and investigated their effects on cell growth; they found that although the genes play essential roles in growth and morphogenesis, they differ in terms of specific functions [67].
Numerous researchers have focused on the effects of chs genes on the cell development and mycelial morphology of industrial filamentous fungi during submerged fermentation. For example, in their investigation of the regulatory effects of chitin synthase on mycelial morphology during submerged fermentation, Sun et al. used an RNAi construct to silence the chs C gene in A. niger and found that this strategy resulted in the shortening of hyphal length, reduction in the proportion of dispersed mycelia, and increase in the compactness of mycelial pellets [69]. In subsequent studies, the same group established that the function of the chs C gene is tightly interrelated with the functioning of the chitin synthase activator (chs3) gene of A. niger, and that the desired mycelial morphology for enhancing citric acid production in submerged fermentation cultures can be obtained by knockdown of the chs3 gene [70]. In addition, disruption of the chs B gene in A. oryzae resulted in a significant reduction in the formation of mycelial clumps and corresponding increases in the number of freely dispersed hyphae and frequency of branching [71]. Conversely, deletion of the csm A (class V) gene resulted in a reduction in branch number in the apical compartment, along with an increase in the average diameter of hyphae [71]. Moreover, Liu et al. established that the chs 4 (class III) gene is essential for the hyphal growth and conidial development of P. chrysogenum, and observed a marked reduction in the diameters of mutant colonies following the knockdown of this gene using RNA silencing [72]. Furthermore, these researchers also established that by disrupting chs 4 gene expression, they could regulate the agglomeration of hyphal elements and diameters of P. chrysogenum mycelial pellets during fermentation [23]. More recently, our research group has been focusing on the morphological changes in M. purpureus during submerged fermentation, and we found that disruption of the biosynthetic pathways of both ergosterol (by deleting the egr 4A and erg 4B genes) and chitin (by knocking out a CHS VI gene) promoted changes in the morphology of the mycelial pellets [8,57].
To summarize, in the relevant studies conducted on chitin synthase to date, researchers have primarily investigated the relationships between cell wall chitin content and pathogenesis or tolerance, based on the deletion of chs genes [54,58,66,73], and evaluated the regulatory effect of different members of the chs family on mycelial morphology in submerged fermentation, with the aim of enhancing the yields of target products [4,57,69,71,72]. Based on the findings of these studies, chitin biosynthesis is closely associated with the cell growth and morphology of filamentous fungi, and different members of the chs family play distinct functional roles in this regard.
In addition to the direct effects of chs genes, the regulation of chitin biosynthesis is also determined to differing extents by other factors, including transcription factors, mitogen-activated protein kinase (MAPK), and calcium (Ca2+). In 2020, Norio Takeshita reported that the oscillation of Ca2+ levels can contribute to the control of chitin biosynthesis associated with the stepwise extension of hyphal tips [74], which is consistent with the findings of earlier studies indicating that intracellular Ca2+ levels are closely associated with the polarized growth of filamentous fungi [75,76]. In addition, the transcription factors CrzA and RlmA, and MPKA, regulate the expression of chs genes, thereby altering the growth phenotypes of A. fumigatus [77]. Besides, He et al. deleted eki gene encoding ethanolamine kinase in Trichoderma reesei, causing the overexpression of chitin synthase genes and increased chitin content; the results also indicate that ethanolamine kinase has a significant effect on cell growth and development in filamentous fungi T. reesei [78].
Many signaling pathways, including the high osmolarity glycerol, protein kinase C (PKC)-MAPK, and Ca2+/calcineurin pathways, are implicated in the regulation of chitin synthase [79], and chitin biosynthesis is mediated by the regulatory factors AbaA, BrlA, and MedA, which control the levels of chsC and chsA transcription during conidiophore development [80,81]. Moreover, deletion of the kexB gene in A. niger resulted in hyper-branching and thickening of cell walls, and upregulated chitin metabolism during submerged fermentation [82]. Based on the evidence accumulated to date, chitin biosynthesis and cell growth are intricate processes regulated via multiple pathways and diverse factors during cell development. Undoubtedly, however, much remains to be discovered; thus, the respective relationships between chitin biosynthesis (chs genes) and mycelial morphologies of diverse filamentous fungi, as well as the regulation of chitin biosynthesis via other pathways, should be further studied.
4. The Application of Morphological Engineering of Industrial Filamentous Fungi
MEMFF can facilitate substantial improvements in the yield and productivity of target metabolites, thereby contributing to the development of industrial-scale production. As shown in Table 2, a diverse range of technologies have been applied in developing MEMFF for enhancing the target production or yield. In this regard, a series of papers have indicated that extractive fermentation via the addition of surfactants can alter the mycelial morphology of Monascus species, thereby significantly enhancing the production of Monascus pigments [83,84,85,86]. Furthermore, based on the integrated control of operational parameters (agitation speed and aeration rate) and overexpression of tyrosine-protein phosphatase, Chen et al. fine-tuned the morphology of A. oryzae to obtain a more compact pellet structure, and larger pellet number, than the original mycelial morphology in submerged fermentation, thereby increasing the production of l-malate to 142.5 g/L in a 30-L bioreactor [87]. Moreover, the pkaC gene has multiple regulatory effects associated with hyphal growth, and the overexpression of this gene modified the mycelial pellet morphology of A. niger during submerged fermentation, thus contributing to a 1.87-fold increase in the concentration of citric acid [19]. More recently, microparticle-enhanced cultivation techniques have been successfully applied in the submerged fermentation cultivation of different fungal genera. For example, the addition of silicate microparticles controlled the morphological development of A. niger. The formation of freely dispersed mycelia promoted 4-fold increases in the concentrations of glucoamylase and fructofuranosidase in shaking flask cultures and production of up to 160 U/mL fructofuranosidase in a 3.0-L stirred tank bioreactor [88]. Similarly, precise control of the morphology of M. purpureus has been achieved by the addition of 4 g/L 5000-mesh talc to a submerged fermentation broth at 24 h. In this fermentation system, yields of up to 554.2 U/mL Monascus yellow pigments were obtained, representing an approximate 113.15% increase compared with the control group [89].

Table 2.
A selection of the technologies applied in metabolic engineering of the morphology of filamentous fungi for enhancing the production of target metabolites.
The following is a summary of the application of MEMFF to enhance the production of fungal target metabolites based on the modification of chitin biosynthesis. Liu et al. reported that in P. chrysogenum, pellets and highly branched hyphae may be the most suitable mycelial morphologies for penicillin production, based on the modified expression of the chs 4 (class III) gene, following which, yields of penicillin increased by 41% [23,72]. More than 20 years ago, the group headed by Mhairi Mclntyre published a comprehensive review of the MEMFF of Aspergillus, in which they described in detail the phenotypic effect of chs genes [10]. Subsequently, this research team developed an MEMFF system for A. oryzae based on disruption of the chsB and csmA (class V) genes, which despite contributing to an increase in hyphal branching, did not enhance α-amylase production [72]. However, deletion of the chsC gene in A. niger significantly modified the mycelial morphology in submerged fermentation, contributing to a 42.6% increase in citric acid production compared with that produced by the wild-type strain [69]. Furthermore, the mycelial morphology of A. niger in submerged cultures can be optimized by silencing the chitin synthase activator (chs3) gene, resulting in a 39.25% increase in the production of citric acid [70]. Notably, our research group has found that the total production of microbial secondary metabolites (Monascus pigments and citrinin) by M. purpureus was significantly reduced in response to the disruption of the CHS VI gene; however, downregulation of the expression levels of different chs genes to appropriate levels can substantially enhance metabolite production [57]. Collectively, these findings indicate that regulation of the mycelial morphologies of filamentous fungi is a complex, multifarious phenomenon, involving multiple genes and pathways. To date, the application of MEMFF based on the disruption of chs genes has been applied primarily with the aim of enhancing the citric acid and penicillin production of A. niger and P. chrysogenum, respectively. However, as proposed by Liu et al., it is reasonable to expect that the manipulation of chs enzymes in other fungal species would be a simple and low-cost strategy for enhancing the production of target metabolites synthesized by industrial filamentous fungi under submerged fermentation conditions [23].
5. Conclusions
Filamentous fungi with certain unique properties have been widely applied in the food and pharmaceutical industries to produce target metabolites based on submerged fermentation cultivation. The yield and productivity of target products synthesized by filamentous fungi under submerged fermentation conditions are closely associated with fungal mycelial morphology, which has a significant influence on the transfer of mass, heat, and momentum. The mycelial morphologies of filamentous fungi are typically diverse and influenced by multiple factors, including environmental conditions, operating parameters, and autologous genes, thereby contributing to the complex relationship between morphology and production. Chitin, a major structural component of the cell wall of filamentous fungi, synthesized by the activity of chitin synthase, has become a high-profile target for investigating the factors affecting fungal morphology, yield, and productivity in submerged fermentation. In this review, we comprehensively summarize the classification and structures of members of the chs family, and describe the biosynthetic pathways of chitin and the associations between chitin biosynthesis, cell growth, and the metabolism of filamentous fungi. We also summarize studies that have focused on the manipulation of fungal chs genes, describing how the disruption of class III and V chs genes can significantly optimize mycelial morphologies during submerged fermentation of species of Aspergillus, thereby enhancing the production of target metabolites. Regulation of the expression levels of other genes and pathways (including the regulatory factors AbaA, BrlA, and MedA, and the PKC-MAPK and Ca2+/calcineurin pathways) can influence the biosynthesis of chitin and cell development, a knowledge of which could be usefully applied in MEMFF to enhance the yields of target products. However, although numerous strategies have been devised to facilitate the control or modification of the mycelial morphologies of filamentous fungi during submerged fermentation, at present, the application of MEMFF based on controlling the expression levels of certain chs genes is not sufficiently comprehensive. Accordingly, further studies are warranted to gain a broader understanding of the regulatory effects of other chs genes on different fungal species and the underlying molecular mechanisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9020205/s1, Table S1: The statistical data of the members of chitin synthase family in different species.
Author Contributions
Conceptualization, J.L.; writing—original draft preparation, Z.G.; writing—review and editing, J.L. and S.Z.; project administration, J.L.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 32101906), The Science and Technology Innovation Program of Hunan Province (2022RC1018), Natural Science Foundation of Hunan Province (No. 2021JJ31146), Changsha key S&T Special Projects (No. kh2003013), Education Department of Scientific Research Project of Hunan Province (No. 20B619).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Ruiz:, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.S.; Giesecke, U.; Reichert, A.; Minas, W. Industrial enzymatic production of cephalosporin-based beta-lactams. Adv. Biochem. Eng. Biotechnol. 2004, 88, 179–215. [Google Scholar] [PubMed]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Grimm, L.H.; Kelly, S.; Krull, R.; Hempel, D.C. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 2005, 69, 375–384. [Google Scholar] [CrossRef]
- Quintanilla, D.; Hagemann, T.; Hansen, K.; Gernaey, K.V. Fungal Morphology in Industrial Enzyme Production--Modelling and Monitoring. Adv Biochem Eng Biotechnol. 2015, 149, 29–54. [Google Scholar]
- Peberdy, J.F. Protein secretion in filamentous fungi—Trying to understand a highly productive black box. Trends Biotechnol. 1994, 12, 50–57. [Google Scholar] [CrossRef]
- Punt, P.J.; van Biezen, N.; Conesa, A.; Albers, A.; Mangnus, J.; van den Hondel, C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002, 20, 200–206. [Google Scholar] [CrossRef]
- Liu, J.; Chai, X.; Guo, T.; Wu, J.; Yang, P.; Luo, Y.; Zhao, H.; Zhao, W.; Nkechi, O.; Dong, J.; et al. Disruption of the Ergosterol Biosynthetic Pathway Results in Increased Membrane Permeability, Causing Overproduction and Secretion of Extracellular Monascus Pigments in Submerged Fermentation. J. Agric. Food Chem. 2019, 67, 13673–13683. [Google Scholar] [CrossRef]
- Liu, J.; Guo, T.; Luo, Y.; Chai, X.; Wu, J.; Zhao, W.; Jiao, P.; Luo, F.; Lin, Q. Enhancement of Monascus pigment productivity via a simultaneous fermentation process and separation system using immobilized-cell fermentation. Bioresour. Technol. 2019, 272, 552–560. [Google Scholar] [CrossRef]
- McIntyre, M.; Müller, C.; Dynesen, J.; Nielsen, J. Metabolic engineering of the morphology of Aspergillus. Adv. Biochem. Eng. Biotechnol. 2001, 73, 103–128. [Google Scholar]
- Wucherpfennig, T.; Kiep, K.A.; Driouch, H.; Wittmann, C.; Krull, R. Morphology and Rheology in Filamentous Cultivations. Adv. Appl. Microbiol. 2010, 72, 89–136. [Google Scholar]
- Kossen, N.W. The morphology of filamentous fungi. Adv. Biochem. Eng. Biotechnol. 2000, 70, 1–33. [Google Scholar] [PubMed]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Mao, J.; Zheng, Y. Effect of microparticle on fermentation process of filamentous microorganisms—A review. Wei Sheng Wu Xue Bao 2015, 55, 258–263. [Google Scholar] [PubMed]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. A quantitative image analysis pipeline for the characterization of filamentous fungal morphologies as a tool to uncover targets for morphology engineering: A case study using aplD in Aspergillus niger. Biotechnol. Biofuels 2019, 12, 149. [Google Scholar] [CrossRef]
- Veiter, L.; Rajamanickam, V.; Herwig, C. The filamentous fungal pellet—Relationship between morphology and productivity. Appl. Microbiol. Biotechnol. 2018, 102, 2997–3006. [Google Scholar] [CrossRef]
- Antecka, A.; Bizukojc, M.; Ledakowicz, S. Modern morphological engineering techniques for improving productivity of filamentous fungi in submerged cultures. World J. Microbiol. Biotechnol. 2016, 32, 193. [Google Scholar] [CrossRef]
- Posch, A.E.; Spadiut, O.; Herwig, C. A novel method for fast and statistically verified morphological characterization of filamentous fungi. Fungal Genet. Biol. 2012, 49, 499–510. [Google Scholar] [CrossRef]
- Zheng, X.; Cairns, T.C.; Ni, X.; Zhang, L.; Zhai, H.; Meyer, V.; Zheng, P.; Sun, J. Comprehensively dissecting the hub regulation of PkaC on high-productivity and pellet macromorphology in citric acid producing Aspergillus niger. Microb. Biotechnol. 2022, 15, 1867–1882. [Google Scholar] [CrossRef]
- Haack, M.B.; Olsson, L.; Hansen, K.; Eliasson Lantz, A. Change in hyphal morphology of Aspergillus oryzae during fed-batch cultivation. Appl. Microbiol. Biotechnol. 2006, 70, 482–487. [Google Scholar] [CrossRef]
- Maumela, P.; Rose, S.; van Rensburg, E.; Chimphango, A.F.A.; Görgens, J.F. Bioprocess Optimisation for High Cell Density Endoinulinase Production from Recombinant Aspergillus niger. Appl. Biochem. Biotechnol. 2021, 193, 3271–3286. [Google Scholar] [CrossRef] [PubMed]
- Spohr, A.; Carlsen, M.; Nielsen, J.; Villadsen, J. α-Amylase production in recombinant Aspergillus oryzae during fed-batch and continuous cultivations. J. Ferment. Bioeng. 1998, 86, 49–56. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, Z.; Wang, P.; Gong, G.; Wang, L.; Zhao, G. Morphological changes induced by class III chitin synthase gene silencing could enhance penicillin production of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2013, 97, 3363–3372. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Zhu, Q.; Hao, S.; Chai, S.; Li, Y.; Jiao, Z.; Shi, J.; Sun, B.; Wang, C. Overexpression of global regulator LaeA increases secondary metabolite production in Monascus purpureus. Appl. Microbiol. Biotechnol. 2020, 104, 3049–3060. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, L.; Shao, Y.; Zhou, Y.; Li, M.; Wang, X.; Chen, F. Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 2016, 120, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Yu, J.H.; Chen, F.; Chen, W. Characterization of the asexual developmental genes brlA and wetA in Monascus ruber M7. Fungal Genet. Biol. 2021, 151, 103564. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Bao, L.; Gao, M.; Chen, M.; Lei, Y.; Liu, G.; Qu, Y. Penicillium decumbens BrlA extensively regulates secondary metabolism and functionally associates with the expression of cellulase genes. Appl. Microbiol. Biotechnol. 2013, 97, 10453–10467. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.; Bao, L.; Gao, L.; Yao, G.; Li, Y.; Yang, Z.; Li, Z.; Zhong, Y.; Li, F.; et al. Putative methyltransferase LaeA and transcription factor CreA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression. Fungal Genet. Biol. 2016, 94, 32–46. [Google Scholar] [CrossRef]
- El Hajj Assaf, C.; Zetina-Serrano, C.; Tahtah, N.; Khoury, A.E.; Atoui, A.; Oswald, I.P.; Puel, O.; Lorber, S. Regulation of Secondary Metabolism in the Penicillium Genus. Int. J. Mol. Sci. 2020, 21, 9462. [Google Scholar] [CrossRef]
- Al Shaqsi, N.H.K.; Al Hoqani, H.A.S.; Hossain, M.A.; Al Sibani, M.A. Optimization of the demineralization process for the extraction of chitin from Omani Portunidae segnis. Biochem. Biophys. Rep. 2020, 23, 100779. [Google Scholar]
- Poerio, A.; Petit, C.; Jehl, J.P.; Arab-Tehrany, E.; Mano, J.F.; Cleymand, F. Extraction and Physicochemical Characterization of Chitin from Cicada orni Sloughs of the South-Eastern French Mediterranean Basin. Molecules 2020, 25, 2543. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Klis, F.M.; Mol, P.; Hellingwerf, K.; Brul, S. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2002, 26, 239–256. [Google Scholar] [CrossRef]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Chattaway, F.W.; Holmes, M.R.; Barlow, A.J. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J. Gen. Microbiol. 1968, 51, 367–376. [Google Scholar] [CrossRef]
- Kanetsuna, F.; Carbonell, L.M.; Moreno, R.E.; Rodriguez, J. Cell wall composition of the yeast and mycelial forms of Paracoccidioides brasiliensis. J. Bacteriol. 1969, 97, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.S.; Elango, N.; Mattia, E.; Au-Young, J.; Robbins, P.W.; Cabib, E. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J. Biol. Chem. 1984, 259, 14966–14972. [Google Scholar] [CrossRef]
- El Knidri, H.; Belaabed, R.; Addaou, A.; Laajeb, A.; Lahsini, A. Extraction, chemical modification and characterization of chitin and chitosan. Int. J. Biol. Macromol. 2018, 120, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Moussian, B. Chitin: Structure, Chemistry and Biology. Adv. Exp. Med. Biol. 2019, 1142, 5–18. [Google Scholar] [PubMed]
- Hassainia, A.; Satha, H.; Boufi, S. Chitin from Agaricus bisporus: Extraction and characterization. Int. J. Biol. Macromol. 2018, 117, 1334–1342. [Google Scholar] [CrossRef]
- Jang, M.K.; Kong, B.G.; Jeong, Y.I.; Lee, C.H.; Nah, J.W. Physicochemical characterization of α-chitin,β-chitin, and γ-chitin separated from natural resources. J. Polym. Sci. Part A-Polym. Chem. 2004, 42, 3423–3432. [Google Scholar] [CrossRef]
- Islam, S.; Bhuiyan, M.A.R.; Islam, M.N. Chitin and Chitosan: Structure, Properties and Applications in Biomedical Engineering. J. Polym. Environ. 2017, 3, 854–866. [Google Scholar] [CrossRef]
- Benchamas, G.; Huang, G.; Huang, S.; Huang, H. Preparation and biological activities of chitosan oligosaccharides. Trends Food Sci. Technol. 2021, 107, 38–44. [Google Scholar] [CrossRef]
- No, H.K.; Meyers, S.P. Application of chitosan for treatment of wastewaters. Rev. Environ. Contam. Toxicol. 2000, 163, 1–27. [Google Scholar] [PubMed]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A.; Heitman, J. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, K.Q. Chitin Synthesis and Degradation in Fungi: Biology and Enzymes. Adv. Exp. Med. Biol. 2019, 1142, 153–167. [Google Scholar]
- Goldman, D.L.; Vicencio, A.G. The chitin connection. mBio 2012, 3, e00056-12. [Google Scholar] [CrossRef]
- Merzendorfer, H. The cellular basis of chitin synthesis in fungi and insects: Common principles and differences. Eur. J. Cell Biol. 2011, 90, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Azuma, M.; Igarashi, K.; Ooshima, H. Analysis of Chitin at the Hyphal Tip of Candida albicans Using Calcofluor White. Biosci. Biotechnol. Biochem. 2005, 69, 1798–1801. [Google Scholar] [CrossRef]
- Roncero, C. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 2002, 41, 367–378. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, H.; Du, Y.; Keyhani, N.O.; Xia, Y.; Jin, K. Members of chitin synthase family in Metarhizium acridum differentially affect fungal growth, stress tolerances, cell wall integrity and virulence. PLoS Pathog. 2019, 15, e1007964. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Gow, N.A.R. Chitin synthesis in human pathogenic fungi. Med. Mycol. 2001, 39, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Larson, T.M.; Kendra, D.F.; Busman, M.; Brown, D.W. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr. Genet. 2011, 57, 177–189. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, Y.; Lei, N.; Wang, K.; Zhu, T. Botrytis cinerea chitin synthase BcChsVI is required for normal growth and pathogenicity. Curr. Genet. 2013, 59, 119–128. [Google Scholar] [CrossRef]
- Takeshita, N.; Yamashita, S.; Ohta, A.; Horiuchi, H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 2006, 59, 1380–1394. [Google Scholar] [CrossRef]
- Shu, M.; Lu, P.; Liu, S.; Zhang, S.; Gong, Z.; Cai, X.; Zhou, B.; Lin, Q.; Liu, J. Disruption of the Chitin Biosynthetic Pathway Results in Significant Changes in the Cell Growth Phenotypes and Biosynthesis of Secondary Metabolites of Monascus purpureus. J. Fungi 2022, 8, 910. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.A.; Yang, J.; Li, G.T.; Qi, L.L.; Zhang, Y.J.; Wang, C.F.; Zhao, W.S.; Xu, J.R.; Peng, Y.L. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002526. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, H.J.; Lee, J.; Kim, K.W.; Yun, S.H.; Shim, W.B.; Lee, Y.W. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 2009, 55, 449–459. [Google Scholar] [CrossRef]
- Santos, B.; Duran, A.; Valdivieso, M.H. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell Biol. 1997, 17, 2485–2496. [Google Scholar] [CrossRef]
- Amnuaykanjanasin, A.; Epstein, L. A class V chitin synthase gene, chsA is essential for conidial and hyphal wall strength in the fungus Colletotrichum graminicola (Glomerella graminicola). Fungal Genet. Biol. 2003, 38, 272–285. [Google Scholar] [CrossRef]
- Fajardo-Somera, R.A.; Jöhnk, B.; Bayram, Ö.; Valerius, O.; Braus, G.H.; Riquelme, M. Dissecting the function of the different chitin synthases in vegetative growth and sexual development in Neurospora crassa. Fungal Genet. Biol. 2015, 75, 30–45. [Google Scholar] [CrossRef]
- Yarden, O.; Yanofsky, C. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 1991, 5, 2420–2430. [Google Scholar] [CrossRef]
- Din, A.B.; Yarden, O. The Neurospora crassa chs-2 gene encodes a non-essential chitin synthase. Microbiology 1994, 140, 2189–2197. [Google Scholar] [CrossRef]
- Chigira, Y.; Abe, K.; Gomi, K.; Nakajima, T. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 2002, 41, 261–267. [Google Scholar] [CrossRef]
- Rogg, L.E.; Fortwendel, J.R.; Juvvadi, P.R.; Lilley, A.; Steinbach, W.J. The chitin synthase genes chsA and chsC are not required for cell wall stress responses in the human pathogen Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2011, 411, 549–554. [Google Scholar] [CrossRef]
- Horiuchi, H. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med. Mycol. 2009, 47 (Suppl. 1), S47–S52. [Google Scholar] [CrossRef]
- Qin, J.; Zhao, P.; Ye, Z.; Sun, L.; Hu, X.; Zhang, J. Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae. J. Fungi 2022, 8, 681. [Google Scholar] [CrossRef]
- Sun, X.; Wu, H.; Zhao, G.; Li, Z.; Wu, X.; Liu, H.; Zheng, Z. Morphological regulation of Aspergillus niger to improve citric acid production by chsC gene silencing. Bioprocess Biosyst. Eng. 2018, 41, 1029–1038. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, H.; Liu, M.; Wang, L.; Yang, R.; Wang, P.; Lu, Z.; Zhou, Y.; Zheng, Z.; Zhao, G. Identification of chitin synthase activator in Aspergillus niger and its application in citric acid fermentation. Appl. Microbiol. Biotechnol. 2022, 106, 6993–7011. [Google Scholar] [CrossRef]
- Müller, C.; McIntyre, M.; Hansen, K.; Nielsen, J. Metabolic engineering of the morphology of Aspergillus oryzae by altering chitin synthesis. Appl. Env. Microbiol. 2002, 68, 1827–1836. [Google Scholar] [CrossRef]
- Liu, H.; Wang, P.; Gong, G.; Wang, L.; Zhao, G.; Zheng, Z. Morphology engineering of Penicillium chrysogenum by RNA silencing of chitin synthase gene. Biotechnol. Lett. 2013, 35, 423–429. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A.R. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Takeshita, N. Control of Actin and Calcium for Chitin Synthase Delivery to the Hyphal Tip of Aspergillus. Curr. Top Microbiol. Immunol. 2020, 425, 113–129. [Google Scholar]
- Brand, A.; Shanks, S.; Duncan, V.M.; Yang, M.; Mackenzie, K.; Gow, N.A. Hyphal orientation of Candida albicans is regulated by a calcium-dependent mechanism. Curr. Biol. 2007, 17, 347–352. [Google Scholar] [CrossRef]
- Kim, H.S.; Czymmek, K.J.; Patel, A.; Modla, S.; Nohe, A.; Duncan, R.; Gilroy, S.; Kang, S. Expression of the Cameleon calcium biosensor in fungi reveals distinct Ca(2+) signatures associated with polarized growth, development, and pathogenesis. Fungal Genet. Biol. 2012, 49, 589–601. [Google Scholar] [CrossRef]
- Ries, L.N.A.; Rocha, M.C.; de Castro, P.A.; Silva-Rocha, R.; Silva, R.N.; Freitas, F.Z.; de Assis, L.J.; Bertolini, M.C.; Malavazi, I.; Goldman, G.H. The Aspergillus fumigatus CrzA Transcription Factor Activates Chitin Synthase Gene Expression during the Caspofungin Paradoxical Effect. mBio 2017, 8, e00705-17. [Google Scholar] [CrossRef]
- He, R.L.; Guo, W.; Zhang, D.Y. An ethanolamine kinase Eki1 affects radial growth and cell wall integrity in Trichoderma reesei. Fems. Microbiol. Lett. 2015, 362, fnv133. [Google Scholar] [CrossRef]
- Rogg, L.E.; Fortwendel, J.R.; Juvvadi, P.R.; Steinbach, W.J. Regulation of expression, activity and localization of fungal chitin synthases. Med. Mycol. 2012, 50, 2–17. [Google Scholar] [CrossRef]
- Park, B.C.; Park, Y.H.; Park, H.M. Activation of chsC transcription by AbaA during asexual development of Aspergillus nidulans. FEMS Microbiol. Lett. 2003, 220, 241–246. [Google Scholar] [CrossRef]
- Fujiwara, M.; Ichinomiya, M.; Motoyama, T.; Horiuchi, H.; Ohta, A.; Takagi, M. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. 2000, 127, 359–366. [Google Scholar] [CrossRef]
- van Leeuwe, T.M.; Arentshorst, M.; Forn-Cuní, G.; Geoffrion, N.; Tsang, A.; Delvigne, F.; Meijer, A.H.; Ram, A.F.J.; Punt, P.J. Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content. Microorganisms 2020, 8, 1918. [Google Scholar] [CrossRef]
- Chen, G.; Wang, M.; Tian, X.; Wu, Z. Analyses of Monascus pigment secretion and cellular morphology in non-ionic surfactant micelle aqueous solution. Microb. Biotechnol. 2018, 11, 409–419. [Google Scholar] [CrossRef]
- Chen, G.; Huang, T.; Bei, Q.; Tian, X.; Wu, Z. Correlation of pigment production with mycelium morphology in extractive fermentation of Monascus anka GIM 3.592. Process Biochem. 2017, 58, 42–50. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, B.B.; Liu, X.D.; Zhang, C.; Chen, L.; Xu, G.R.; Cheung, P.C.K. Enhanced production of natural yellow pigments from Monascus purpureus by liquid culture: The relationship between fermentation conditions and mycelial morphology. J. Biosci. Bioeng. 2017, 124, 452–458. [Google Scholar] [CrossRef]
- Lv, J.; Qian, G.F.; Chen, L.; Liu, H.; Xu, H.X.; Xu, G.R.; Zhang, B.B.; Zhang, C. Efficient Biosynthesis of Natural Yellow Pigments by Monascus purpureus in a Novel Integrated Fermentation System. J. Agric. Food Chem. 2018, 66, 918–925. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, J.; Ding, Q.; Luo, Q.; Liu, L. Morphology engineering of Aspergillus oryzae for l-malate production. Biotechnol. Bioeng. 2019, 116, 2662–2673. [Google Scholar] [CrossRef]
- Driouch, H.; Sommer, B.; Wittmann, C. Morphology engineering of Aspergillus niger for improved enzyme production. Biotechnol. Bioeng. 2010, 105, 1058–1068. [Google Scholar]
- Huang, J.; Guan, H.W.; Huang, Y.Y.; Lai, K.S.; Chen, H.Y.; Xue, H.; Zhang, B.B. Evaluating the effects of microparticle addition on mycelial morphology, natural yellow pigments productivity, and key genes regulation in submerged fermentation of Monascus purpureus. Biotechnol. Bioeng. 2021, 118, 2503–2513. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).