Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
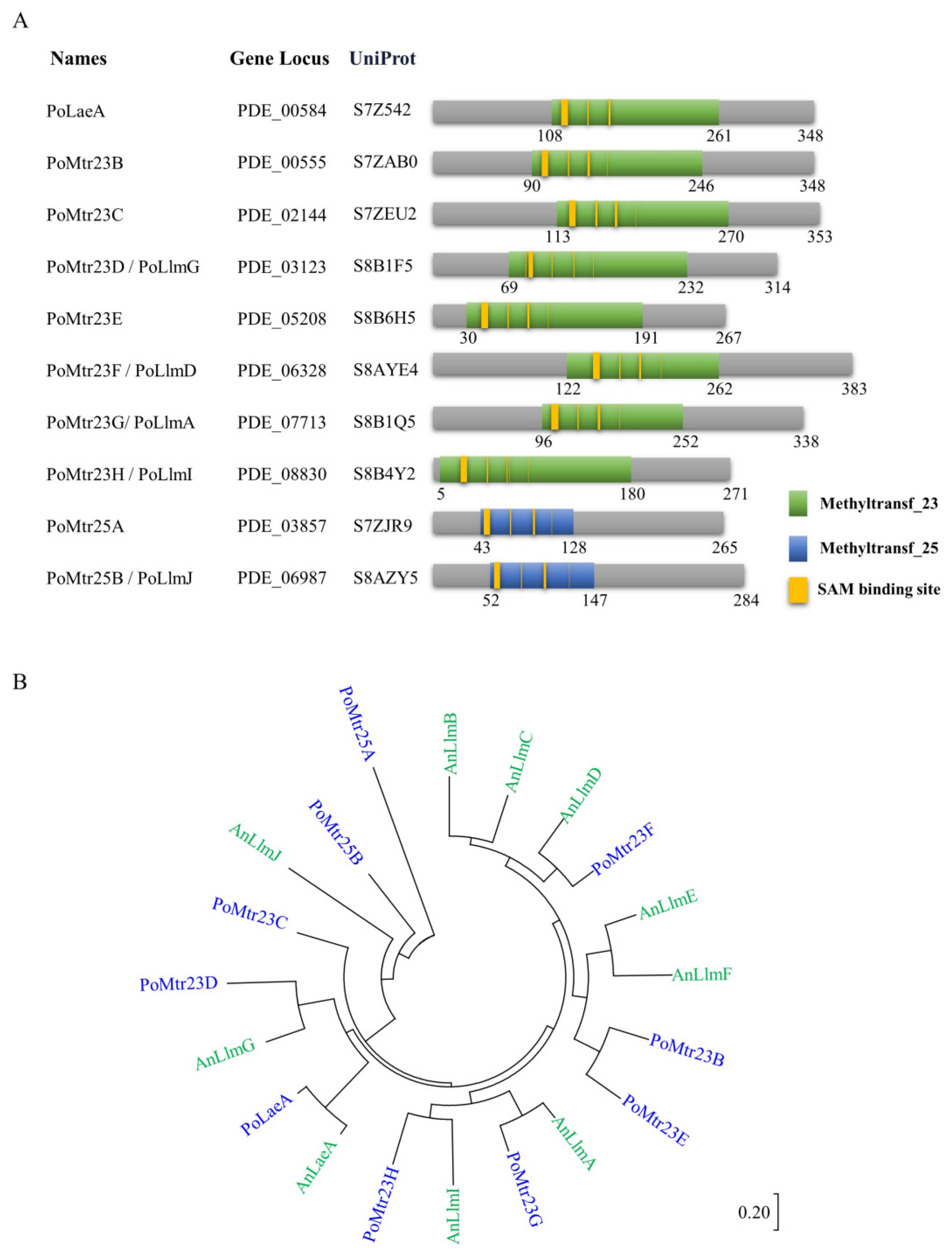
2.2. Phylogenetic Tree Analysis and Domain Architecture Analysis
2.3. Construction of Different Mutants
2.4. Fungal Colony, Conidiation, and Biomass Analysis
2.5. Determination of Enzyme Activities
2.6. Quantitative Reverse Transcription PCR
2.7. Subcellular Localization Observation
2.8. Tandem Affinity Purification and Mass Spectrometry
3. Results
3.1. Naming the Putative Methyltransferases in P. oxalicum
3.2. Various Methyltransferase Genes Display Different Transcriptional Levels
3.3. The Differences in Phenotype Caused by the Deletion of Each Methyltransferase Gene Are Not Notable
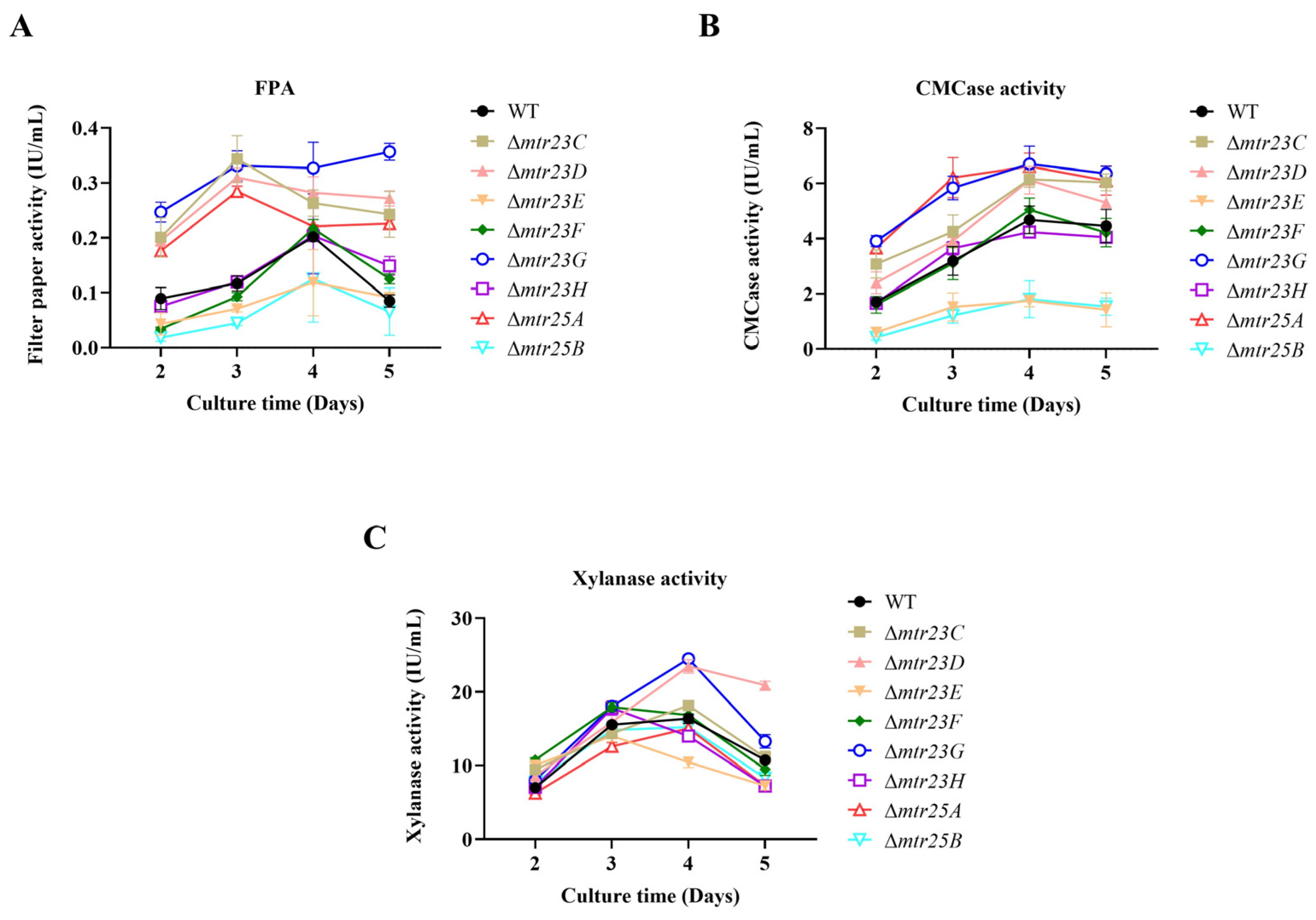
3.4. Various Mutants Have Different Patterns in Cellulolytic Enzyme Production
3.5. Various Mutants Have Different Patterns in the Expression of the Cellulolytic Enzyme Gene
3.6. Some Methyltransferases Affected the Expression of Transcription Factors
3.7. Preliminary Characterization of the Protein PoMtr25A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Liu, G.; Qu, Y. Engineering of filamentous fungi for efficient conversion of lignocellulose: Tools, recent advances and prospects. Biotechnol. Adv. 2019, 37, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, L.; Qin, Y.; Zou, G.; Li, Z.; Yan, X.; Wei, X.; Chen, M.; Chen, L.; Zheng, K.; et al. Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci. Rep. 2013, 3, 1569. [Google Scholar] [CrossRef] [PubMed]
- Mattam, A.J.; Chaudhari, Y.B.; Velankar, H.R. Factors regulating cellulolytic gene expression in filamentous fungi: An overview. Microb. Cell Fact. 2022, 21, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qu, Y.; Qin, Y. Expression and chromatin structures of cellulolytic enzyme gene regulated by heterochromatin protein 1. Biotechnol. Biofuels 2016, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.V.; Pham, K.T.; Nakayashiki, H. Substrate-induced transcriptional activation of the MoCel7C cellulase gene is associated with methylation of histone H3 at lysine 4 in the rice blast fungus Magnaporthe oryzae. Appl. Environ. Microbiol. 2013, 79, 6823–6832. [Google Scholar] [CrossRef]
- Xin, Q.; Gong, Y.; Lv, X.; Chen, G.; Liu, W. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr. Microbiol. 2013, 67, 580–589. [Google Scholar] [CrossRef]
- Seiboth, B.; Karimi, R.A.; Phatale, P.A.; Linke, R.; Hartl, L.; Sauer, D.G.; Smith, K.M.; Baker, S.E.; Freitag, M.; Kubicek, C.P. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol. Microbiol. 2012, 84, 1150–1164. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Sarikaya-Bayram, Ö.; Palmer, J.M.; Keller, N.; Braus, G.H.; Bayram, Ö. One Juliet and four Romeos: VeA and its methyltransferases. Front. Microbiol. 2015, 6, 1. [Google Scholar] [CrossRef]
- Abad, A.; Fernández-Molina, J.V.; Bikandi, J.; Ramírez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Pontón, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Karimi Aghcheh, R.; Druzhinina, I.S.; Kubicek, C.P. The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS ONE 2013, 8, e67144. [Google Scholar]
- Zhang, X.; Zhu, Y.; Bao, L.; Gao, L.; Yao, G.; Li, Y.; Yang, Z.; Li, Z.; Zhong, Y.; Li, F.; et al. Putative methyltransferase LaeA and transcription factor CreA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression. Fungal Genet. Biol. 2016, 94, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Ortiz-Urquiza, A.; Keyhani, N.O. A putative methyltransferase, mtrA, contributes to development, spore viability, protein secretion and virulence in the entomopathogenic fungus Beauveria bassiana. Microbiology 2014, 160 Pt 11, 2526–2537. [Google Scholar] [CrossRef]
- Zhao, Z.; Gu, S.; Liu, D.; Liu, D.; Chen, B.; Li, J.; Tian, C. The putative methyltransferase LaeA regulates mycelium growth and cellulase production in Myceliophthora thermophila. Biotechnol. Biofuels Bioprod. 2023, 16, 58. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, X.; Zhang, X.; Bao, L.; Zhu, Y.; Qu, Y.; Zhao, J.; Qin, Y. The Different Roles of Penicillium oxalicum LaeA in the Production of Extracellular Cellulase and β-xylosidase. Front. Microbiol. 2016, 7, 2091. [Google Scholar] [CrossRef]
- Palmer, J.M.; Theisen, J.M.; Duran, R.M.; Grayburn, W.S.; Calvo, A.M.; Keller, N.P. Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 2013, 9, e1003193. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Zhu, Y.; Yang, L.; Li, Y.; Qu, J.; Wang, L.; Zhao, J.; Qu, Y.; Qin, Y. Penicillium oxalicum putative methyltransferase Mtr23B has similarities and differences with LaeA in regulating conidium development and glycoside hydrolase gene expression. Fungal Genet. Biol. 2020, 143, 103445. [Google Scholar] [CrossRef]
- Grau, M.F.; Entwistle, R.; Oakley, C.E.; Wang, C.C.C.; Oakley, B.R. Overexpression of an LaeA-like Methyltransferase Upregulates Secondary Metabolite Production in Aspergillus nidulans. ACS Chem. Biol. 2019, 14, 1643–1651. [Google Scholar] [CrossRef]
- Bi, Q.; Wu, D.; Zhu, X.; Gillian Turgeon, B. Cochliobolus heterostrophus Llm1—A Lae1-like methyltransferase regulates T-toxin production, virulence, and development. Fungal Genet. Biol. 2013, 51, 21–33. [Google Scholar] [CrossRef]
- Zhang, G.; Yan, P.; Leng, D.; Shang, L.; Zhang, C.; Wu, Z.; Wang, Z. Functional Roles of LaeA-like Genes in Fungal Growth, Cellulase Activity, and Secondary Metabolism in Pleurotus ostreatus. J. Fungi 2022, 8, 902. [Google Scholar] [CrossRef]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [CrossRef]
- Patananan, A.N.; Palmer, J.M.; Garvey, G.S.; Keller, N.P.; Clarke, S.G. A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J. Biol. Chem. 2013, 288, 14032–14045. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kubodera, T.; Yamashita, N.; Nishimura, A. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2002, 66, 404–406. [Google Scholar] [CrossRef]
- Li, Z.; Yao, G.; Wu, R.; Gao, L.; Kan, Q.; Liu, M.; Yang, P.; Liu, G.; Qin, Y.; Song, X.; et al. Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in Penicillium oxalicum. PLoS Genet. 2015, 11, e1005509. [Google Scholar] [CrossRef]
- Nakayashiki, H.; Hanada, S.; Nguyen, B.Q.; Kadotani, N.; Tosa, Y.; Mayama, S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 2005, 42, 275–283. [Google Scholar] [CrossRef]
- Yao, G.; Li, Z.; Gao, L.; Wu, R.; Kan, Q.; Liu, G.; Qu, Y. Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum. Biotechnol. Biofuels 2015, 8, 71. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Y.; Zhu, Z.; Zhao, K.; Liu, G.; Wang, L.; Qu, Y.; Zhao, J.; Qin, Y. Normal transcription of cellulolytic enzyme genes relies on the balance between the methylation of H3K36 and H3K4 in Penicillium oxalicum. Biotechnol. Biofuels 2019, 12, 198. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, Z.; Xu, S.; Zhao, Q.; Liu, G.; Song, X.; Qu, Y.; Qin, Y. The interaction between the histone acetyltransferase complex Hat1-Hat2 and transcription factor AmyR provides a molecular brake to regulate amylase gene expression. Mol. Microbiol. 2023, 119, 471–491. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 2005, 4, 1265–1272. [Google Scholar] [CrossRef]
- Oda, K.; Kakizono, D.; Yamada, O.; Iefuji, H.; Akita, O.; Iwashita, K. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006, 72, 3448–3457. [Google Scholar] [CrossRef]
- Kunitake, E.; Kobayashi, T. Conservation and diversity of the regulators of cellulolytic enzyme genes in Ascomycete fungi. Curr. Genet. 2017, 63, 951–958. [Google Scholar] [CrossRef]
- Orejas, M.; MacCabe, A.P.; Pérez González, J.A.; Kumar, S.; Ramón, D. Carbon catabolite repression of the Aspergillus nidulans xlnA gene. Mol. Microbiol. 1999, 31, 177–184. [Google Scholar] [CrossRef]
- Ilmén, M.; Thrane, C.; Penttilä, M. The glucose repressor gene cre1 of Trichoderma: Isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 1996, 251, 451–460. [Google Scholar]
- Rassinger, A.; Gacek-Matthews, A.; Strauss, J.; Mach, R.L.; Mach-Aigner, A.R. Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 2018, 5, 15. [Google Scholar] [CrossRef]
- Hashimoto, W.; Arai, H.; Mizutani, O.; Yamada, O.; Shintani, T.; Gomi, K. Expression profiles of amylolytic genes in AmyR and CreA transcription factor deletion mutants of the black koji mold Aspergillus luchuensis. J. Biosci. Bioeng. 2021, 132, 321–326. [Google Scholar] [CrossRef]
- Sun, J.; Glass, N.L. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS ONE 2011, 6, e25654. [Google Scholar] [CrossRef]
- Shin, K.S.; Kwon, N.J.; Kim, Y.H.; Park, H.S.; Kwon, G.S.; Yu, J.H. Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 2009, 8, 738–746. [Google Scholar] [CrossRef]
- White, S.; McIntyre, M.; Berry, D.R.; McNeil, B. The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 2002, 22, 1–14. [Google Scholar] [CrossRef]
- Gao, L.; He, X.; Guo, Y.; Wu, Z.; Zhao, J.; Liu, G.; Qu, Y. Combinatorial Engineering of Transcriptional Activators in Penicillium oxalicum for Improved Production of Corn-Fiber-Degrading Enzymes. J. Agric. Food Chem. 2021, 69, 2539–2548. [Google Scholar] [CrossRef]
- Fekete, E.; Karaffa, L.; Karimi Aghcheh, R.; Németh, Z.; Fekete, E.; Orosz, A.; Paholcsek, M.; Stágel, A.; Kubicek, C.P. The transcriptome of lae1 mutants of Trichoderma reesei cultivated at constant growth rates reveals new targets of LAE1 function. BMC Genom. 2014, 15, 447. [Google Scholar] [CrossRef]
- Perrin, R.M.; Fedorova, N.D.; Bok, J.W.; Cramer, R.A.; Wortman, J.R.; Kim, H.S.; Nierman, W.C.; Keller, N.P. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007, 3, e50. [Google Scholar] [CrossRef]
- Collier, L.A.; Ghosh, A.; Borkovich, K.A. Heterotrimeric G-Protein Signaling Is Required for Cellulose Degradation in Neurospora crassa. mBio 2020, 11, e02419–e02420. [Google Scholar] [CrossRef]
- Lin, L.; Chen, Y.; Li, J.; Wang, S.; Sun, W.; Tian, C. Disruption of non-anchored cell wall protein NCW-1 promotes cellulase production by increasing cellobiose uptake in Neurospora crassa. Biotechnol. Lett. 2017, 39, 545–551. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Karimi Aghcheh, R.; Németh, Z.; Atanasova, L.; Fekete, E.; Paholcsek, M.; Sándor, E.; Aquino, B.; Druzhinina, I.S.; Karaffa, L.; Kubicek, C.P. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE 2014, 9, e112799. [Google Scholar]
- Karuppiah, V.; Zhixiang, L.; Liu, H.; Vallikkannu, M.; Chen, J. Co-culture of Vel1-overexpressed Trichoderma asperellum and Bacillus amyloliquefaciens: An eco-friendly strategy to hydrolyze the lignocellulose biomass in soil to enrich the soil fertility, plant growth and disease resistance. Microb. Cell Fact. 2021, 20, 57. [Google Scholar] [CrossRef]
- Reyes, A.A.; Marcum, R.D.; He, Y. Structure and Function of Chromatin Remodelers. J. Mol. Biol. 2021, 433, 166929. [Google Scholar]
- Cao, Y.; Zheng, F.; Zhang, W.; Meng, X.; Liu, W. Trichoderma reesei XYR1 recruits SWI/SNF to facilitate cellulase gene expression. Mol. Microbiol. 2019, 112, 1145–1162. [Google Scholar] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Ma, K.; Zhang, X.; Song, X.; Qin, Y. Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes. J. Fungi 2023, 9, 1118. https://doi.org/10.3390/jof9111118
Liu Z, Ma K, Zhang X, Song X, Qin Y. Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes. Journal of Fungi. 2023; 9(11):1118. https://doi.org/10.3390/jof9111118
Chicago/Turabian StyleLiu, Zhongjiao, Kexuan Ma, Xiujun Zhang, Xin Song, and Yuqi Qin. 2023. "Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes" Journal of Fungi 9, no. 11: 1118. https://doi.org/10.3390/jof9111118
APA StyleLiu, Z., Ma, K., Zhang, X., Song, X., & Qin, Y. (2023). Different Putative Methyltransferases Have Different Effects on the Expression Patterns of Cellulolytic Genes. Journal of Fungi, 9(11), 1118. https://doi.org/10.3390/jof9111118





