The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Isolation of Polymorphonuclear Cells from Human Peripheral Blood
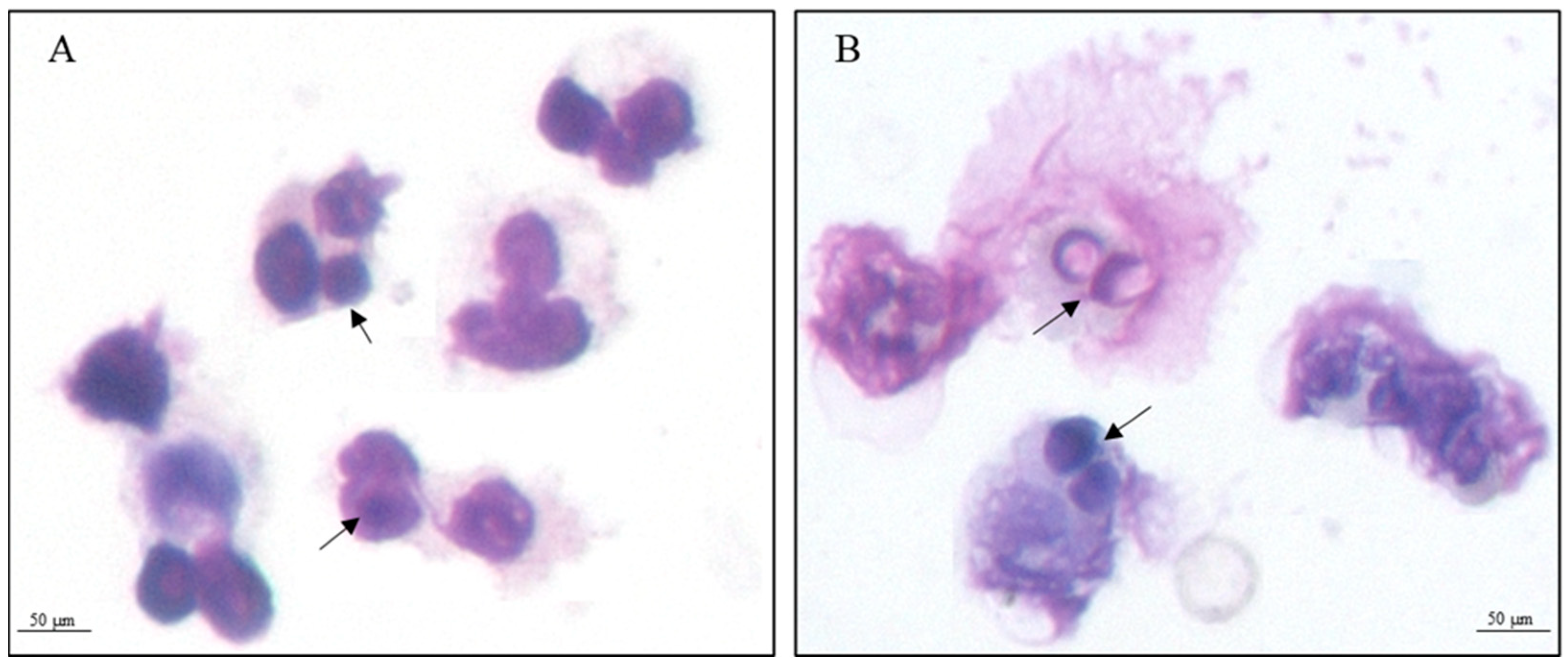
2.3. Internalization Assay
2.4. Preparation of Protein Extract
2.5. Digestion of Proteins Extracts for Mass Spectrometry
2.6. Chromatography and Mass Spectrometry
2.7. Raw Data Processing and Protein Identification Analysis
2.8. Statistical Analyses and Graphics Construction
3. Results
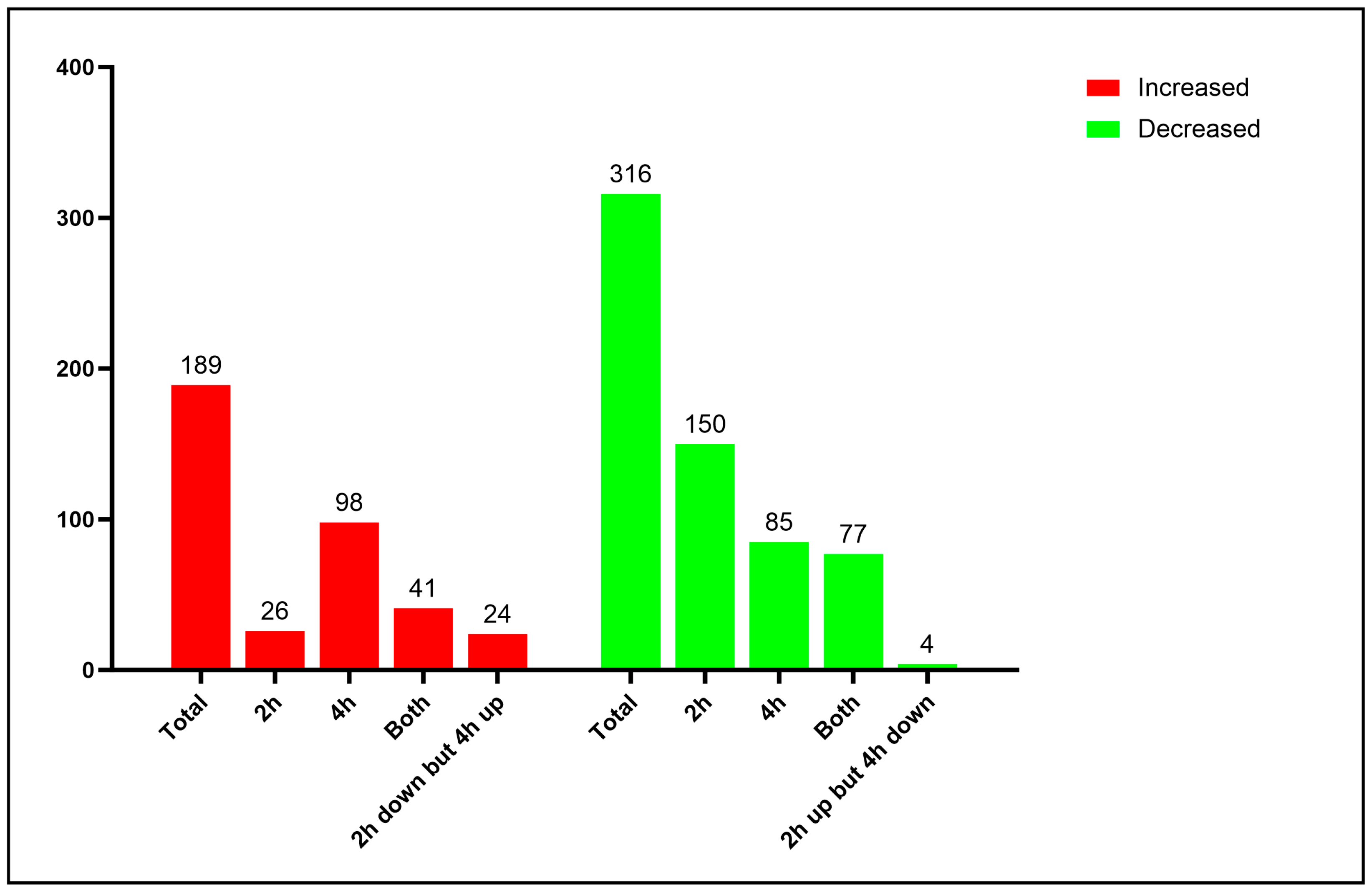
3.1. Proteomic Analysis of Yeast Cells Interacting with Neutrophils
3.2. Interaction with Human Neutrophils Promotes Increase in Proteins of the Oxidative Stress Response in P. lutzii
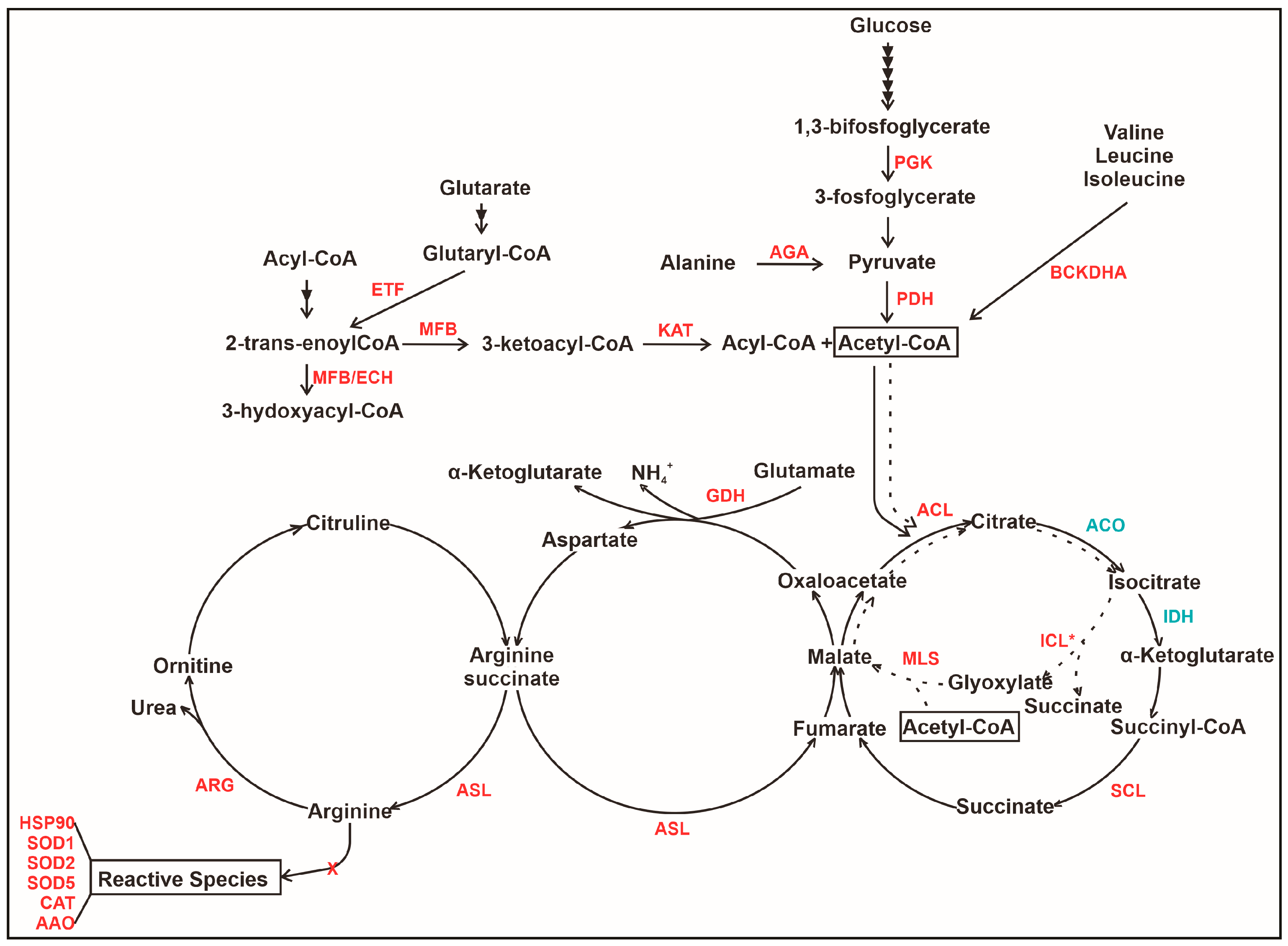
3.3. The Interaction of P. lutzii with Neutrophils Promotes Increase in Proteins Related to Acetyl-CoA Formation
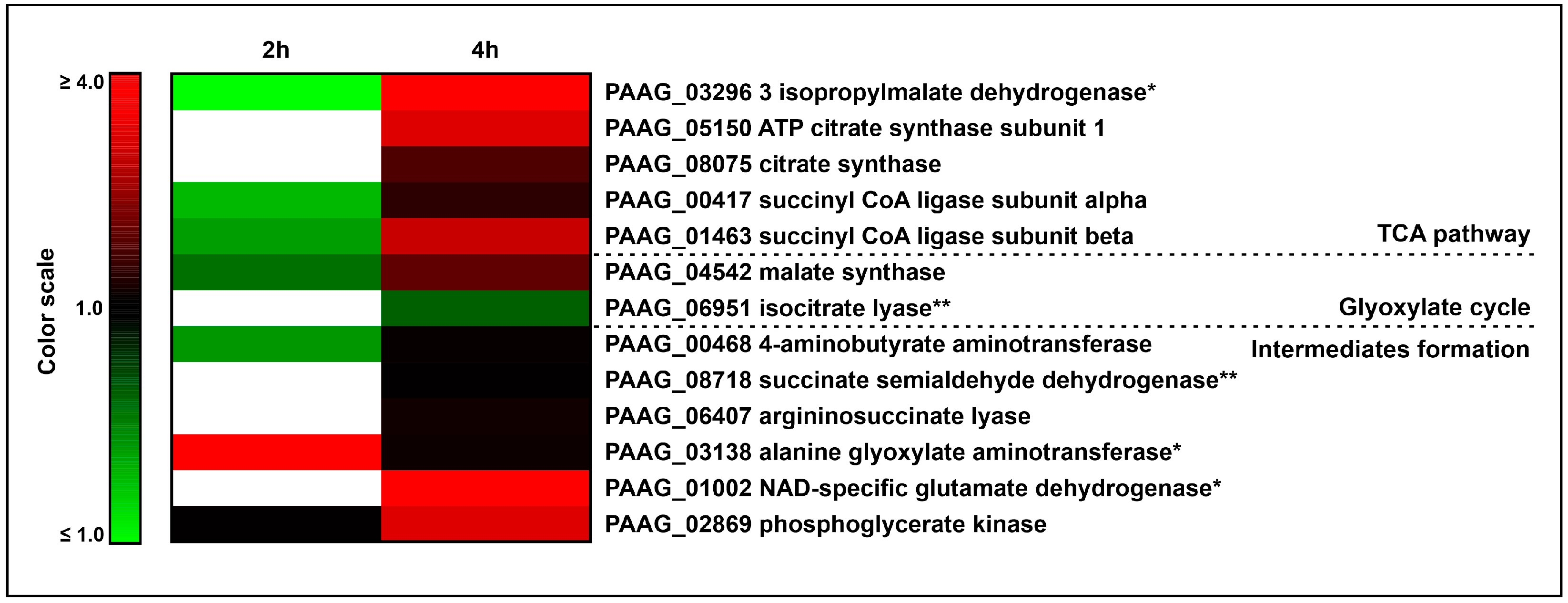
3.4. The Interaction of P. lutzii with Neutrophils Promotes Increase in Enzymes of TCA and Glyoxylate Pathway and Its Intermediates Formation
3.5. An Overview of Metabolic Changes in P. lutzii Interacting with Neutrophils
3.6. Descriptive Analysis of Some Neutrophil Proteins Identified upon Interaction with P. lutzii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Franco, M. Host-Parasite Relationships in Paracoccidioidomycosis. J. Med. Vet. Mycol. 1987, 25, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Hagen, F.; Puccia, R.; Hahn, R.C.; de Camargo, Z.P. Paracoccidioides and Paracoccidioidomycosis in the 21st Century. Mycopathologia 2023, 188, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Shikanai-Yasuda, M.A.; Mendes, R.P.; Colombo, A.L.; de Queiroz-Telles, F.; Kono, A.S.G.; Paniago, A.M.M.; Nathan, A.; do Valle, A.C.F.; Bagagli, E.; Benard, G.; et al. Brazilian Guidelines for the Clinical Management of Paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017, 50, 715–740. [Google Scholar] [CrossRef] [PubMed]
- de Castro, L.F.; Ferreira, M.C.; da Silva, R.M.; de Souza Lima Blotta, M.H.; Longhi, L.N.A.; Mamoni, R.L. Characterization of the Immune Response in Human Paracoccidioidomycosis. J. Infect. 2013, 67, 470–485. [Google Scholar] [CrossRef]
- Fortes, M.R.P.; Miot, H.A.; Kurokawa, C.S.; Marques, M.E.A.; Marques, S.A. Immunology of Paracoccidioidomycosis. An. Bras. Dermatol. 2011, 86, 516–524. [Google Scholar] [CrossRef]
- Appelberg, R.; Castro, A.G.; Silva, M.T. Neutrophils as Effector Cells of T-Cell-Mediated, Acquired Immunity in Murine Listeriosis. Immunology 1994, 83, 302–307. [Google Scholar]
- Tateda, K.; Moore, T.A.; Deng, J.C.; Newstead, M.W.; Zeng, X.; Matsukawa, A.; Swanson, M.S.; Yamaguchi, K.; Standiford, T.J. Early Recruitment of Neutrophils Determines Subsequent T1/T2 Host Responses in a Murine Model of Legionella pneumophila Pneumonia. J. Immunol. 2001, 166, 3355–3361. [Google Scholar] [CrossRef]
- Pedrosa, J.; Saunders, B.M.; Appelberg, R.; Orme, I.M.; Silva, M.T.; Cooper, A.M. Neutrophils Play a Protective Nonphagocytic Role in Systemic Mycobacterium tuberculosis Infection of Mice. Infect. Immun. 2000, 68, 577–583. [Google Scholar] [CrossRef]
- Romani, L.; Mencacci, A.; Cenci, E.; Del Sero, G.; Bistoni, F.; Puccetti, P. An Immunoregulatory Role for Neutrophils in CD4+ T Helper Subset Selection in Mice with Candidiasis. J. Immunol. 1997, 158, 2356–2362. [Google Scholar] [CrossRef]
- Miranda, F.J.B.; Rocha, B.C.; Pereira, M.C.A.; Pereira, L.M.N.; de Souza, E.H.M.; Marino, A.P.; Costa, P.A.C.; Vasconcelos-Santos, D.V.; Antonelli, L.R.V.; Gazzinelli, R.T. Toxoplasma gondii-Induced Neutrophil Extracellular Traps Amplify the Innate and Adaptive Response. mBio 2021, 12, e01307-21. [Google Scholar] [CrossRef]
- de Buhr, N.; Bonilla, M.C.; Jimenez-Soto, M.; von Köckritz-Blickwede, M.; Dolz, G. Extracellular Trap Formation in Response to Trypanosoma cruzi Infection in Granulocytes Isolated From Dogs and Common Opossums, Natural Reservoir Hosts. Front. Microbiol. 2018, 9, 996. [Google Scholar] [CrossRef]
- Kennedy, M.A. A Brief Review of the Basics of Immunology: The Innate and Adaptive Response. Vet. Clin. N. Am. Small Anim. Pract. 2010, 40, 369–379. [Google Scholar] [CrossRef]
- Amulic, B.; Cazalet, C.; Hayes, G.L.; Metzler, K.D.; Zychlinsky, A. Neutrophil Function: From Mechanisms to Disease. Annu. Rev. Immunol. 2012, 30, 459–489. [Google Scholar] [CrossRef]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A.R. An Integrated Model of the Recognition of Candida albicans by the Innate Immune System. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Giusiano, G. The Trojan Horse Model in Paracoccidioides: A Fantastic Pathway to Survive Infecting Human Cells. Front. Cell Infect. Microbiol. 2021, 10, 605679. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Mejía, S.P.; Cano, L.E.; López, J.A.; Hernandez, O.; González, Á. Human Neutrophils Produce Extracellular Traps against Paracoccidioides brasiliensis. Microbiology 2015, 161, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Acorci, M.J.; Dias-Melicio, L.A.; Golim, M.A.; Bordon-Graciani, A.P.; Peraçoli, M.T.S.; Soares, Â.M.V.C. Inhibition of Human Neutrophil Apoptosis by Paracoccidioides brasiliensis: Role of Interleukin-8. Scand. J. Immunol. 2009, 69, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.F.R.; Filgueira, A.L.; de Souza, W. A Morphological and Cytochemical Study of the Interaction between Paracoccidiodes brasiliensis and Neutrophils. Microsc. Microanal. 2004, 10, 215–223. [Google Scholar] [CrossRef]
- Netto, C.F.; Vegas, V.S.; Sciannaméa, I.M.; Guarnieri, D.B. Antígeno polissacarídico do Paracoccidioides brasiliensis. Estudo do tempo de cultivo do P. brasiliensis, necessario ao preparo do antigeno. Rev. Inst. Med. Trop. Sao Paulo 1969, 11, 177–181. [Google Scholar] [PubMed]
- Fradin, C.; Kretschmar, M.; Nichterlein, T.; Gaillardin, C.; D’Enfert, C.; Hube, B. Stage-Specific Gene Expression of Candida albicans in Human Blood. Mol. Microbiol. 2003, 47, 1523–1543. [Google Scholar] [CrossRef] [PubMed]
- Lacerda Pigosso, L.; Baeza, L.C.; Vieira Tomazett, M.; Batista Rodrigues Faleiro, M.; Brianezi Dignani de Moura, V.M.; Melo Bailão, A.; Borges, C.L.; Alves Parente Rocha, J.; Rocha Fernandes, G.; Gauthier, G.M.; et al. Paracoccidioides brasiliensis Presents Metabolic Reprogramming and Secretes a Serine Proteinase during Murine Infection. Virulence 2017, 8, 1417–1434. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Murad, A.M.; Souza, G.H.M.F.; Garcia, J.S.; Rech, E.L. Detection and Expression Analysis of Recombinant Proteins in Plant-Derived Complex Mixtures Using nanoUPLC-MSE. J. Sep. Sci. 2011, 34, 2618–2630. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Denny, R.; Dorschel, C.A.; Gorenstein, M.; Kass, I.J.; Li, G.-Z.; McKenna, T.; Nold, M.J.; Richardson, K.; Young, P.; et al. Quantitative Proteomic Analysis by Accurate Mass Retention Time Pairs. Anal. Chem. 2005, 77, 2187–2200. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.-Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute Quantification of Proteins by LCMSE: A Virtue of Parallel Ms Acquisition * S. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef]
- Geromanos, S.J.; Vissers, J.P.C.; Silva, J.C.; Dorschel, C.A.; Li, G.-Z.; Gorenstein, M.V.; Bateman, R.H.; Langridge, J.I. The Detection, Correlation, and Comparison of Peptide Precursor and Product Ions from Data Independent LC-MS with Data Dependant LC-MS/MS. Proteomics 2009, 9, 1683–1695. [Google Scholar] [CrossRef]
- Poirier, Y.; Antonenkov, V.D.; Glumoff, T.; Hiltunen, J.K. Peroxisomal β-Oxidation—A Metabolic Pathway with Multiple Functions. Biochim. Biophys. Acta BBA Mol. Cell Res. 2006, 1763, 1413–1426. [Google Scholar] [CrossRef]
- Miller, S.P.; Chen, R.; Karschnia, E.J.; Romfo, C.; Dean, A.; LaPorte, D.C. Locations of the Regulatory Sites for Isocitrate Dehydrogenase Kinase/Phosphatase *. J. Biol. Chem. 2000, 275, 833–839. [Google Scholar] [CrossRef]
- Niemiec, M.J.; Grumaz, C.; Ermert, D.; Desel, C.; Shankar, M.; Lopes, J.P.; Mills, I.G.; Stevens, P.; Sohn, K.; Urban, C.F. Dual Transcriptome of the Immediate Neutrophil and Candida albicans Interplay. BMC Genom. 2017, 18, 696. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Smolarz, M.; Seweryn-Ozog, K.; Satala, D.; Zawrotniak, M.; Wronowska, E.; Bochenska, O.; Kozik, A.; Nobbs, A.H.; Gogol, M.; et al. Proteinous Components of Neutrophil Extracellular Traps Are Arrested by the Cell Wall Proteins of Candida albicans during Fungal Infection, and Can Be Used in the Host Invasion. Cells 2021, 10, 2736. [Google Scholar] [CrossRef] [PubMed]
- Fradin, C.; De Groot, P.; MacCallum, D.; Schaller, M.; Klis, F.; Odds, F.C.; Hube, B. Granulocytes Govern the Transcriptional Response, Morphology and Proliferation of Candida albicans in Human Blood. Mol. Microbiol. 2005, 56, 397–415. [Google Scholar] [CrossRef]
- Cox, G.M.; Harrison, T.S.; McDade, H.C.; Taborda, C.P.; Heinrich, G.; Casadevall, A.; Perfect, J.R. Superoxide Dismutase Influences the Virulence of Cryptococcus neoformans by Affecting Growth within Macrophages. Infect. Immun. 2003, 71, 173–180. [Google Scholar] [CrossRef]
- Youseff, B.H.; Holbrook, E.D.; Smolnycki, K.A.; Rappleye, C.A. Extracellular Superoxide Dismutase Protects Histoplasma Yeast Cells from Host-Derived Oxidative Stress. PLoS Pathog. 2012, 8, e1002713. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, D.; Muñoz, J.F.; Lopez, Á.; Urán, M.; Herrera, J.; Borges, C.L.; Restrepo, Á.; Soares, C.M.; Taborda, C.P.; Almeida, A.J.; et al. Identification and Analysis of the Role of Superoxide Dismutases Isoforms in the Pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 2016, 10, e0004481. [Google Scholar] [CrossRef] [PubMed]
- de Arruda Grossklaus, D.; Bailão, A.M.; Vieira Rezende, T.C.; Borges, C.L.; de Oliveira, M.A.P.; Parente, J.A.; de Almeida Soares, C.M. Response to Oxidative Stress in Paracoccidioides Yeast Cells as Determined by Proteomic Analysis. Microbes Infect. 2013, 15, 347–364. [Google Scholar] [CrossRef]
- Parente-Rocha, J.A.; Parente, A.F.A.; Baeza, L.C.; Bonfim, S.M.R.C.; Hernandez, O.; McEwen, J.G.; Bailão, A.M.; Taborda, C.P.; Borges, C.L.; Soares, C.M.D.A. Macrophage Interaction with Paracoccidioides brasiliensis Yeast Cells Modulates Fungal Metabolism and Generates a Response to Oxidative Stress. PLoS ONE 2015, 10, e0137619. [Google Scholar] [CrossRef]
- Matthews, R.; Burnie, J. The Role of Hsp90 in Fungal Infection. Immunol. Today 1992, 13, 345–348. [Google Scholar] [CrossRef]
- Moura, Á.N.D.; de Oliveira, D.S.L.; Paredes, V.; Rocha, L.B.; de Oliveira, F.F.M.; Lessa, G.M.; Riasco-Palacios, J.F.; Casadevall, A.; Albuquerque, P.; Felipe, M.S.S.; et al. Paracoccidioides HSP90 Can Be Found in the Cell Surface and Is a Target for Antibodies with Therapeutic Potential. J. Fungi 2020, 6, 193. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional Response of Candida albicans upon Internalization by Macrophages. Eukaryot. Cell 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Landgraf, T.N.; Costa, M.V.; Oliveira, A.F.; Ribeiro, W.C.; Panunto-Castelo, A.; Fernandes, F.F. Involvement of Dihydrolipoyl Dehydrogenase in the Phagocytosis and Killing of Paracoccidioides brasiliensis by Macrophages. Front. Microbiol. 2017, 8, 1803. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.L.E.; Oliveira, M.A.P.; Silva, L.O.S.; Inácio, M.M.; Bailão, A.M.; Parente-Rocha, J.A.; Cruz-Leite, V.R.M.; Paccez, J.D.; de Almeida Soares, C.M.; Weber, S.S.; et al. Immunoproteomic Approach of Extracellular Antigens From Paracoccidioides Species Reveals Exclusive B-Cell Epitopes. Front. Microbiol. 2020, 10, 2968. [Google Scholar] [CrossRef]
- Weber, S.S.; Parente, A.F.A.; Borges, C.L.; Parente, J.A.; Bailão, A.M.; Soares, C.M.D.A. Analysis of the Secretomes of Paracoccidioides Mycelia and Yeast Cells. PLoS ONE 2012, 7, e52470. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-L.; Cannon, R.D.; Gallo, S.M.; Villas-Bôas, S.G. A Metabolomic Study of the Effect of Candida albicans Glutamate Dehydrogenase Deletion on Growth and Morphogenesis. NPJ Biofilms Microbiomes 2019, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Sanyal, K. P330 Role of a D-Amino Acid Oxidase (DAO) in an Efficient Human Commensal Candida albicans. Med. Mycol. 2022, 60, myac072P330. [Google Scholar] [CrossRef]
- Calcedo, R.; Ramirez-Garcia, A.; Abad, A.; Rementeria, A.; Pontón, J.; Hernando, F.L. Phosphoglycerate Kinase and Fructose Bisphosphate Aldolase of Candida albicans as New Antigens Recognized by Human Salivary IgA. Rev. Iberoam. Micol. 2012, 29, 172–174. [Google Scholar] [CrossRef]
- Boone, T.J.; Tyrrell, G.J. Identification of the Actin and Plasminogen Binding Regions of Group B Streptococcal Phosphoglycerate Kinase. J. Biol. Chem. 2012, 287, 29035–29044. [Google Scholar] [CrossRef]
- Stie, J.; Bruni, G.; Fox, D. Surface-Associated Plasminogen Binding of Cryptococcus neoformans Promotes Extracellular Matrix Invasion. PLoS ONE 2009, 4, e5780. [Google Scholar] [CrossRef]
- Miles, L.A.; Dahlberg, C.M.; Plescia, J.; Felez, J.; Kato, K.; Plow, E.F. Role of Cell-Surface Lysines in Plasminogen Binding to Cells: Identification of Alpha-Enolase as a Candidate Plasminogen Receptor. Biochemistry 1991, 30, 1682–1691. [Google Scholar] [CrossRef]
- Alloush, H.M.; López-Ribot, J.L.; Masten, B.J.; Chaffin, W.L. 3-Phosphoglycerate Kinase: A Glycolytic Enzyme Protein Present in the Cell Wall of Candida albicans. Microbiology 1997, 143 (Pt. 2), 321–330. [Google Scholar] [CrossRef]
- Chaves, E.G.A.; Weber, S.S.; Báo, S.N.; Pereira, L.A.; Bailão, A.M.; Borges, C.L.; Soares, C.M.d.A. Analysis of Paracoccidioides Secreted Proteins Reveals Fructose 1,6-Bisphosphate Aldolase as a Plasminogen-Binding Protein. BMC Microbiol. 2015, 15, 53. [Google Scholar] [CrossRef]
- Alden, K.M.; Jezewski, A.J.; Beattie, S.R.; Fox, D.; Krysan, D.J. Genetic Interaction Analysis Reveals That Cryptococcus neoformans Utilizes Multiple Acetyl-CoA-Generating Pathways during Infection. mBio 2022, 13, e0127922. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, E.J.; Hu, G.; Fries, B.; Caza, M.; Wang, J.; Gsponer, J.; Gates-Hollingsworth, M.A.; Kozel, T.R.; De Repentigny, L.; Kronstad, J.W. A Defect in ATP-Citrate Lyase Links Acetyl-CoA Production, Virulence Factor Elaboration and Virulence in Cryptococcus neoformans. Mol. Microbiol. 2012, 86, 1404–1423. [Google Scholar] [CrossRef] [PubMed]
- Orasch, T.; Dietl, A.-M.; Shadkchan, Y.; Binder, U.; Bauer, I.; Lass-Flörl, C.; Osherov, N.; Haas, H. The Leucine Biosynthetic Pathway Is Crucial for Adaptation to Iron Starvation and Virulence in Aspergillus fumigatus. Virulence 2019, 10, 925–934. [Google Scholar] [CrossRef]
- Do, E.; Hu, G.; Caza, M.; Oliveira, D.; Kronstad, J.W.; Jung, W.H. Leu1 Plays a Role in Iron Metabolism and Is Required for Virulence in Cryptococcus neoformans. Fungal Genet. Biol. 2015, 75, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chaves, E.G.A.; Parente-Rocha, J.A.; Baeza, L.C.; Araújo, D.S.; Borges, C.L.; de Oliveira, M.A.P.; de Almeida Soares, C.M. Proteomic Analysis of Paracoccidioides brasiliensis During Infection of Alveolar Macrophages Primed or Not by Interferon-Gamma. Front. Microbiol. 2019, 10, 96. [Google Scholar] [CrossRef]
- Felipe, M.S.S.; Andrade, R.V.; Arraes, F.B.M.; Nicola, A.M.; Maranhão, A.Q.; Torres, F.A.G.; Silva-Pereira, I.; Poças-Fonseca, M.J.; Campos, E.G.; Moraes, L.M.P.; et al. Transcriptional Profiles of the Human Pathogenic Fungus Paracoccidioides brasiliensis in Mycelium and Yeast Cells. J. Biol. Chem. 2005, 280, 24706–24714. [Google Scholar] [CrossRef]
- Araújo, D.S.; Pereira, M.; Portis, I.G.; dos Santos Junior, A.d.C.M.; Fontes, W.; de Sousa, M.V.; do Prado Assunção, L.; Baeza, L.C.; Bailão, A.M.; Ricart, C.A.O.; et al. Metabolic Peculiarities of Paracoccidioides brasiliensis Dimorphism as Demonstrated by iTRAQ Labeling Proteomics. Front. Microbiol. 2019, 10, 555. [Google Scholar] [CrossRef]
- Derengowski, L.S.; Tavares, A.H.; Silva, S.; Procópio, L.S.; Felipe, M.S.S.; Silva-Pereira, I. Upregulation of Glyoxylate Cycle Genes upon Paracoccidioides brasiliensis Internalization by Murine Macrophages and in Vitro Nutritional Stress Condition. Med. Mycol. 2008, 46, 125–134. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The Glyoxylate Cycle Is Required for Fungal Virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef]
- Rude, T.H.; Toffaletti, D.L.; Cox, G.M.; Perfect, J.R. Relationship of the Glyoxylate Pathway to the Pathogenesis of Cryptococcus neoformans. Infect. Immun. 2002, 70, 5684–5694. [Google Scholar] [CrossRef]
- Fan, W.; Kraus, P.R.; Boily, M.-J.; Heitman, J. Cryptococcus neoformans Gene Expression during Murine Macrophage Infection. Eukaryot. Cell 2005, 4, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Lima, P.; Casaletti, L.; Bailão, A.M.; de Vasconcelos, A.T.R.; da Rocha Fernandes, G.; de Almeida Soares, C.M. Transcriptional and Proteomic Responses to Carbon Starvation in Paracoccidioides. PLoS Negl. Trop. Dis. 2014, 8, e2855. [Google Scholar] [CrossRef]
- Araújo, F.S.; Coelho, L.M.; do Carmo Silva, L.; da Silva Neto, B.R.; Parente-Rocha, J.A.; Bailão, A.M.; de Oliveira, C.M.A.; da Rocha Fernandes, G.; Hernández, O.; Ochoa, J.G.M.; et al. Effects of Argentilactone on the Transcriptional Profile, Cell Wall and Oxidative Stress of Paracoccidioides spp. PLoS Negl. Trop. Dis. 2016, 10, e0004309. [Google Scholar] [CrossRef] [PubMed]
- Soehnlein, O.; Lindbom, L. Neutrophil-Derived Azurocidin Alarms the Immune System. J. Leukoc. Biol. 2009, 85, 344–351. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil Elastase and Myeloperoxidase Regulate the Formation of Neutrophil Extracellular Traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. A Myeloperoxidase-Containing Complex Regulates Neutrophil Elastase Release and Actin Dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef]
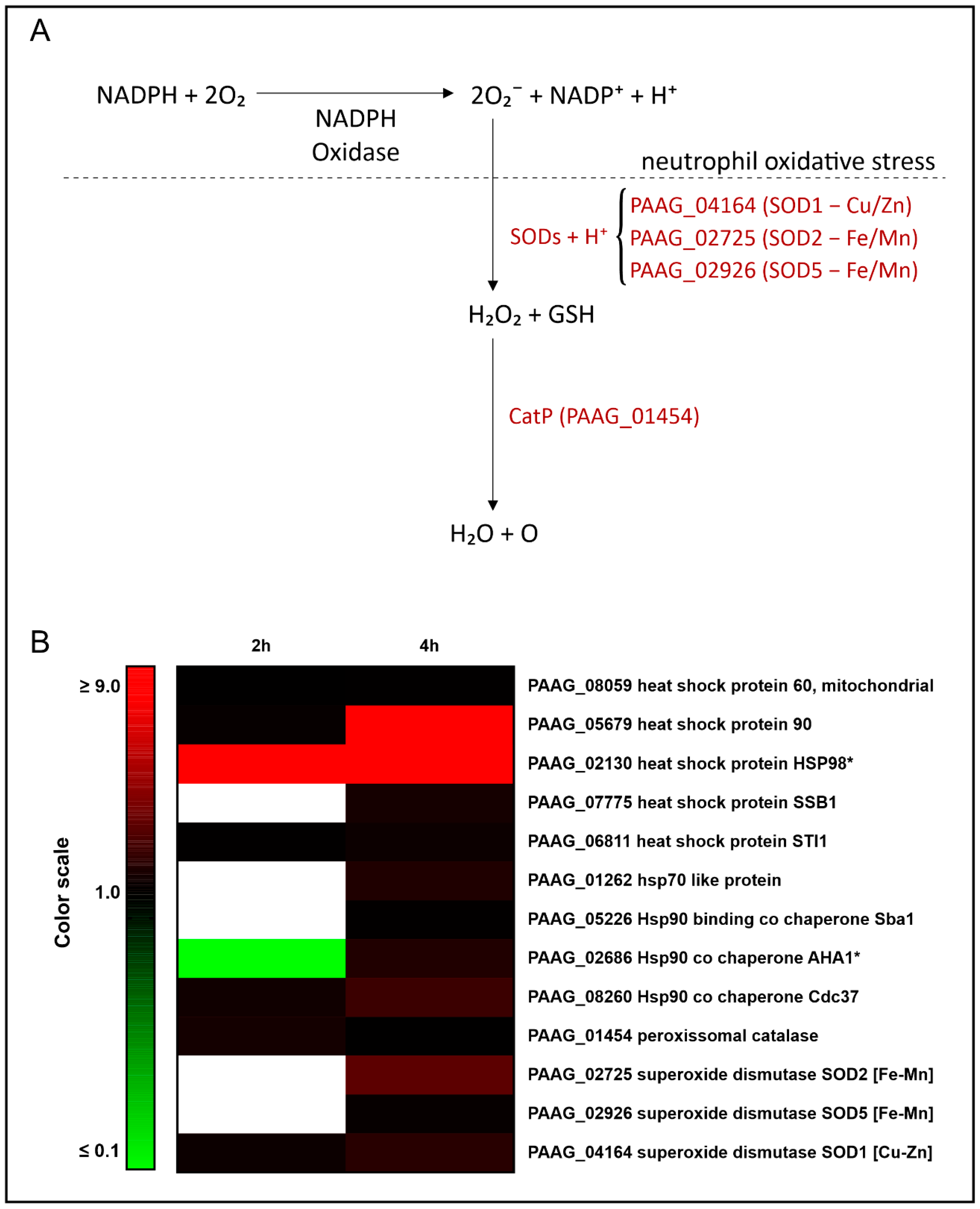

| Accession a | Description b | Expression 2 h c | Expression 4 h c |
|---|---|---|---|
| PAAG_08059 | heat shock protein 60, mitochondrial | # | 1.26 |
| PAAG_05679 | heat shock protein 90 | 1.40 | 8.76 |
| PAAG_02130 | heat shock protein HSP98 | * | * |
| PAAG_07775 | heat shock protein SSB1 | # | 1.84 |
| PAAG_06811 | heat shock protein STI1 | 1.27 | 1.58 |
| PAAG_01262 | hsp70-like protein | # | 2.08 |
| PAAG_05226 | Hsp90-binding co chaperone Sba | # | 1.27 |
| PAAG_02686 | Hsp90 co chaperone AHA1 | # | 2.10 |
| PAAG_08260 | Hsp90 co chaperone Cdc37 | 1.70 | 2.86 |
| PAAG_01454 | peroxisomal catalase | 1.79 | # |
| PAAG_02725 | superoxide dismutase SOD2 [Fe-Mn] | # | 3.78 |
| PAAG_02926 | superoxide dismutase SOD5 [Fe-Mn] | # | 1.42 |
| PAAG_04164 | superoxide dismutase SOD1 [Cu-Zn] | 1.57 | 2.34 |
| Accession a | Description b | Expression 2 h c | Expression 4 h c |
|---|---|---|---|
| PAAG_06224 | carnitine O acetyltransferase | 0.68 ** | |
| PAAG_08859 | peroxisomal multifunctional enzyme | 1.43 | * |
| PAAG_06329 | 3 hydroxybutyryl CoA dehydrogenase | # | 1.12 |
| PAAG_06309 | enoyl CoA hydratase | 1.27 | 1.32 |
| PAAG_02664 | 3 ketoacyl CoA thiolase | # | 1.62 |
| PAAG_01310 | 2-oxoisovalerate dehydrogenase subunit alpha | # | 1.65 |
| PAAG_05984 | glutaryl CoA dehydrogenase | # | 1.38 |
| PAAG_03330 | dihydrolipoyl dehydrogenase | # | 1.26 |
| PAAG_01534 | pyruvate dehydrogenase E1 component subunit beta | # | 1.55 |
| PAAG_00050 | pyruvate dehydrogenase protein X component | # | 1.15 ** |
| PAAG_02769 | pyruvate dehydrogenase protein X component | * | * |
| Accession a | Description b | Expression 2 h c | Expression 4 h c |
|---|---|---|---|
| Tricarboxylic-acid pathway | |||
| PAAG_03296 | 3-isopropylmalate dehydratase | # | * |
| PAAG_05150 | ATP citrate synthase subunit 1 | # | 3.97 |
| PAAG_08075 | citrate synthase | # | 2.05 |
| PAAG_00417 | succinyl-CoA ligase subunit alpha | # | 1.65 |
| PAAG_01463 | succinyl CoA ligase subunit beta | # | 3.67 |
| Glyoxylate cycle | |||
| PAAG_04542 | malate synthase | 0.78 | 2.29 |
| PAAG_06951 | isocitrate lyase | # | 0.81 ** |
| Intermediates formation | |||
| PAAG_00468 | 4 aminobutyrate aminotransferase | # | 1.21 |
| PAAG_08718 | succinate semialdehyde dehydrogenase | 1.14 ** | |
| PAAG_06407 | argininosuccinate lyase | # | 1.32 |
| PAAG_03138 | alanine glyoxylate aminotransferase | * | 1.26 |
| PAAG_01002 | NAD specific glutamate dehydrogenase | # | * |
| PAAG_02869 | phosphoglycerate kinase | # | 3.97 |
| Acession a | Description b | Amount (Fmol) | Amount (Ngrams) |
|---|---|---|---|
| H4_HUMAN | Histone H4 | 647.15 | 7.36 |
| TRFL_HUMAN | Lactotransferrin | 404.55 | 32.39 |
| H2B1B_HUMAN | Histone H2B type 1 B | 379.01 | 5.29 |
| HSP7C_HUMAN | Heat shock cognate 71 kDa protein | 328.01 | 23.33 |
| H2A2C_HUMAN | Histone H2A type 2-C | 319.81 | 4.47 |
| PERM_HUMAN | Myeloperoxidase | 234.63 | 19.91 |
| ELNE_HUMAN | Neutrophil elastase | 225.46 | 6.57 |
| RL40_HUMAN | Ubiquitin 60S ribosomal protein L40 | 215.02 | 3.23 |
| SREK1_HUMAN | Splicing regulatory glutamine/lysine-rich protein 1 | 207.21 | 12.32 |
| LYSC_HUMAN | Lysozyme C | 180.92 | 3.07 |
| H33_HUMAN | Histone H3.3 | 144.39 | 2.22 |
| DEF1_HUMAN | Neutrophil defensin 1 | 131.28 | 1.38 |
| H32_HUMAN | Histone H3.2 | 81.10 | 1.25 |
| CAP7_HUMAN | Azurocidin | 78.46 | 2.15 |
| SPIN1_HUMAN | Spindlin-1 | 44.74 | 1.33 |
| 1433Z_HUMAN | 14-3-3 protein zeta/delta | 39.23 | 1.10 |
| PNKP_HUMAN | Bifunctional polynucleotide phosphatase/kinase | 28.24 | 1.63 |
| 1433B_HUMAN | 14-3-3 protein beta/alpha | 25.68 | 0.72 |
| HSP72_HUMAN | Heat shock related 70 kDa protein 2 | 15.92 | 1.12 |
| PYGB_HUMAN | Glycogen phosphorylase, brain form | 6.72 | 0.65 |
| NELFE_HUMAN | Negative elongation factor E | 4.85 | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, L.O.S.; Baeza, L.C.; Pigosso, L.L.; Silva, K.S.F.e.; Pereira, M.; de Carvalho Júnior, M.A.B.; de Almeida Soares, C.M. The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils. J. Fungi 2023, 9, 1088. https://doi.org/10.3390/jof9111088
Silva LOS, Baeza LC, Pigosso LL, Silva KSFe, Pereira M, de Carvalho Júnior MAB, de Almeida Soares CM. The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils. Journal of Fungi. 2023; 9(11):1088. https://doi.org/10.3390/jof9111088
Chicago/Turabian StyleSilva, Lana O’Hara Souza, Lilian Cristiane Baeza, Laurine Lacerda Pigosso, Kleber Santiago Freitas e Silva, Maristela Pereira, Marcos Antonio Batista de Carvalho Júnior, and Célia Maria de Almeida Soares. 2023. "The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils" Journal of Fungi 9, no. 11: 1088. https://doi.org/10.3390/jof9111088
APA StyleSilva, L. O. S., Baeza, L. C., Pigosso, L. L., Silva, K. S. F. e., Pereira, M., de Carvalho Júnior, M. A. B., & de Almeida Soares, C. M. (2023). The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils. Journal of Fungi, 9(11), 1088. https://doi.org/10.3390/jof9111088








