Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts
Abstract
1. Introduction
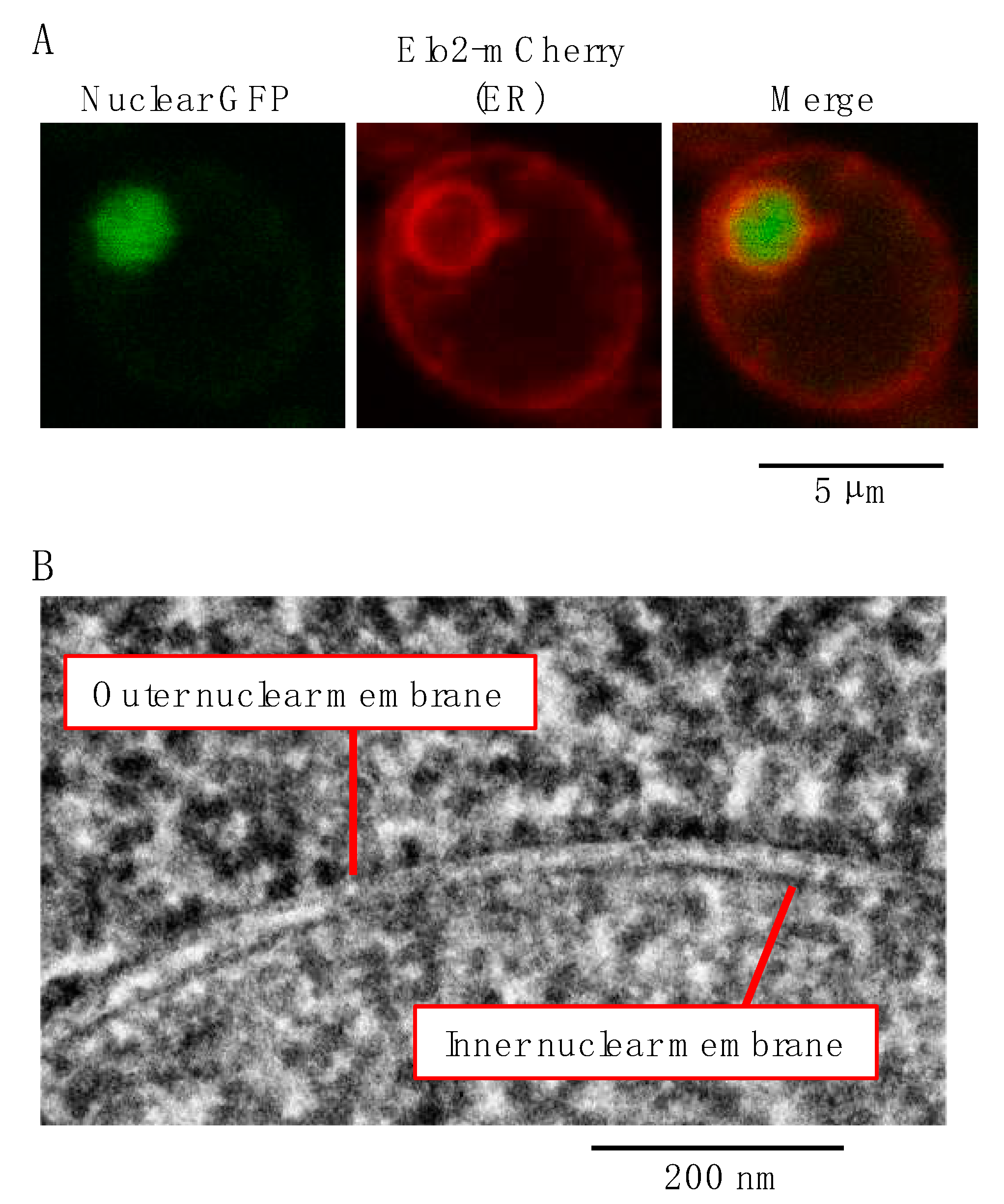
2. The Endoplasmic Reticulum (ER)
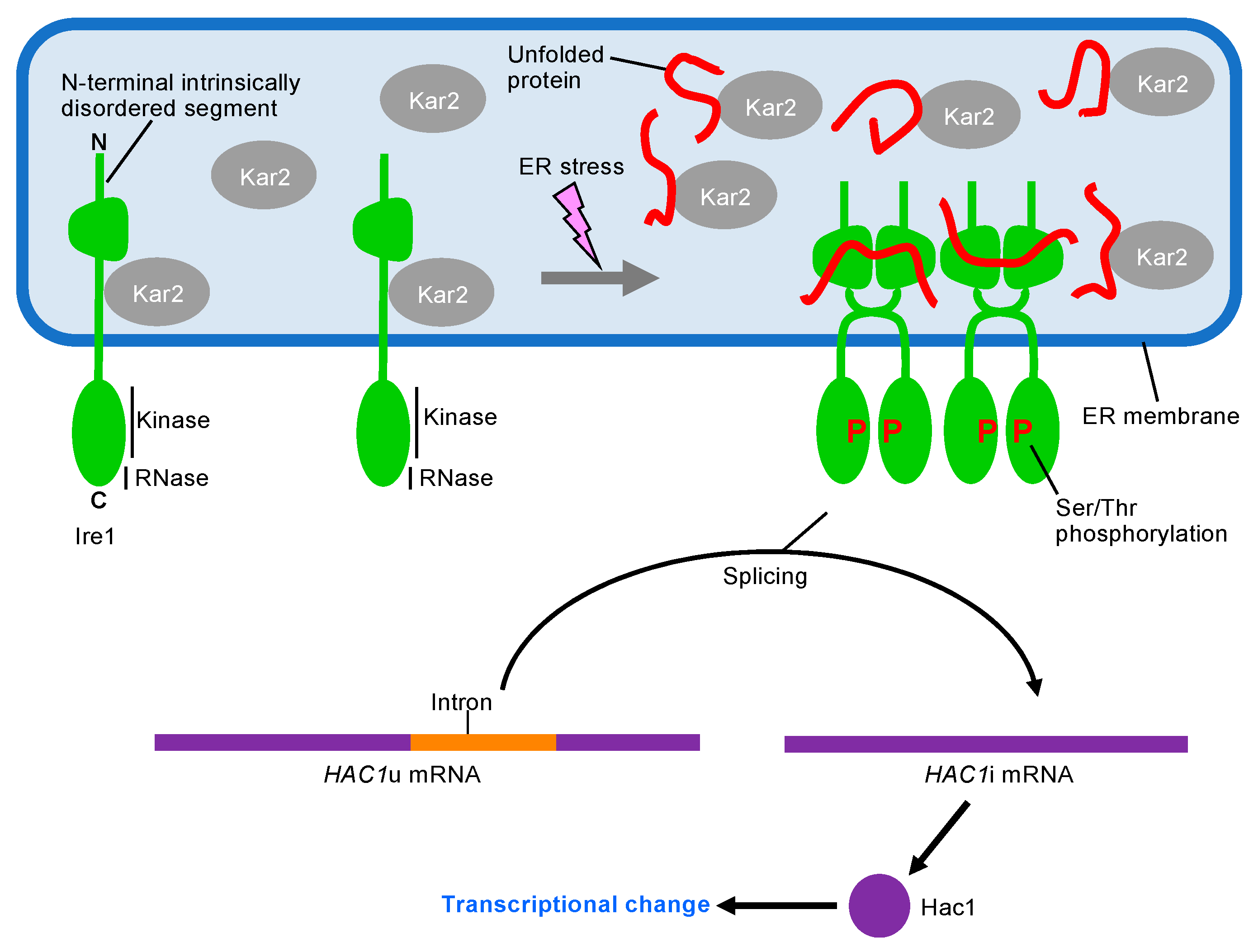
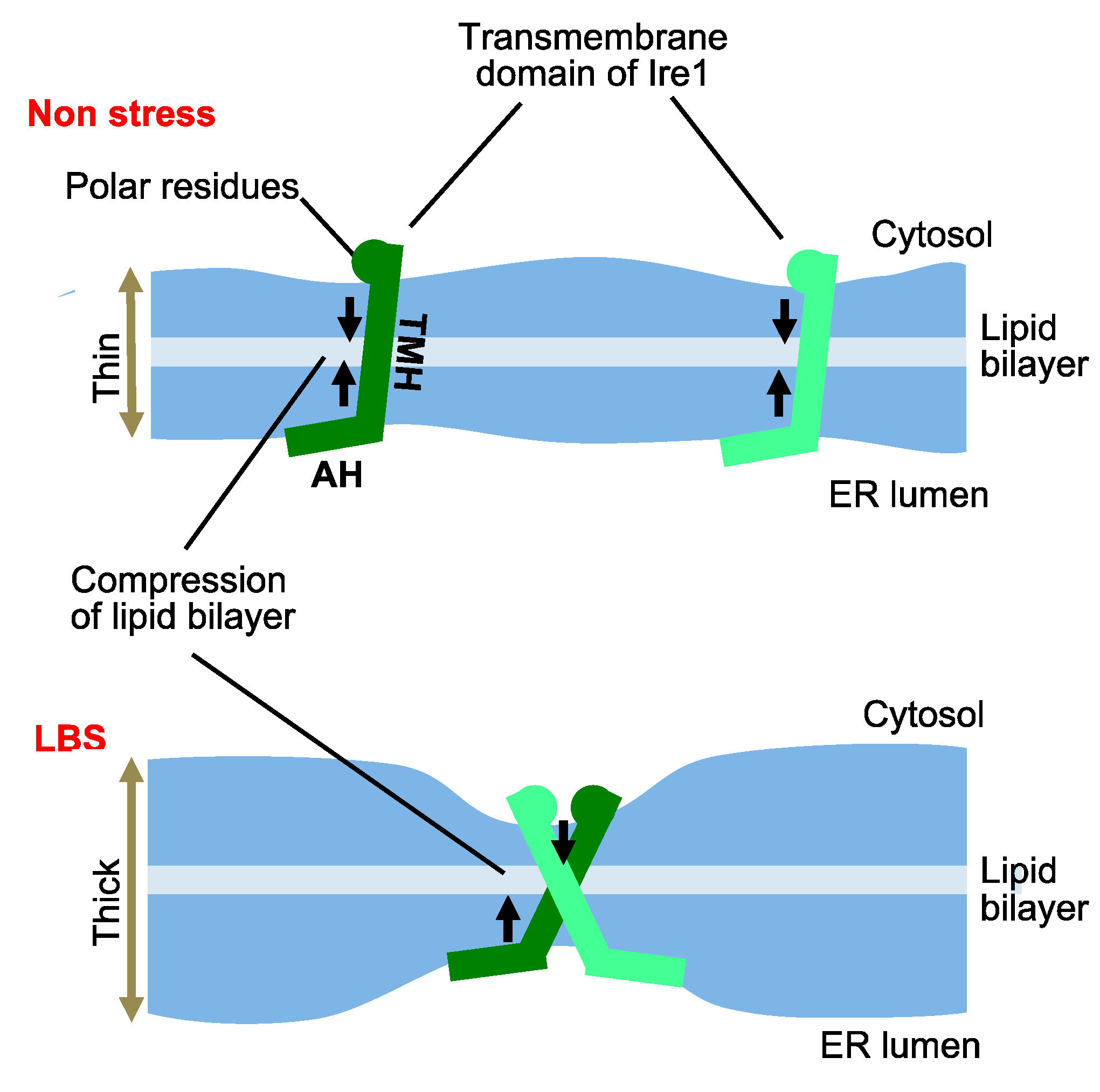
3. UPR Inducing and Repressing Mechanisms in S. cerevisiae Cells
4. Scenes in which the UPR Is Provoked in S. cerevisiae Cells
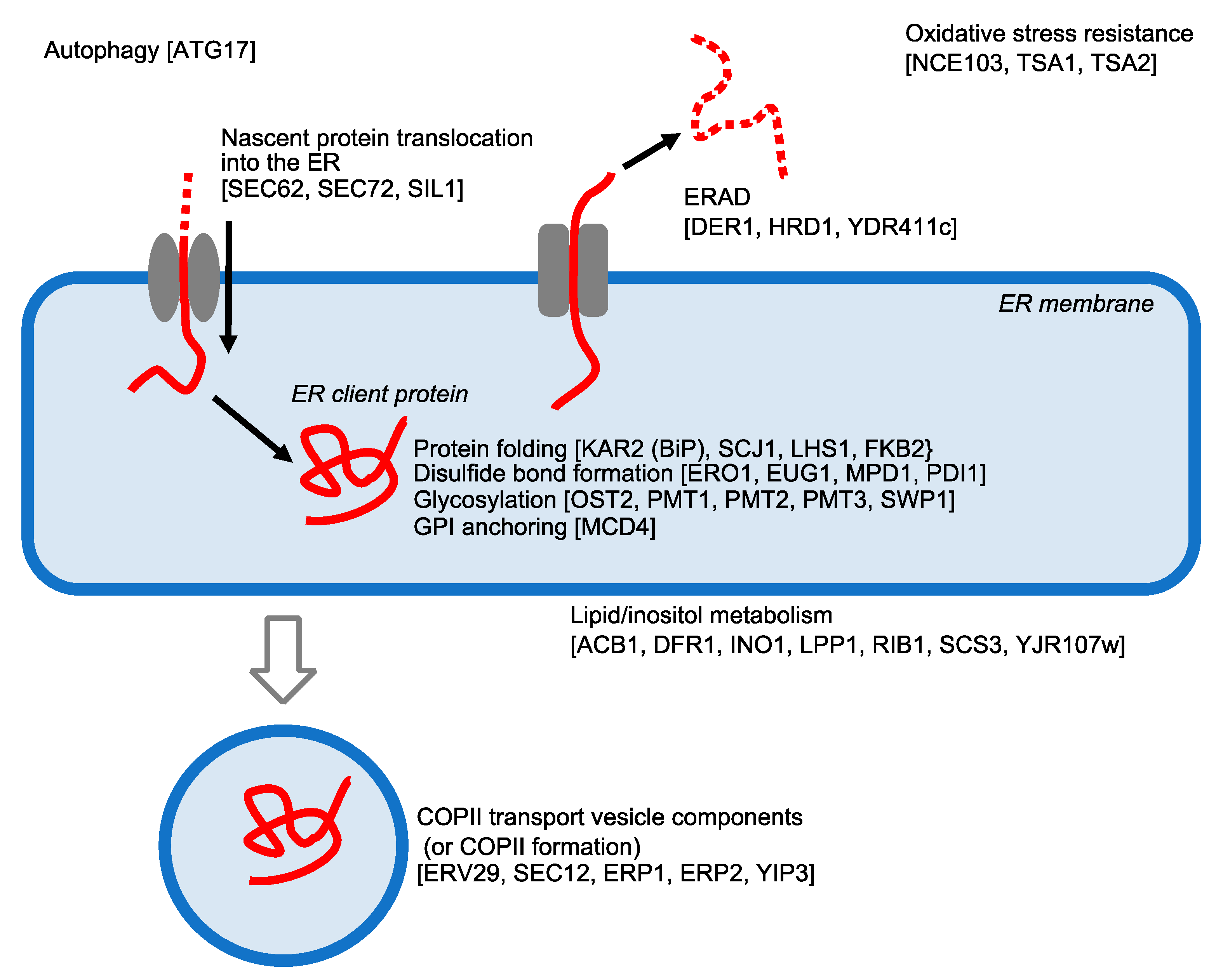
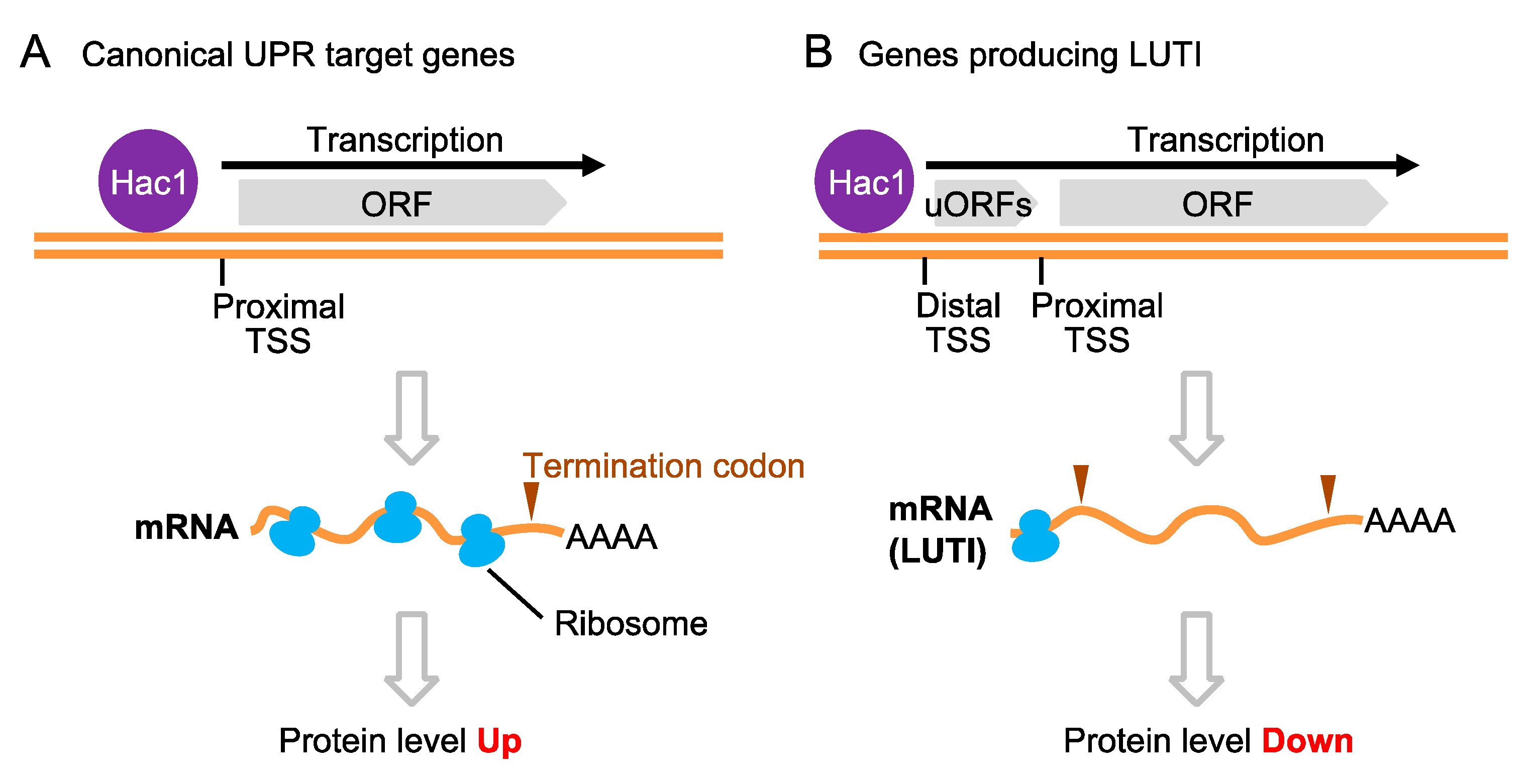
5. UPR Target Genes in S. cerevisiae Cells
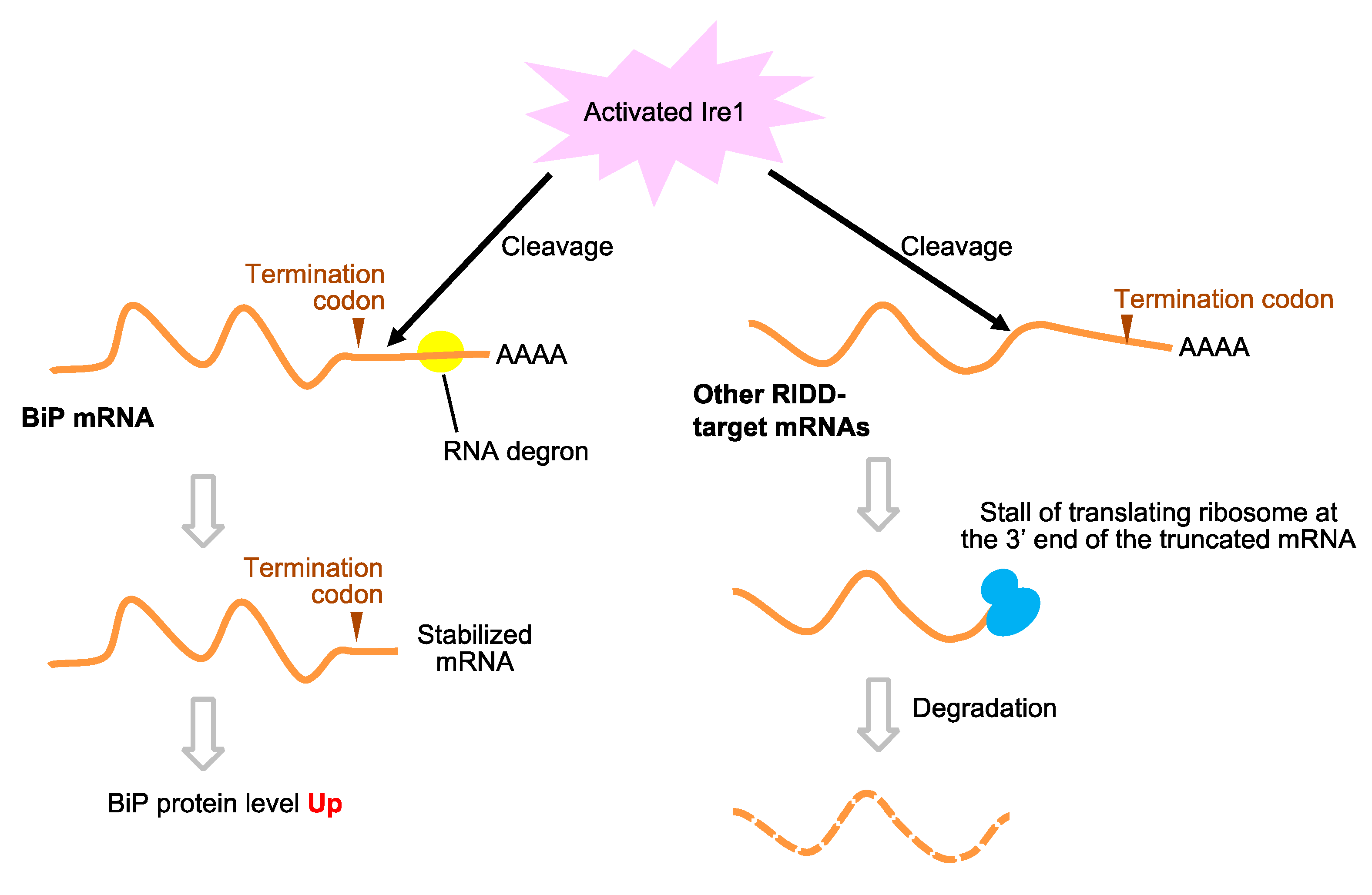
6. The UPR Signaling Pathway in Other Yeast and Fungal Species
7. Engineering of the UPR for Application Purpose
8. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aquilano, K.; Zhou, B.; Brestoff, J.R.; Lettieri-Barbato, D. Multifaceted mitochondrial quality control in brown adipose tissue. Trends Cell Biol. 2022, 33, 517–529. [Google Scholar] [CrossRef]
- Ohsawa, S.; Oku, M.; Yurimoto, H.; Sakai, Y. Regulation of peroxisome homeostasis by post-translational modification in the methylotrophic yeast Komagataella phaffii. Front. Cell Dev. Biol. 2022, 10, 887806. [Google Scholar] [CrossRef]
- Gasser, B.; Prielhofer, R.; Marx, H.; Maurer, M.; Nocon, J.; Steiger, M.; Puxbaum, V.; Sauer, M.; Mattanovich, D. Pichia pastoris: Protein production host and model organism for biomedical research. Future Microbiol. 2013, 8, 191–208. [Google Scholar] [CrossRef]
- Türkanoğlu Özçelik, A.; Yılmaz, S.; Inan, M. Pichia pastoris promoters. Methods Mol. Biol. 2019, 1923, 97–112. [Google Scholar] [CrossRef]
- Preuss, D.; Mulholland, J.; Kaiser, C.A.; Orlean, P.; Albright, C.; Rose, M.D.; Robbins, P.W.; Botstein, D. Structure of the yeast endoplasmic reticulum: Localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 1991, 7, 891–911. [Google Scholar] [CrossRef]
- Venditti, R.; Wilson, C.; De Matteis, M.A. Exiting the ER: What we know and what we don’t. Trends Cell Biol. 2014, 24, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Misra, L.M.; Vogel, J.P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 1989, 57, 1211–1221. [Google Scholar] [CrossRef]
- Normington, K.; Kohno, K.; Kozutsumi, Y.; Gething, M.J.; Sambrook, J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 1989, 57, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Behnke, J.; Feige, M.J.; Hendershot, L.M. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J. Mol. Biol. 2015, 427, 1589–1608. [Google Scholar] [CrossRef] [PubMed]
- Matlack, K.E.; Misselwitz, B.; Plath, K.; Rapoport, T.A. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 1999, 97, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Krshnan, L.; van de Weijer, M.L.; Carvalho, P. Endoplasmic reticulum-associated protein degradation. Cold Spring Harb. Perspect. Biol. 2022, 14, a041247. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.C. Oxidative protein folding fidelity and redoxtasis in the endoplasmic reticulum. Trends Biochem. Sci. 2023, 48, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Graef, M. Lipid droplet-mediated lipid and protein homeostasis in budding yeast. FEBS Lett. 2018, 592, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Strayle, J.; Pozzan, T.; Rudolph, H.K. Steady-state free Ca(2+) in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 1999, 18, 4733–4743. [Google Scholar] [CrossRef]
- Kozutsumi, Y.; Segal, M.; Normington, K.; Gething, M.J.; Sambrook, J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 1988, 332, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Mori, K.; Ma, W.; Gething, M.J.; Sambrook, J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 1993, 74, 743–756. [Google Scholar] [CrossRef]
- Cox, J.S.; Walter, P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Peschek, J.; Walter, P. tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay. Elife 2019, 8, e44199. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Uppala, J.K.; Sathe, L.; Hammond, C.I.; Anshu, A.; Pokkuluri, P.R.; Turk, B.E.; Dey, M. Phosphorylation of Pal2 by the protein kinases Kin1 and Kin2 modulates HAC1 mRNA splicing in the unfolded protein response in yeast. Sci. Signal. 2021, 14, eaaz4401. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Ishiwata-Kimata, Y.; Ito, T.; Hirata, A.; Suzuki, T.; Oikawa, D.; Takeuchi, M.; Kohno, K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007, 179, 75–86. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef]
- Ishiwata-Kimata, Y.; Yamamoto, Y.H.; Takizawa, K.; Kohno, K.; Kimata, Y. F-actin and a type-II myosin are required for efficient clustering of the ER stress sensor Ire1. Cell Struct. Funct. 2013, 38, 135–143. [Google Scholar] [CrossRef]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Zhang, C.; Shokat, K.M.; Stroud, R.M.; Walter, P. The unfolded protein response signals through high-order assembly of Ire1. Nature 2009, 457, 687–693. [Google Scholar] [CrossRef]
- Aragón, T.; van Anken, E.; Pincus, D.; Serafimova, I.M.; Korennykh, A.V.; Rubio, C.A.; Walter, P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 2009, 457, 736–740. [Google Scholar] [CrossRef] [PubMed]
- van Anken, E.; Pincus, D.; Coyle, S.; Aragón, T.; Osman, C.; Lari, F.; Gómez Puerta, S.; Korennykh, A.V.; Walter, P. Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1. Elife 2014, 3, e05031. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Kimata, Y.I.; Shimizu, Y.; Abe, H.; Farcasanu, I.C.; Takeuchi, M.; Rose, M.D.; Kohno, K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 2003, 14, 2559–2569. [Google Scholar] [CrossRef]
- Kimata, Y.; Oikawa, D.; Shimizu, Y.; Ishiwata-Kimata, Y.; Kohno, K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 2004, 167, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Mathuranyanon, R.; Tsukamoto, T.; Takeuchi, A.; Ishiwata-Kimata, Y.; Tsuchiya, Y.; Kohno, K.; Kimata, Y. Tight regulation of the unfolded protein sensor Ire1 by its intramolecularly antagonizing subdomain. J. Cell Sci. 2015, 128, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Shamu, C.E.; Walter, P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996, 15, 3028–3039. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata-Kimata, Y.; Promlek, T.; Kohno, K.; Kimata, Y. BiP-bound and nonclustered mode of Ire1 evokes a weak but sustained unfolded protein response. Genes Cells 2013, 18, 288–301. [Google Scholar] [CrossRef]
- Lee, K.P.; Dey, M.; Neculai, D.; Cao, C.; Dever, T.E.; Sicheri, F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 2008, 132, 89–100. [Google Scholar] [CrossRef]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Stroud, R.M.; Zhang, C.; Shokat, K.M.; Walter, P. Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol. 2011, 9, 48. [Google Scholar] [CrossRef]
- Le, Q.G.; Ishiwata-Kimata, Y.; Phuong, T.H.; Fukunaka, S.; Kohno, K.; Kimata, Y. The ADP-binding kinase region of Ire1 directly contributes to its responsiveness to endoplasmic reticulum stress. Sci. Rep. 2021, 11, 4506. [Google Scholar] [CrossRef]
- Chawla, A.; Chakrabarti, S.; Ghosh, G.; Niwa, M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J. Cell Biol. 2011, 193, 41–50. [Google Scholar] [CrossRef]
- Rubio, C.; Pincus, D.; Korennykh, A.; Schuck, S.; El-Samad, H.; Walter, P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 2011, 193, 171–184. [Google Scholar] [CrossRef]
- Pincus, D.; Chevalier, M.W.; Aragón, T.; van Anken, E.; Vidal, S.E.; El-Samad, H.; Walter, P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010, 8, e1000415. [Google Scholar] [CrossRef]
- Matabishi-Bibi, L.; Challal, D.; Barucco, M.; Libri, D.; Babour, A. Termination of the unfolded protein response is guided by ER stress-induced HAC1 mRNA nuclear retention. Nat. Commun. 2022, 13, 6331. [Google Scholar] [CrossRef]
- Rüegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Sathe, L.; Bolinger, C.; Mannan, M.A.; Dever, T.E.; Dey, M. Evidence that base-pairing interaction between intron and mRNA leader sequences inhibits initiation of HAC1 mRNA translation in yeast. J. Biol. Chem. 2015, 290, 21821–21832. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R.; Aboulhouda, S.; Weinberg, D.E. The fail-safe mechanism of post-transcriptional silencing of unspliced. Elife 2016, 5, e20069. [Google Scholar] [CrossRef]
- Tehfe, A.; Roseshter, T.; Wei, Y.; Xia, X. Does Saccharomyces cerevisiae require specific post-translational silencing against leaky translation of Hac1up? Microorganisms 2021, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Paira, S.; Das, B. Nuclear mRNA degradation tunes the gain of the unfolded protein response in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 1139–1156. [Google Scholar] [CrossRef] [PubMed]
- Uppala, J.K.; Bhattacharjee, S.; Dey, M. Vps34 and TOR kinases coordinate HAC1 mRNA translation in the presence or absence of Ire1-dependent splicing. Mol. Cell. Biol. 2021, 41, e0066220. [Google Scholar] [CrossRef]
- Promlek, T.; Ishiwata-Kimata, Y.; Shido, M.; Sakuramoto, M.; Kohno, K.; Kimata, Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 2011, 22, 3520–3532. [Google Scholar] [CrossRef]
- Cox, J.S.; Chapman, R.E.; Walter, P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 1997, 8, 1805–1814. [Google Scholar] [CrossRef]
- Ho, N.; Yap, W.S.; Xu, J.; Wu, H.; Koh, J.H.; Goh, W.W.B.; George, B.; Chong, S.C.; Taubert, S.; Thibault, G. Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J. Cell Biol. 2020, 219, e201909165. [Google Scholar] [CrossRef]
- Halbleib, K.; Pesek, K.; Covino, R.; Hofbauer, H.F.; Wunnicke, D.; Hänelt, I.; Hummer, G.; Ernst, R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell 2017, 67, 673–684. [Google Scholar] [CrossRef]
- Väth, K.; Mattes, C.; Reinhard, J.; Covino, R.; Stumpf, H.; Hummer, G.; Ernst, R. Cysteine cross-linking in native membranes establishes the transmembrane architecture of Ire1. J. Cell Biol. 2021, 220, e202011078. [Google Scholar] [CrossRef]
- Korennykh, A.V.; Korostelev, A.A.; Egea, P.F.; Finer-Moore, J.; Stroud, R.M.; Zhang, C.; Shokat, K.M.; Walter, P. Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 2011, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Pineau, L.; Colas, J.; Dupont, S.; Beney, L.; Fleurat-Lessard, P.; Berjeaud, J.M.; Bergès, T.; Ferreira, T. Lipid-induced ER stress: Synergistic effects of sterols and saturated fatty acids. Traffic 2009, 10, 673–690. [Google Scholar] [CrossRef]
- Micoogullari, Y.; Basu, S.S.; Ang, J.; Weisshaar, N.; Schmitt, N.D.; Abdelmoula, W.M.; Lopez, B.; Agar, J.N.; Agar, N.; Hanna, J. Dysregulation of very-long-chain fatty acid metabolism causes membrane saturation and induction of the unfolded protein response. Mol. Biol. Cell 2020, 31, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Thibault, G.; Shui, G.; Kim, W.; McAlister, G.C.; Ismail, N.; Gygi, S.P.; Wenk, M.R.; Ng, D.T. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell 2012, 48, 16–27. [Google Scholar] [CrossRef]
- Boumann, H.A.; Gubbens, J.; Koorengevel, M.C.; Oh, C.S.; Martin, C.E.; Heck, A.J.; Patton-Vogt, J.; Henry, S.A.; de Kruijff, B.; de Kroon, A.I. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: Evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol. Biol. Cell 2006, 17, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata-Kimata, Y.; Le, Q.G.; Kimata, Y. Induction and aggravation of the endoplasmic-reticulum stress by membrane-lipid metabolic intermediate phosphatidyl-N-monomethylethanolamine. Front. Cell Dev. Biol. 2021, 9, 743018. [Google Scholar] [CrossRef]
- Tran, D.M.; Takagi, H.; Kimata, Y. Categorization of endoplasmic reticulum stress as accumulation of unfolded proteins or membrane lipid aberrancy using yeast Ire1 mutants. Biosci. Biotechnol. Biochem. 2019, 83, 326–329. [Google Scholar] [CrossRef]
- Phuong, H.T.; Ishiwata-Kimata, Y.; Kimata, Y. An ER-accumulated mutant of yeast Pma1 causes membrane-related stress to induce the unfolded protein response. Biochem. Biophys. Res. Commun. 2023, 667, 58–63. [Google Scholar] [CrossRef]
- Miyagawa, K.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Ethanol stress impairs protein folding in the endoplasmic reticulum and activates Ire1 in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2014, 78, 1389–1391. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Tapia, E.; Querol, A.; Pérez-Torrado, R. Membrane fluidification by ethanol stress activates unfolded protein response in yeasts. Microb. Biotechnol. 2018, 11, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.M.; Ishiwata-Kimata, Y.; Mai, T.C.; Kubo, M.; Kimata, Y. The unfolded protein response alongside the diauxic shift of yeast cells and its involvement in mitochondria enlargement. Sci. Rep. 2019, 9, 12780. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.G.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Cadmium impairs protein folding in the endoplasmic reticulum and induces the unfolded protein response. FEMS Yeast Res. 2016, 16, fow049. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, S.R.; Fazio, E.N.; Etedali-Zadeh, P.; Genereaux, J.; Duennwald, M.L.; Lajoie, P. A functional unfolded protein response is required for chronological aging in Saccharomyces cerevisiae. Curr. Genet. 2020, 66, 263–277. [Google Scholar] [CrossRef]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Kimata, Y.; Ishiwata-Kimata, Y.; Yamada, S.; Kohno, K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells 2006, 11, 59–69. [Google Scholar] [CrossRef]
- Fordyce, P.M.; Pincus, D.; Kimmig, P.; Nelson, C.S.; El-Samad, H.; Walter, P.; DeRisi, J.L. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl. Acad. Sci. USA 2012, 109, E3084–E3093. [Google Scholar] [CrossRef]
- Friedlander, R.; Jarosch, E.; Urban, J.; Volkwein, C.; Sommer, T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2000, 2, 379–384. [Google Scholar] [CrossRef]
- Schuck, S.; Prinz, W.A.; Thorn, K.S.; Voss, C.; Walter, P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 2009, 187, 525–536. [Google Scholar] [CrossRef]
- Bernales, S.; Schuck, S.; Walter, P. ER-phagy: Selective autophagy of the endoplasmic reticulum. Autophagy 2007, 3, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, N.; Mori, K. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells 2004, 9, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Van Dalfsen, K.M.; Hodapp, S.; Keskin, A.; Otto, G.M.; Berdan, C.A.; Higdon, A.; Cheunkarndee, T.; Nomura, D.K.; Jovanovic, M.; Brar, G.A. Global proteome remodeling during ER stress involves Hac1-driven expression of long undecoded transcript isoforms. Dev. Cell 2018, 46, 219–235. [Google Scholar] [CrossRef]
- Matsuki, Y.; Saito, T.; Nakano, Y.; Hashimoto, S.; Matsuo, Y.; Inada, T. Crucial role of leaky initiation of uORF3 in the downregulation of HNT1 by ER stress. Biochem. Biophys. Res. Commun. 2020, 528, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Patil, C.K.; DeRisi, J.; Walter, P. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol. 2005, 6, R3. [Google Scholar] [CrossRef]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef]
- Tam, A.B.; Koong, A.C.; Niwa, M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014, 9, 850–858. [Google Scholar] [CrossRef]
- Fauzee, Y.N.B.M.; Yoshida, Y.; Kimata, Y. Endoplasmic stress sensor Ire1 is involved in cytosolic/nuclear protein quality control in Pichia pastoris cells independent of HAC1. Front. Microbiol. 2023, 14, 1157146. [Google Scholar] [CrossRef]
- Bashir, S.; Banday, M.; Qadri, O.; Bashir, A.; Hilal, N.; Nida-I-Fatima; Rader, S.; Fazili, K.M. The molecular mechanism and functional diversity of UPR signaling sensor IRE1. Life Sci. 2021, 265, 118740. [Google Scholar] [CrossRef]
- Li, Z.; Howell, S.H. Review: The two faces of IRE1 and their role in protecting plants from stress. Plant Sci. 2021, 303, 110758. [Google Scholar] [CrossRef]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Maurel, M.; Chevet, E.; Tavernier, J.; Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014, 39, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kimmig, P.; Diaz, M.; Zheng, J.; Williams, C.C.; Lang, A.; Aragón, T.; Li, H.; Walter, P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife 2012, 1, e00048. [Google Scholar] [CrossRef] [PubMed]
- Guydosh, N.R.; Kimmig, P.; Walter, P.; Green, R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in. Elife 2017, 6, e29216. [Google Scholar] [CrossRef]
- Li, W.; Okreglak, V.; Peschek, J.; Kimmig, P.; Zubradt, M.; Weissman, J.S.; Walter, P. Engineering ER-stress dependent non-conventional mRNA splicing. Elife 2018, 7, e35388. [Google Scholar] [CrossRef]
- Li, W.; Crotty, K.; Garrido Ruiz, D.; Voorhies, M.; Rivera, C.; Sil, A.; Mullins, R.D.; Jacobson, M.P.; Peschek, J.; Walter, P. Protomer alignment modulates specificity of RNA substrate recognition by Ire1. Elife 2021, 10, e67425. [Google Scholar] [CrossRef]
- Zhao, D.; Zou, C.X.; Liu, X.M.; Jiang, Z.D.; Yu, Z.Q.; Suo, F.; Du, T.Y.; Dong, M.Q.; He, W.; Du, L.L. A UPR-induced soluble ER-phagy receptor acts with VAPs to confer ER stress resistance. Mol. Cell 2020, 79, 963–977. [Google Scholar] [CrossRef]
- Whyteside, G.; Nor, R.M.; Alcocer, M.J.; Archer, D.B. Activation of the unfolded protein response in Pichia pastoris requires splicing of a HAC1 mRNA intron and retention of the C-terminal tail of Hac1p. FEBS Lett. 2011, 585, 1037–1041. [Google Scholar] [CrossRef][Green Version]
- Fauzee, Y.N.B.M.; Taniguchi, N.; Ishiwata-Kimata, Y.; Takagi, H.; Kimata, Y. The unfolded protein response in Pichia pastoris without external stressing stimuli. FEMS Yeast Res. 2020, 20, foaa053. [Google Scholar] [CrossRef]
- Moon, H.Y.; Cheon, S.A.; Kim, H.; Agaphonov, M.O.; Kwon, O.; Oh, D.B.; Kim, J.Y.; Kang, H.A. Hansenula polymorpha Hac1p is critical to protein N-glycosylation activity modulation, as revealed by functional and transcriptomic analyses. Appl. Environ. Microbiol. 2015, 81, 6982–6993. [Google Scholar] [CrossRef]
- Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The unfolded protein response pathway in the yeast Kluyveromyces lactis. A Comparative view among yeast species. Cells 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Cheon, S.A.; Kang, H.A.; Kim, J.Y. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010, 27, 443–452. [Google Scholar] [CrossRef]
- Wimalasena, T.T.; Enjalbert, B.; Guillemette, T.; Plumridge, A.; Budge, S.; Yin, Z.; Brown, A.J.; Archer, D.B. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 2008, 45, 1235–1247. [Google Scholar] [CrossRef] [PubMed]
- Iracane, E.; Donovan, P.D.; Ola, M.; Butler, G.; Holland, L.M. Identification of an exceptionally long intron in the HAC1 gene of Candida parapsilosis. mSphere 2018, 3, e00532-18. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, M.; Valkonen, M.; Penttilä, M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003, 47, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Lang, E.A.S.; Sanches, P.R.; Peres, N.T.A.; Oliveira, V.M.; Fachin, A.L.; Rossi, A.; Martinez-Rossi, N.M. HacA governs virulence traits and adaptive stress responses in Trichophyton rubrum. Front. Microbiol. 2020, 11, 193. [Google Scholar] [CrossRef]
- Mulder, H.J.; Nikolaev, I. HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot. Cell 2009, 8, 665–675. [Google Scholar] [CrossRef]
- Tanaka, M.; Shintani, T.; Gomi, K. Unfolded protein response is required for Aspergillus oryzae growth under conditions inducing secretory hydrolytic enzyme production. Fungal Genet. Biol. 2015, 85, 1–6. [Google Scholar] [CrossRef]
- Yokota, J.I.; Shiro, D.; Tanaka, M.; Onozaki, Y.; Mizutani, O.; Kakizono, D.; Ichinose, S.; Shintani, T.; Gomi, K.; Shintani, T. Cellular responses to the expression of unstable secretory proteins in the filamentous fungus Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2017, 101, 2437–2446. [Google Scholar] [CrossRef]
- Graf, A.; Gasser, B.; Dragosits, M.; Sauer, M.; Leparc, G.G.; Tüchler, T.; Kreil, D.P.; Mattanovich, D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genom. 2008, 9, 390. [Google Scholar] [CrossRef]
- Ata, Ö.; Ergün, B.G.; Fickers, P.; Heistinger, L.; Mattanovich, D.; Rebnegger, C.; Gasser, B. What makes Komagataella phaffii non-conventional? FEMS Yeast Res. 2021, 21, foab059. [Google Scholar] [CrossRef] [PubMed]
- Sircaik, S.; Román, E.; Bapat, P.; Lee, K.K.; Andes, D.R.; Gow, N.A.R.; Nobile, C.J.; Pla, J.; Panwar, S.L. The protein kinase Ire1 impacts pathogenicity of Candida albicans by regulating homeostatic adaptation to endoplasmic reticulum stress. Cell. Microbiol. 2021, 23, e13307. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Zavala, B.; Krüger, I.; Dunker, C.; Jacobsen, I.D.; Morschhäuser, J. The protein kinase Ire1 has a Hac1-independent essential role in iron uptake and virulence of Candida albicans. PLoS Pathog. 2022, 18, e1010283. [Google Scholar] [CrossRef]
- Richie, D.L.; Hartl, L.; Aimanianda, V.; Winters, M.S.; Fuller, K.K.; Miley, M.D.; White, S.; McCarthy, J.W.; Latgé, J.P.; Feldmesser, M.; et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009, 5, e1000258. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Krishnan, K.; Richie, D.L.; Aimanianda, V.; Hartl, L.; Grahl, N.; Powers-Fletcher, M.V.; Zhang, M.; Fuller, K.K.; Nierman, W.C.; et al. HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog. 2011, 7, e1002330. [Google Scholar] [CrossRef]
- Guirao-Abad, J.P.; Weichert, M.; Askew, D.S. Cell death induction in Aspergillus fumigatus: Accentuating drug toxicity through inhibition of the unfolded protein response (UPR). Curr. Res. Microb. Sci. 2022, 3, 100119. [Google Scholar] [CrossRef] [PubMed]
- Cross, B.C.; Bond, P.J.; Sadowski, P.G.; Jha, B.K.; Zak, J.; Goodman, J.M.; Silverman, R.H.; Neubert, T.A.; Baxendale, I.R.; Ron, D.; et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. USA 2012, 109, E869–E878. [Google Scholar] [CrossRef]
- Guirao-Abad, J.P.; Weichert, M.; Albee, A.; Deck, K.; Askew, D.S. A human IRE1 inhibitor blocks the unfolded protein response in the pathogenic fungus Aspergillus fumigatus and suggests noncanonical functions within the pathway. mSphere 2020, 5, e00879-20. [Google Scholar] [CrossRef]
- Cheon, S.A.; Jung, K.W.; Chen, Y.L.; Heitman, J.; Bahn, Y.S.; Kang, H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002177. [Google Scholar] [CrossRef]
- Cheon, S.A.; Jung, K.W.; Bahn, Y.S.; Kang, H.A. The unfolded protein response (UPR) pathway in Cryptococcus. Virulence 2014, 5, 341–350. [Google Scholar] [CrossRef]
- Jung, K.W.; Lee, K.T.; Averette, A.F.; Hoy, M.J.; Everitt, J.; Heitman, J.; Bahn, Y.S. Evolutionarily conserved and divergent roles of unfolded protein response (UPR) in the pathogenic Cryptococcus species complex. Sci. Rep. 2018, 8, 8132. [Google Scholar] [CrossRef]
- Guerfal, M.; Ryckaert, S.; Jacobs, P.P.; Ameloot, P.; Van Craenenbroeck, K.; Derycke, R.; Callewaert, N. The HAC1 gene from Pichia pastoris: Characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb. Cell Factories 2010, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Raschmanová, H.; Weninger, A.; Knejzlík, Z.; Melzoch, K.; Kovar, K. Engineering of the unfolded protein response pathway in Pichia pastoris: Enhancing production of secreted recombinant proteins. Appl. Microbiol. Biotechnol. 2021, 105, 4397–4414. [Google Scholar] [CrossRef] [PubMed]
- Bankefa, O.E.; Wang, M.; Zhu, T.; Li, Y. Hac1p homologues from higher eukaryotes can improve the secretion of heterologous proteins in the yeast Pichia pastoris. Biotechnol. Lett. 2018, 40, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Zahrl, R.J.; Prielhofer, R.; Burgard, J.; Mattanovich, D.; Gasser, B. Synthetic activation of yeast stress response improves secretion of recombinant proteins. Nat. Biotechnol. 2023, 73, 19–28. [Google Scholar] [CrossRef]
- de Ruijter, J.C.; Frey, A.D. Analysis of antibody production in Saccharomyces cerevisiae: Effects of ER protein quality control disruption. Appl. Microbiol. Biotechnol. 2015, 99, 9061–9071. [Google Scholar] [CrossRef][Green Version]
- Valkonen, M.; Penttilä, M.; Saloheimo, M. Effects of inactivation and constitutive expression of the unfolded- protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 2065–2072. [Google Scholar] [CrossRef]
- Bao, C.; Li, J.; Chen, H.; Sun, Y.; Wang, G.; Chen, G.; Zhang, S. Expression and function of an Hac1-regulated multi-copy xylanase gene in Saccharomyces cerevisiae. Sci. Rep. 2020, 10, 11686. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar] [CrossRef]
- Nguyen, P.T.M.; Ishiwata-Kimata, Y.; Kimata, Y. Fast-growing Saccharomyces cerevisiae cells with a constitutive unfolded protein response and their potential for lipidic molecule production. Appl. Environ. Microbiol. 2022, 88, e0108322. [Google Scholar] [CrossRef]
- Lin, Y.; Feng, Y.; Zheng, L.; Zhao, M.; Huang, M. Improved protein production in yeast using cell engineering with genes related to a key factor in the unfolded protein response. Metab. Eng. 2023, 77, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Zhang, L.; Zhu, S.; Yuan, W.; Hang, J.; Yin, D.; Tang, X.; Zheng, J.; Wang, Z.; Sun, J. Overexpression of the transcription factor HAC1 improves nerolidol production in engineered yeast. Enzym. Microb. Technol. 2020, 134, 109485. [Google Scholar] [CrossRef] [PubMed]
- Thibault, G.; Ismail, N.; Ng, D.T. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 20597–20602. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishiwata-Kimata, Y.; Kimata, Y. Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts. J. Fungi 2023, 9, 989. https://doi.org/10.3390/jof9100989
Ishiwata-Kimata Y, Kimata Y. Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts. Journal of Fungi. 2023; 9(10):989. https://doi.org/10.3390/jof9100989
Chicago/Turabian StyleIshiwata-Kimata, Yuki, and Yukio Kimata. 2023. "Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts" Journal of Fungi 9, no. 10: 989. https://doi.org/10.3390/jof9100989
APA StyleIshiwata-Kimata, Y., & Kimata, Y. (2023). Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts. Journal of Fungi, 9(10), 989. https://doi.org/10.3390/jof9100989




