Cryphonectria parasitica Detections in England, Jersey, and Guernsey during 2020–2023 Reveal Newly Affected Areas and Infections by the CHV1 Mycovirus
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling and Isolation
2.2. Fungal Detection by Real-Time PCR from Plant Material
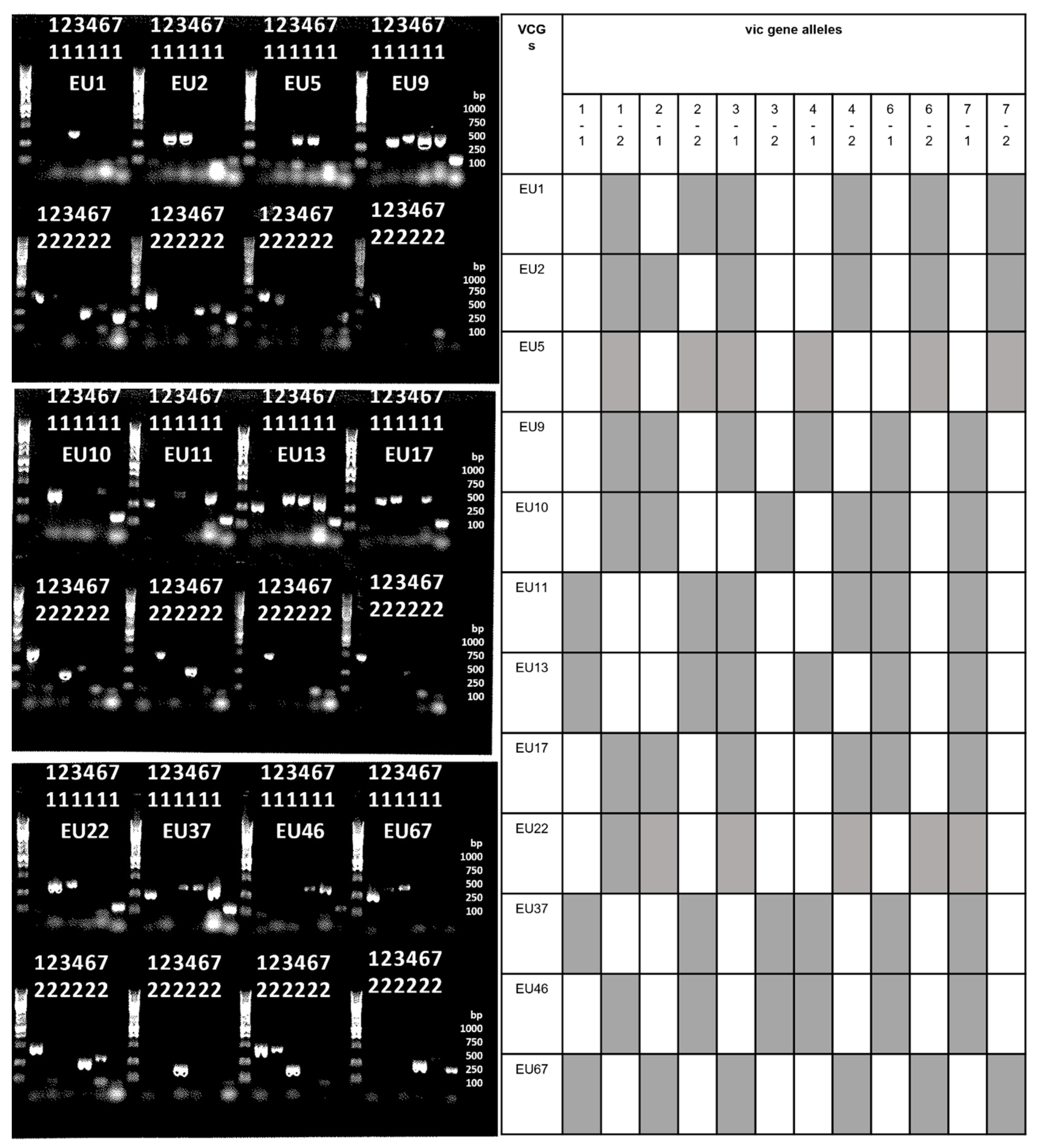
2.3. Vegetative Incompatibility Genes (Vic) Profiles
2.4. Mating Types
2.5. Cryphonectria Hypovirus 1 (CHV1) Concentration and Sequencing
3. Results
3.1. Isolations
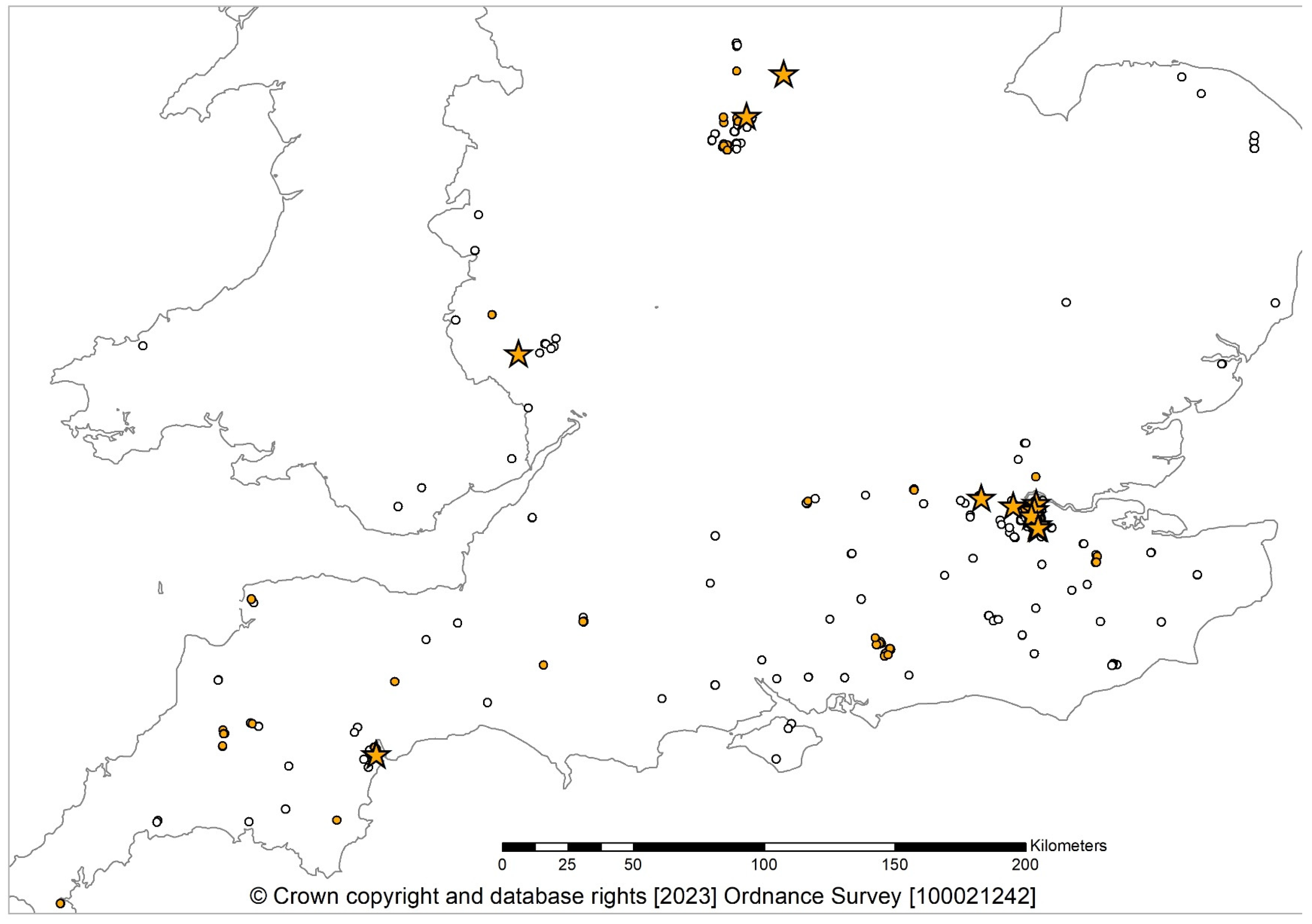
3.2. Fungal Detections
3.3. VCGs
3.4. Perithecia Occurrence
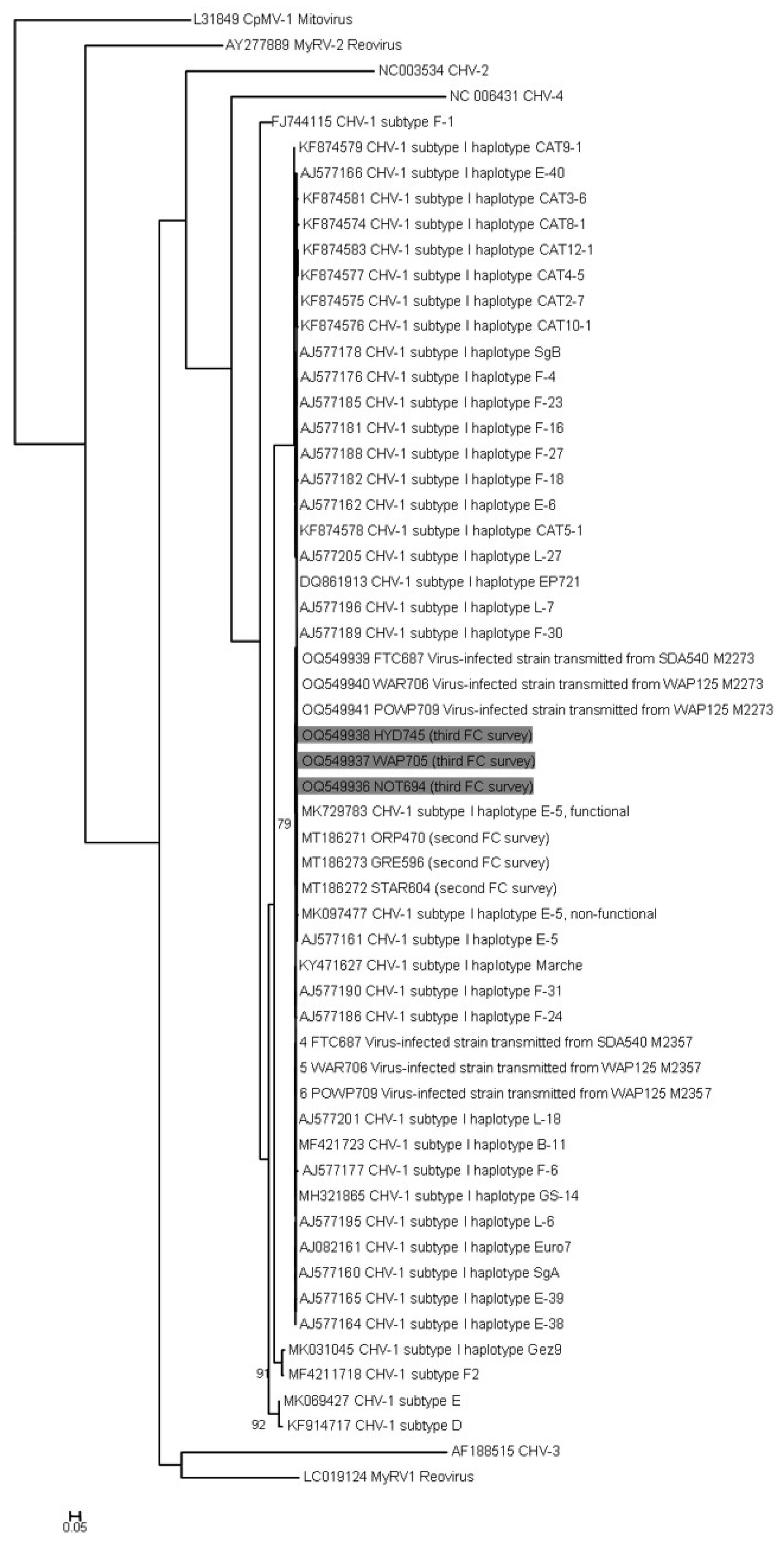
3.5. Hypovirus Detection and Sequencing
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EPPO. Cryphonectria parasitica. EPPO Bull. 2005, 35, 295–298. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Dynek, J.N.; Hillman, B.I.; Milgroom, M.G. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res. 2007, 111, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Roane, M.K.; Griffin, G.J.; Elkins, J.R. Chestnut Blight, Other Endothia Diseases, and the Genus Endothia; APS Monograph Series 53; APS Press: St. Paul, MN, USA, 1986. [Google Scholar]
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Robin, C.; Heiniger, U. Chestnut blight in Europe: Diversity of Cryphonectria parasitica, hypovirulence and biocontrol. For. Snow Landsc. Res. 2001, 76, 361–367. [Google Scholar]
- Hunter, G.; Wylder, B.; Jones, B.; Webber, J.F. First finding of Cryphonectria parasitica causing chestnut blight on Castanea sativa trees in England. New Dis. Rep. 2013, 27, 1. [Google Scholar] [CrossRef]
- Forestry Commission. Sweet Chestnut Blight (Cryphonectria parasitica). 2018. Available online: https://www.forestresearch.gov.uk/tools-and-resources/fthr/pest-and-disease-resources/sweet-chestnut-blight-cryphonectria-parasitica/ (accessed on 10 September 2023).
- Pérez-Sierra, A.; Romon-Ochoa, P.; Gorton, C.; Lewis, A.; Rees, H.; van der Linde, S.; Webber, J. High vegetative compatibility diversity of Cryphonectria parasitica infecting sweet chestnut (Castanea sativa) in Britain indicates multiple pathogen introductions. Plant Pathol. 2019, 68, 727–737. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Gorton, C.; Lewis, A.; van der Linde, S.; Webber, J.; Pérez-Sierra, A. Hypovirulent effect of the Cryphonectria Hypovirus 1 in British isolates of Cryphonectria parasitica. Pest Manag. Sci. 2020, 76, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Romon-Ochoa, P.; Kranjec Orlovic, J.; Gorton, C.; Lewis, A.; van der Linde, S.; Pérez-Sierra, A. New detections of chestnut blight in Great Britain during 2019–2020 reveal high Cryphonectria parasitica diversity and limited spread of the disease. Plant Pathol. 2021, 71, 793–804. [Google Scholar] [CrossRef]
- McGuire, I.C.; Marra, R.E.; Milgroom, M.G. Mating-type heterokaryosis and selfing in Cryphonectria parasitica. Fungal Genet. Biol. 2004, 41, 521–533. [Google Scholar] [CrossRef]
- Lione, G.; Brescia, F.; Giordano, L.; Gonthier, P. Effects of seasonality and climate on the propagule deposition patterns of the chestnut blight pathogen Cryphonectria parasitica in orchards of the Alpine district of North Western Italy. Agriculture 2022, 12, 644. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Lione, G.; Giordano, L.; Turina, M.; Gonthier, P. Hail-induced infections of the chestnut blight pathogen Cryphonectria parasitica depend on wound size and may lead to severe diebacks. Phytopathology 2020, 110, 1280–1293. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Forster, B. Chestnut gall wasp (Dryocosmus kuriphilus) infestations: New opportunities for the chestnut blight fungus Cryphonectria parasitica? New Dis. Rep. 2011, 23, 35. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.; van der Linde, S.; Romon-Ochoa, P.; Jones, B.; Gorton, C. First report of Cryphonectria parasitica on abandoned galls of Dryocosmus kuriphilus on sweet chestnut in the UK. New Dis. Rep. 2020, 41, 34. [Google Scholar] [CrossRef]
- Hillman, B.I.; Suzuki, N. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 2004, 63, 423–472. [Google Scholar] [PubMed]
- Fahima, T.; Kazmierczak, P.; Hansen, D.R.; Pfeiffer, P.; van Alfen, N.K. Membrane-associated replication of an un-encapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 1993, 195, 81–89. [Google Scholar]
- Chandelier, A.; Massot, M.; Fabreguettes, O.; Gischer, F.; Teng, F.; Robin, C. Early detection of Cryphonectria parasitica by real-time PCR. Eur. J. Plant Pathol. 2019, 153, 135–152. [Google Scholar] [CrossRef]
- Cornejo, C.; Sever, B.; Kupper, Q.; Prospero, S.; Rigling, D. A multiplexed genotyping assay to determine vegetative incompatibility and mating type in Cryphonectria parasitica. Eur. J. Plant Pathol. 2019, 155, 81–91. [Google Scholar] [CrossRef]
- Cortesi, P.; Milgroom, M.G. Genetics of vegetative incompatibility in Cryphonectria parasitica. Appl. Environ. Microbiol. 1998, 64, 2988–2994. [Google Scholar] [CrossRef]
- Gobbin, D.; Hoegger, P.J.; Heiniger, U.; Rigling, D. Sequence variation and evolution of Cryphonectria hypovirus 1 (CHV1) in Europe. Virus Res. 2003, 97, 39–46. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Glez-Peña, D.; Gomez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented format conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likehood phylogenies: Assessing the performance of PhyML 3. 0. Syst. Biol. 2006, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Romon-Ochoa, P.; Forster, J.; Chitty, R.; Gorton, C.; Lewis, A.; Eacock, A.; Kupper, Q.; Rigling, D.; Pérez-Sierra, A. Canker development and biocontrol potential of CHV1 infected English isolates of Cryphonectria parasitica is dependent on the virus concentration and the compatibility of the fungal inoculums. Viruses 2022, 14, 2678. [Google Scholar] [CrossRef] [PubMed]
- Milgroom, M.G.; Sotirovski, K.; Spica, D.; Davis, J.E.; Brewer, M.T.; Milev, M.; Cortesi, P. Clonal population structure of the chestnut blight fungus in expanding ranges in southeastern Europe. Mol. Ecol. 2008, 17, 4446–4458. [Google Scholar] [CrossRef] [PubMed]
- Perlerou, C.; Diamandis, S. Identification and geographic distribution of vegetative compatibility types of Cryphonectria parasitica and occurrence of hypovirulence in Greece. For. Pathol. 2006, 36, 413–421. [Google Scholar] [CrossRef]
- Radocz, L. Chestnut blight and the hypovirulence in the Carpathian basin. Acta Hortic. 1998, 494, 501–508. [Google Scholar] [CrossRef]
- Sotirovski, K.; Papazova-Anakieva, I.; Grünwald, N.J.; Milgroom, M.G. Low diversity of vegetative compatibility types and mating type of Cryphonectria parasitica in the southern Balkans. Plant Pathol. 2004, 53, 325–333. [Google Scholar] [CrossRef]
- Milgroom, M.G.; Cortesi, P. Analysis of population structure of the chestnut blight fungus based on vegetative incompatibility genotypes. Proc. Natl. Acad. Sci. USA 1999, 96, 10518–10523. [Google Scholar] [CrossRef]
- Trestic, T.; Uscuplic, M.; Colinas, C.; Rolland, G.; Giraud, A.; Robin, C. Vegetative compatibility type diversity of Cryphonectria parasitica populations in Bosnia-Herzegovina, Spain and France. For. Snow Landsc. Res. 2001, 76, 391–396. [Google Scholar]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Curkovic-Perica, M. Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol. 2011, 60, 752–761. [Google Scholar] [CrossRef]
- Jezic, M.; Mlinarec, J.; Vukovic, R.; Katanic, Z.; Krstin, L.; Nuskern, L.; Poljak, I.; Idzojtic, M.; Tkalec, M.; Curkovic-Perica, M. Changes in Cryphonectria parasitica populations affect natural biological control of chestnut blight. Phytopathology 2018, 108, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Tarcali, G.; Radocz, L. Occurrence of fungus Cryphonectria parasitica (Murr.) Barr on oak trees in the Carpathian-basin. Folia Oecologica 2006, 33, 129–132. [Google Scholar]
- Bazzigher, G. Selection of blight-resistant chestnut trees in Switzerland. Eur. J. For. Pathol. 1981, 11, 199–207. [Google Scholar] [CrossRef]
- Rigling, D.; Borst, N.; Cornejo, C.; Supatashvili, A.; Prospero, S. Genetic and phenotypic characterization of Cryphonectria hypovirus 1 from Eurasian Georgia. Viruses 2018, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Allemann, C.; Hoegger, P.; Heiniger, U.; Rigling, D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 1999, 8, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Robin, C.; Lanz, S.; Soutrenon, A.; Rigling, D. Dominance of natural over released biological control agents of the chestnut blight fungus Cryphonectria parasitica in south-eastern France is associated with fitness-related traits. Biol. Control 2010, 53, 55–61. [Google Scholar] [CrossRef]
- Milgroom, M.G.; Cortesi, P. Biological control of chestnut blight with hypovirulence: A critical analysis. Annu. Rev. Phytopathol. 2004, 42, 311–338. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Smith, O.; Lewis, A.; Kupper, Q.; Shamsi, W.; Rigling, D.; Pérez-Sierra, A.; Ward, L. Temperature effects on the Cryphonectria hypovirus 1 accumulation and recovery within its fungal host, the chestnut blight pathogen Cryphonectria parasitica. Viruses 2023, 15, 1260. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Lewis, A.; Gorton, C.; van der Linde, S.; Pérez-Sierra, A. Effects of growth medium, temperature and mycelium age on CHV-1 accumulation and transmission. For. Ecol. Manag. 2023, 529, 120705. [Google Scholar] [CrossRef]

| County | No. Positive Sites/Total Sites | No. Molecular Detections | Cultured Isolates | No. Isolates per VCG | H’ | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EU1 | EU2 | EU5 | EU9 | EU10 | EU11 | EU13 | EU17 | EU22 | EU37 | EU46 | EU67 | |||||
| Derbyshire | 1/6 | 3 | 3 | 3 | 0 | |||||||||||
| Devon | 4/8 | 13 | 13 | 1 | 11 (1) | 1 | 0.51 | |||||||||
| Kent | 3/5 | 26 | 26 | 1 | 4 | 14 | 7 | 1.06 | ||||||||
| Nottinghamshire | 2/2 | 6 | 6 | 6 (1) | 0 | |||||||||||
| Herefordshire | 1/1 | 3 | 3 | 3 | 0 | |||||||||||
| Leicestershire | 1/1 | 1 | 1 | 1 | 0 | |||||||||||
| London | 10/16 | 45 | 45 | 1 | 39 | 1 | 1 | 1 | 2 (1) | 0.57 | ||||||
| West Sussex | 1/9 | 2 | 2 | 1 | 1 | 0.69 | ||||||||||
| Island of Jersey | 9/20 | 9 | 9 | 1 | 8 | 0.35 | ||||||||||
| Island of Guernsey | 3/4 | 4 | 4 | 1 | 1 | 1 | 1 | 1.38 | ||||||||
| Total | 35/72 | 112 | 112 | 2 | 13 | 1 | 19 | 39 | 1 | 1 | 15 | 1 | 10 | 9 | 1 | |
| VCG | Derbyshire | Devon | Kent | Nottinghamshire | Herefordshire | Leicestershire | London | West Sussex | Island of Jersey | Island of Guernsey | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | MAT-1 | MAT-2 | |
| EU1 | 1 | 1 | ||||||||||||||||||
| EU2 | 3 | 1 | 4 | 3 | 1 | 1 | ||||||||||||||
| EU5 | 1 | |||||||||||||||||||
| EU9 | 11 | 6 | 1 | 1 | ||||||||||||||||
| EU10 | 39 | |||||||||||||||||||
| EU11 | 1 | |||||||||||||||||||
| EU13 | 1 | |||||||||||||||||||
| EU17 | 14 | 1 | ||||||||||||||||||
| EU22 | 1 | |||||||||||||||||||
| EU37 | 7 | 2 | 1 | |||||||||||||||||
| EU46 | 8 | 1 | ||||||||||||||||||
| EU67 | 1 | |||||||||||||||||||
| Total | 3 | - | 2 | 11 | 22 | 4 | 6 | - | 3 | - | 1 | - | 3 | 42 | - | 2 | - | 9 | 1 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romon-Ochoa, P.; Samal, P.; Gorton, C.; Lewis, A.; Chitty, R.; Eacock, A.; Krzywinska, E.; Crampton, M.; Pérez-Sierra, A.; Biddle, M.; et al. Cryphonectria parasitica Detections in England, Jersey, and Guernsey during 2020–2023 Reveal Newly Affected Areas and Infections by the CHV1 Mycovirus. J. Fungi 2023, 9, 1036. https://doi.org/10.3390/jof9101036
Romon-Ochoa P, Samal P, Gorton C, Lewis A, Chitty R, Eacock A, Krzywinska E, Crampton M, Pérez-Sierra A, Biddle M, et al. Cryphonectria parasitica Detections in England, Jersey, and Guernsey during 2020–2023 Reveal Newly Affected Areas and Infections by the CHV1 Mycovirus. Journal of Fungi. 2023; 9(10):1036. https://doi.org/10.3390/jof9101036
Chicago/Turabian StyleRomon-Ochoa, Pedro, Pankajini Samal, Caroline Gorton, Alex Lewis, Ruth Chitty, Amy Eacock, Elzbieta Krzywinska, Michael Crampton, Ana Pérez-Sierra, Mick Biddle, and et al. 2023. "Cryphonectria parasitica Detections in England, Jersey, and Guernsey during 2020–2023 Reveal Newly Affected Areas and Infections by the CHV1 Mycovirus" Journal of Fungi 9, no. 10: 1036. https://doi.org/10.3390/jof9101036
APA StyleRomon-Ochoa, P., Samal, P., Gorton, C., Lewis, A., Chitty, R., Eacock, A., Krzywinska, E., Crampton, M., Pérez-Sierra, A., Biddle, M., Jones, B., & Ward, L. (2023). Cryphonectria parasitica Detections in England, Jersey, and Guernsey during 2020–2023 Reveal Newly Affected Areas and Infections by the CHV1 Mycovirus. Journal of Fungi, 9(10), 1036. https://doi.org/10.3390/jof9101036






