Lignin-Modifying Enzymes in Scedosporium Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolates and Culture Conditions
2.2. Enzymatic Assays
2.2.1. Detection of Peroxidase Activity
2.2.2. Detection of Oxidase Activity
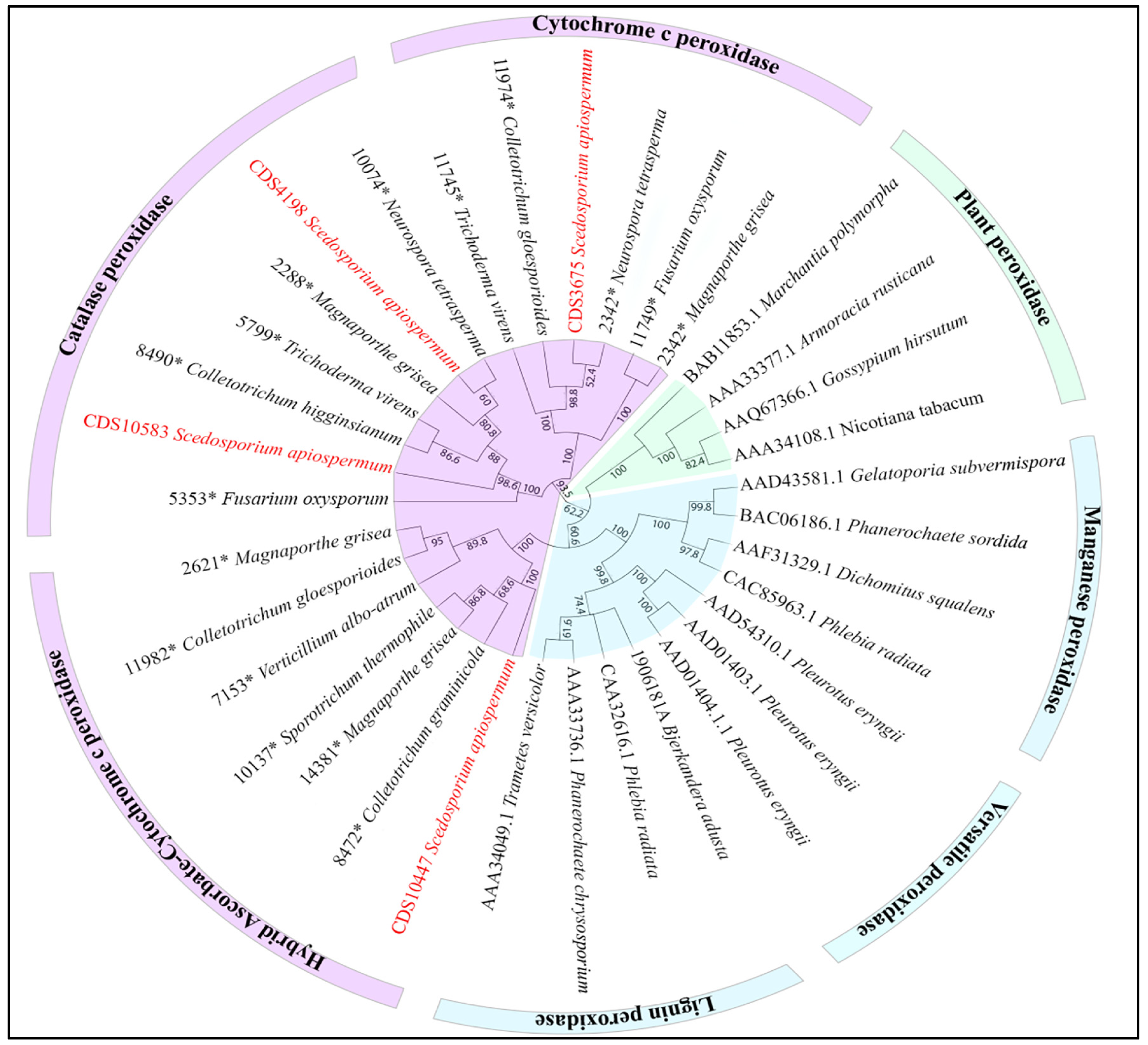
2.3. Genome Mining
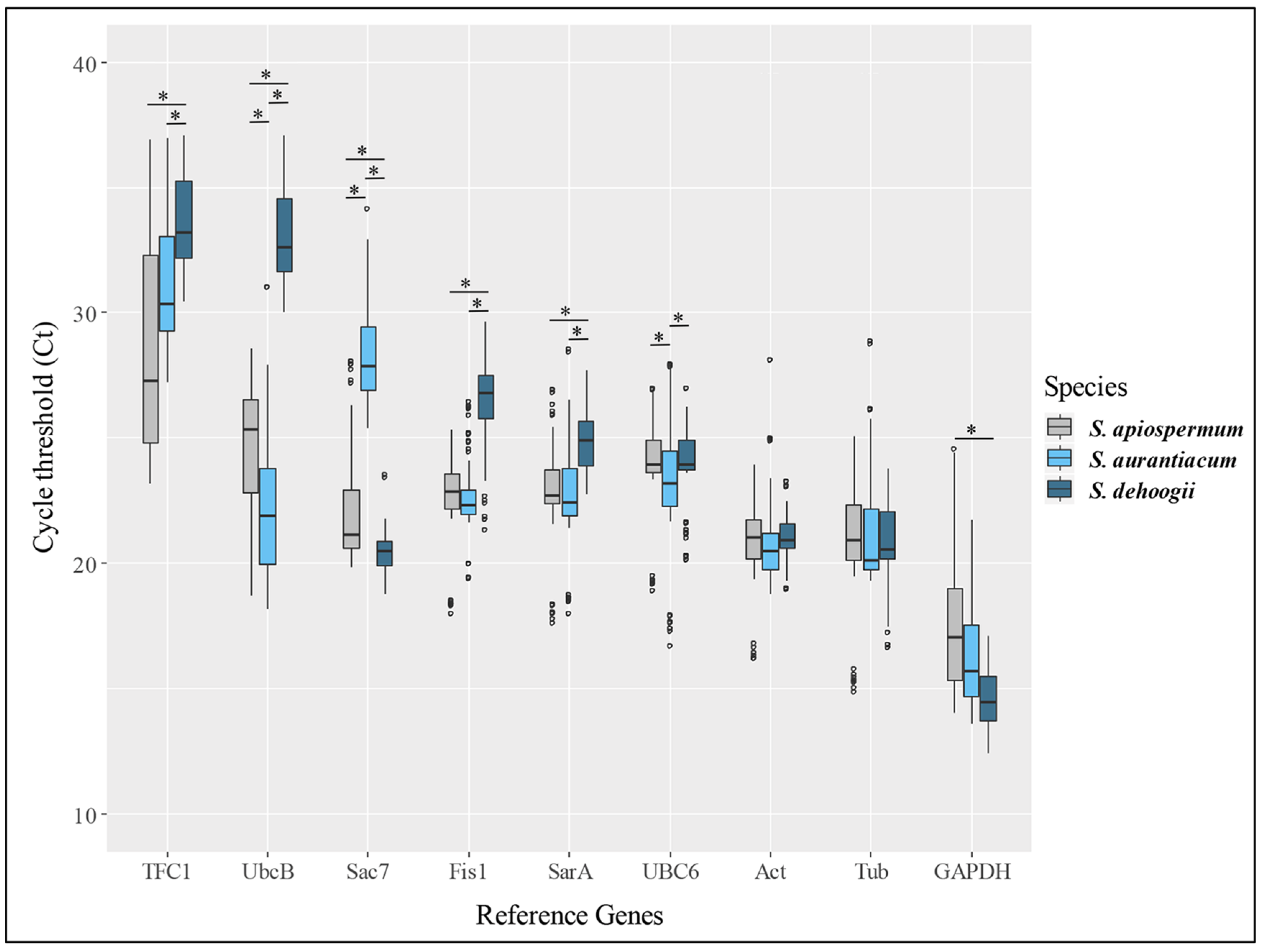
2.4. RNA Isolation, Reverse Transcription and Real-Time Quantitative PCR
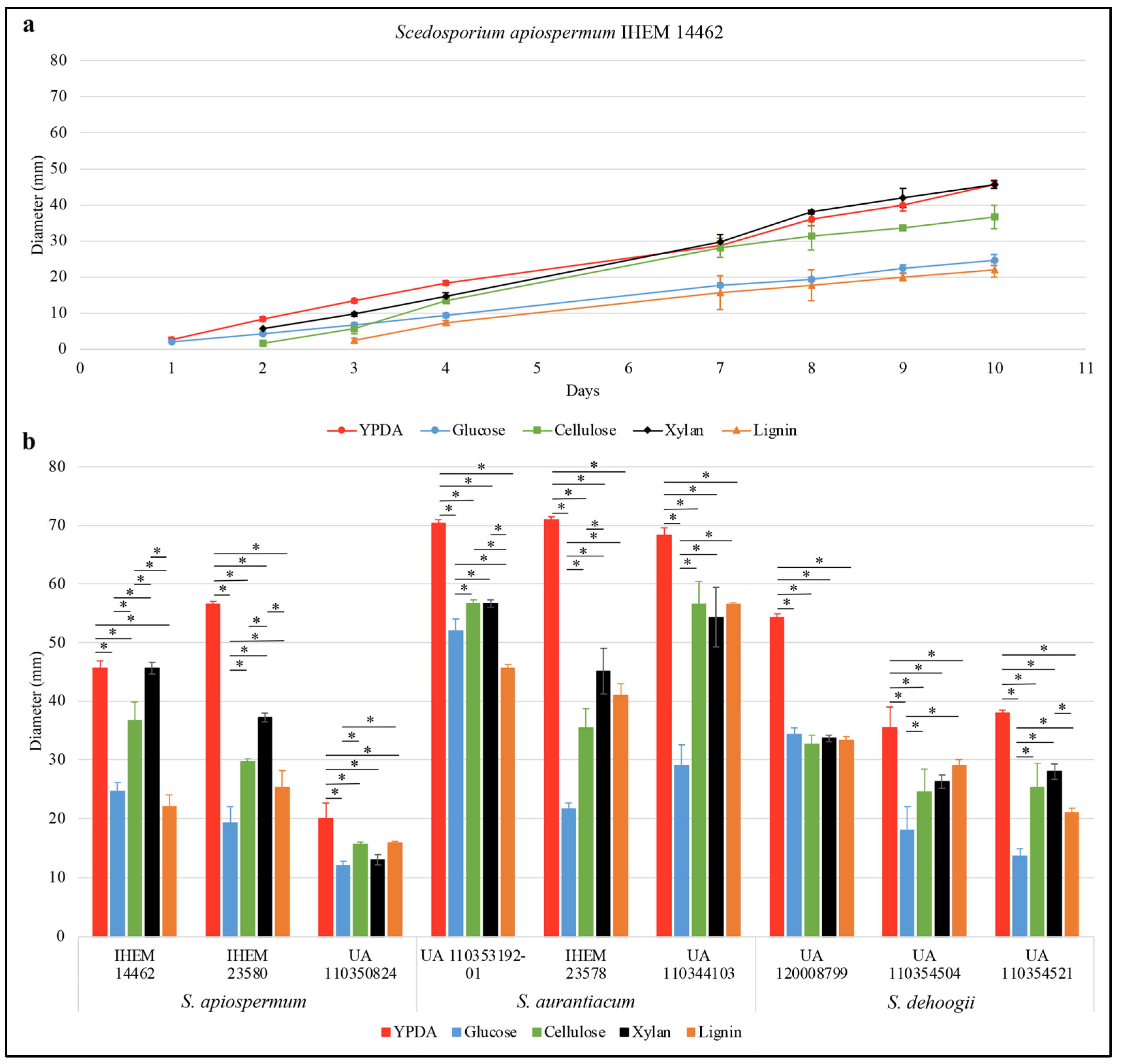
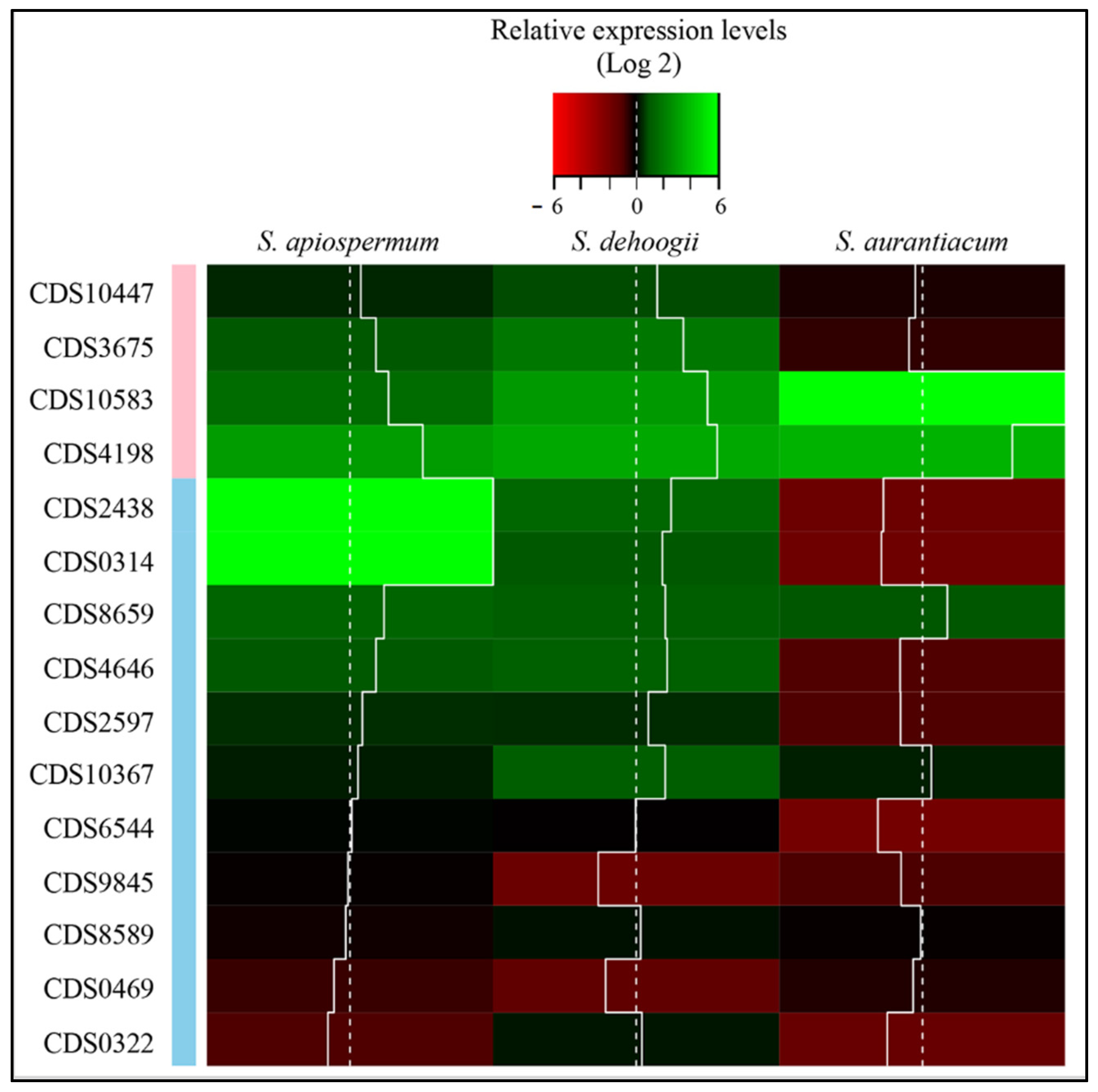
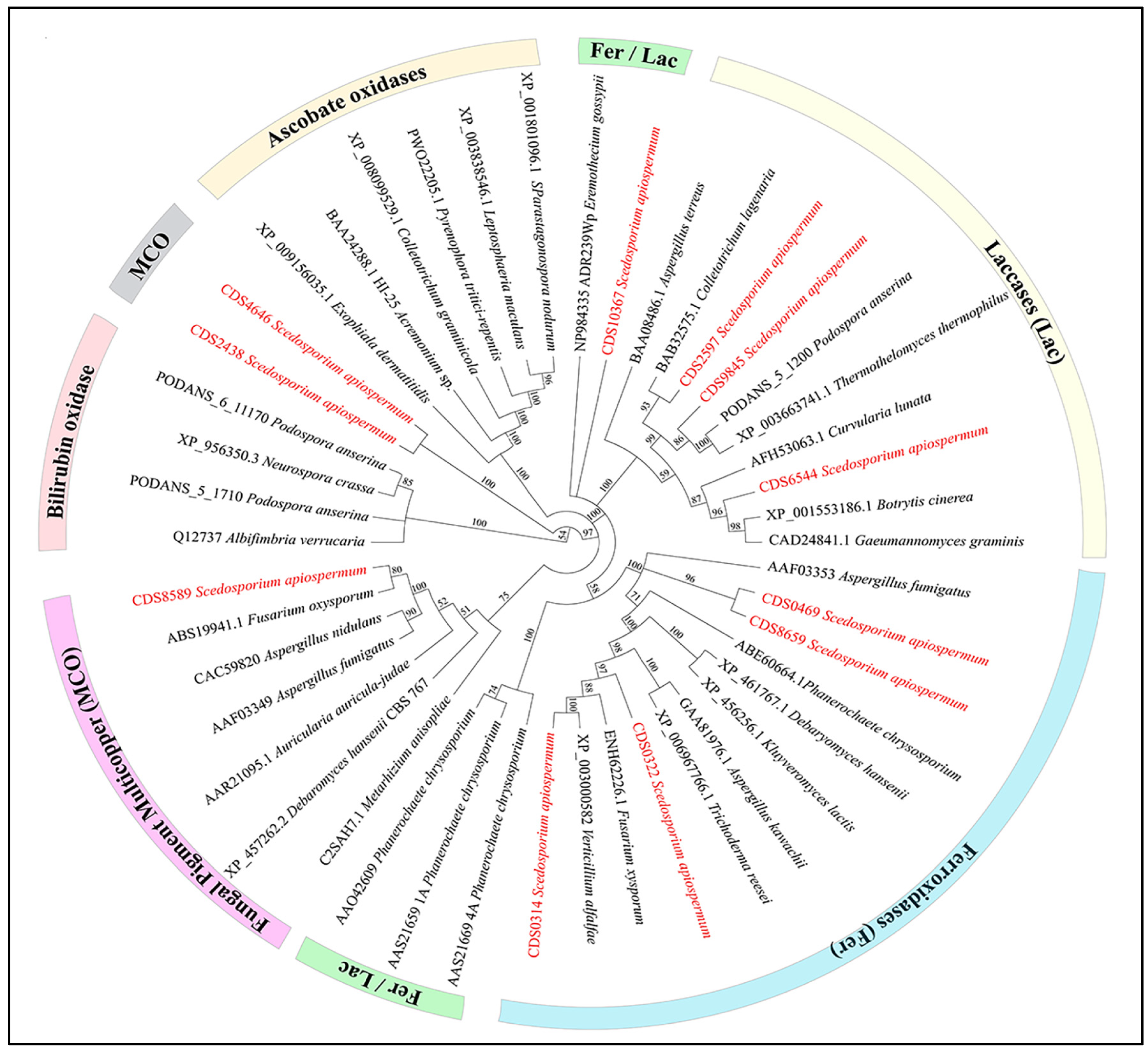
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hatakka, A. Biodegradation of lignin. In Biopolymers. Biology, Chemistry, Biotechnology, Applications. Vol 1. Lignin, Humic Substances and Coal; Wiley-Blackwell: New York, NY, USA, 2001; pp. 129–180. [Google Scholar]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez, A.; del Río, J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar]
- Ruiz-Dueñas, F.J.; Martínez, Á.T. Microbial degradation of lignin: How a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2009, 2, 164–177. [Google Scholar] [CrossRef] [Green Version]
- Savory, J.G. Damage to wood caused by microorganisms. J. Appl. Bacteriol. 1954, 17, 213–218. [Google Scholar] [CrossRef]
- Savory, J.G. Breakdown of timber by Ascomycetes and Fungi Imperfecti. Ann. Appl. Biol. 1954, 41, 336–347. [Google Scholar] [CrossRef]
- Duncan, C.G.; Eslyn, W.E. Wood-decaying Ascomycetes and Fungi Imperfecti. Mycologia 1966, 58, 642. [Google Scholar] [CrossRef]
- Ferraz, A.; Baeza, J.; Duránt, N. Softwood biodegradation by an Ascomycete Chrysonilia sitophila (TFB 27441 strain). Lett. Appl. Microbiol. 1991, 13, 82–86. [Google Scholar] [CrossRef]
- Norris, D.M. Degradation of C-labeled lignins and C-labeled aromatic acids by Fusarium solani. Appl. Environ. Microbiol. 1980, 40, 376–380. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, A.; Carnicero, A.; Perestelo, F.; de la Fuente, G.; Milstein, O.; Falcón, M.A. Effect of Penicillium chrysogenum on lignin transformation. Appl. Environ. Microbiol. 1994, 60, 2971–2976. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; Perestelo, F.; Carnicero, A.; Regalado, V.; Perez, R.; Fuente, G.; Falcon, M.A. Degradation of natural lignins and lignocellulosic substrates by soil-inhabiting Fungi Imperfecti. FEMS Microbiol. Ecol. 1996, 21, 213–219. [Google Scholar] [CrossRef]
- Liers, C.; Ullrich, R.; Steffen, K.T.; Hatakka, A.; Hofrichter, M. Mineralization of 14C-labelled synthetic lignin and extracellular enzyme activities of the wood-colonizing Ascomycetes Xylaria hypoxylon and Xylaria polymorpha. Appl. Microbiol. Biotechnol. 2006, 69, 573–579. [Google Scholar] [CrossRef]
- Kluczek-Turpeinen, B.; Tuomela, M.; Hatakka, A.; Hofrichter, M. Lignin degradation in a compost environment by the Deuteromycete Paecilomyces inflatus. Appl. Microbiol. Biotechnol. 2003, 61, 374–379. [Google Scholar] [CrossRef]
- Rougeron, A.; Schuliar, G.; Leto, J.; Sitterlé, E.; Landry, D.; Bougnoux, M.-E.; Kobi, A.; Bouchara, J.-P.; Giraud, S. Human-impacted areas of France are environmental reservoirs of the Pseudallescheria boydii/Scedosporium apiospermum species complex. Environ. Microbiol. 2015, 17, 1039–1048. [Google Scholar] [CrossRef]
- Kaltseis, J.; Rainer, J.; De Hoog, G.S.; Kaltseis, J.; Rainer, J.; De Hoog, G.S. Ecology of Pseudallescheria and Scedosporium species in human-dominated and natural environments and their distribution in clinical samples. Med. Mycol. 2009, 47, 398–405. [Google Scholar] [CrossRef] [Green Version]
- Harun, A.; Gilgado, F.; Chen, S.C.-A.; Meyer, W. Abundance of Pseudallescheria/Scedosporium species in the Australian urban environment suggests a possible source for scedosporiosis including the colonization of airways in cystic fibrosis. Med. Mycol. 2010, 48, S70–S76. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Marvin-Sikkema, F.D.; Lahpoor, G.A.; Gottschall, J.C.; Prins, R.A.; Guého, E. Ecology and physiology of the emerging opportunistic fungi Pseudallescheria boydii and Scedosporium prolificans. Mycoses 1994, 37, 71–78. [Google Scholar] [CrossRef]
- Cajthaml, T.; Svobodová, K. Biodegradation of aromatic pollutants by ligninolytic fungal strains. In Microbial Degradation of Xenobiotics; Singh, S.N., Ed.; Environmental Science and Engineering; Springer: Berlin/Heidelberg, Germany, 2012; pp. 291–316. ISBN 978-3-642-23788-1. [Google Scholar]
- Cajthaml, T. Biodegradation of endocrine-disrupting compounds by ligninolytic fungi: Mechanisms involved in the degradation: Ligninolytic fungi degrade EDCs via various ways. Environ. Microbiol. 2015, 17, 4822–4834. [Google Scholar] [CrossRef]
- Korniłłowicz-Kowalska, T.; Rybczyńska, K. Screening of microscopic fungi and their enzyme activities for decolorization and biotransformation of some aromatic compounds. Int. J. Environ. Sci. Technol. 2015, 12, 2673–2686. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.; Giraud, S.; Schuliar, G.; Rougeron, A.; Bouchara, J.-P. Scedo-Select III: A new semi-selective culture medium for detection of the Scedosporium apiospermum species complex. Med. Mycol. 2015, 53, 512–519. [Google Scholar] [CrossRef]
- Naranjo, L.; Urbina, H.; Sisto, A.D.; Leon, V. Isolation of autochthonous non-white rot fungi with potential for enzymatic upgrading of Venezuelan extra-heavy crude oil. Biocatal. Biotransform. 2007, 25, 341–349. [Google Scholar] [CrossRef] [Green Version]
- Romano, M.; Baralle, F.E.; Patriarca, P. Expression and characterization of recombinant human eosinophil peroxidase: Impact of the R286H substitution on the biosynthesis and activity of the enzyme. Eur. J. Biochem. 2000, 267, 3704–3711. [Google Scholar] [CrossRef]
- Shrestha, P.; Joshi, B.; Joshi, J.; Malla, R.; Sreerama, L. Isolation and physicochemical characterization of laccase from Ganoderma lucidum-CDBT1 isolated from its native habitat in Nepal. BioMed Res. Int. 2016, 2016, 3238909. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Theiler, G.; Zamocky, M.; Cosio, C.; Rouhier, N.; Teixera, F.; Margis-Pinheiro, M.; Ioannidis, V.; Penel, C.; Falquet, L.; et al. PeroxiBase: The peroxidase database. Phytochemistry 2007, 68, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kues, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Vandesompele, J.; Preter, K.D.; Roy, N.V.; Paepe, A.D. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]
- Bohle, K.; Jungebloud, A.; Göcke, Y.; Dalpiaz, A.; Cordes, C.; Horn, H.; Hempel, D.C. Selection of reference genes for normalisation of specific gene quantification data of Aspergillus niger. J. Biotechnol. 2007, 132, 353–358. [Google Scholar] [CrossRef]
- Teste, M.-A.; Duquenne, M.; François, J.M.; Parrou, J.-L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [Green Version]
- Llanos, A.; François, J.; Parrou, J.-L. Tracking the best reference genes for RT-qPCR data normalization in filamentous fungi. BMC Genom. 2015, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Poirier, W.; Ravenel, K.; Bouchara, J.P.; Giraud, S. Lower funneling pathways in Scedopsorium species. Front. Microbiol. 2021, 12, 630753. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M.; Gasselhuber, B.; Furtmüller, P.G.; Obinger, C. Turning points in the evolution of peroxidase–catalase superfamily: Molecular phylogeny of hybrid heme peroxidases. Cell. Mol. Life Sci. 2014, 71, 4681–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. In Industrial Applications; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 319–340. ISBN 978-3-642-11457-1. [Google Scholar]
- Regalado, V.; Perestelo, F.; Rodríguez, A.; Carnicero, A.; Sosa, F.J.; De la Fuente, G.; Falcón, M.A. Activated oxygen species and two extracellular enzymes: Laccase and aryl-alcohol oxidase, novel for the lignin-degrading fungus Fusarium proliferatum. Appl. Microbiol. Biotechnol. 1999, 51, 388–390. [Google Scholar] [CrossRef]
- Kirk, P.W. A comparison of saline tolerance and sporulation in marine and clinical isolates of Allescheria boydii Shear. Mycopath. Mycol. Appl. 1967, 33, 65–75. [Google Scholar] [CrossRef]
- Azevedo, E.; Caeirao, M.F.; Rebelo, R.; Barata, M. Biodiversity and characterization of marine mycota from Portuguese waters. Anim. Biodivers. Conserv. 2011, 34, 205–215. [Google Scholar] [CrossRef]
- Gilgado, F.; Cano, J.; Gené, J.; Guarro, J. Molecular phylogeny of the Pseudallescheria boydii species complex: Proposal of two new species. J. Clin. Microbiol. 2005, 43, 4930–4942. [Google Scholar] [CrossRef] [Green Version]
- Nirma, C.; Eparvier, V.; Stien, D. Antifungal agents from Pseudallescheria boydii SNB-CN73 isolated from a Nasutitermes sp. termite. J. Nat. Prod. 2013, 76, 988–991. [Google Scholar] [CrossRef]
- Wu, Q.; Jiang, N.; Bo Han, W.; Ning Mei, Y.; Ming Ge, H.; Kai Guo, Z.; Seik Weng, N.; Xiang Tan, R. Antibacterial epipolythiodioxopiperazine and unprecedented sesquiterpene from Pseudallescheria boydii, a beetle (Coleoptera)-associated fungus. Org. Biomol. Chem. 2014, 12, 9405–9412. [Google Scholar] [CrossRef]
- Rojas-Jiménez, K.; Hernández, M. Isolation of fungi and bacteria associated with the guts of tropical wood-feeding Coleoptera and determination of their lignocellulolytic activities. Int. J. Microbiol. 2015, 2015, 285018. [Google Scholar] [CrossRef] [Green Version]
- Prenafeta-Boldú, F.X.; Summerbell, R.; Sybren de Hoog, G. Fungi growing on aromatic hydrocarbons: Biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 2006, 30, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.; Varela, A.; Leclercq, C.C.; Núñez, O.; Větrovský, T.; Renaut, J.; Baldrian, P.; Silva Pereira, C. Specialisation events of fungal metacommunities exposed to a persistent organic pollutant are suggestive of augmented pathogenic potential. Microbiome 2018, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Penn, C.D.; Daniel, S.L. Salicylate degradation by the fungal plant pathogen Sclerotinia sclerotiorum. Curr. Microbiol. 2013, 67, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Sbaghi, M.; Jeandet, P.; Bessis, R.; Leroux, P. Degradation of stilbene-type phytoalexins in relation to the pathogenicity of Botrytis cinerea to grapevines. Plant Pathol. 1996, 45, 139–144. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poirier, W.; Bouchara, J.-P.; Giraud, S. Lignin-Modifying Enzymes in Scedosporium Species. J. Fungi 2023, 9, 105. https://doi.org/10.3390/jof9010105
Poirier W, Bouchara J-P, Giraud S. Lignin-Modifying Enzymes in Scedosporium Species. Journal of Fungi. 2023; 9(1):105. https://doi.org/10.3390/jof9010105
Chicago/Turabian StylePoirier, Wilfried, Jean-Philippe Bouchara, and Sandrine Giraud. 2023. "Lignin-Modifying Enzymes in Scedosporium Species" Journal of Fungi 9, no. 1: 105. https://doi.org/10.3390/jof9010105
APA StylePoirier, W., Bouchara, J.-P., & Giraud, S. (2023). Lignin-Modifying Enzymes in Scedosporium Species. Journal of Fungi, 9(1), 105. https://doi.org/10.3390/jof9010105





