Pseudo-Chromosomal Genome Assembly in Combination with Comprehensive Transcriptome Analysis in Agaricus bisporus Strain KMCC00540 Reveals Mechanical Stimulus Responsive Genes Associated with Browning Effect
Abstract
1. Introduction
2. Materials and Methods
2.1. A. bisporus Strain and DNA Isolation
2.2. Genome Sequencing and Assembly
2.3. Comparative Genomic Analysis
2.4. Gene Prediction of Assembled Genome
2.5. RNA Extraction and mRNA-Sequencing
2.6. mRNA-Sequencing Analysis and Functional Annotation
3. Results
3.1. De Novo Genome Assembly of Agaricus Bisporus Strain KMCC00540
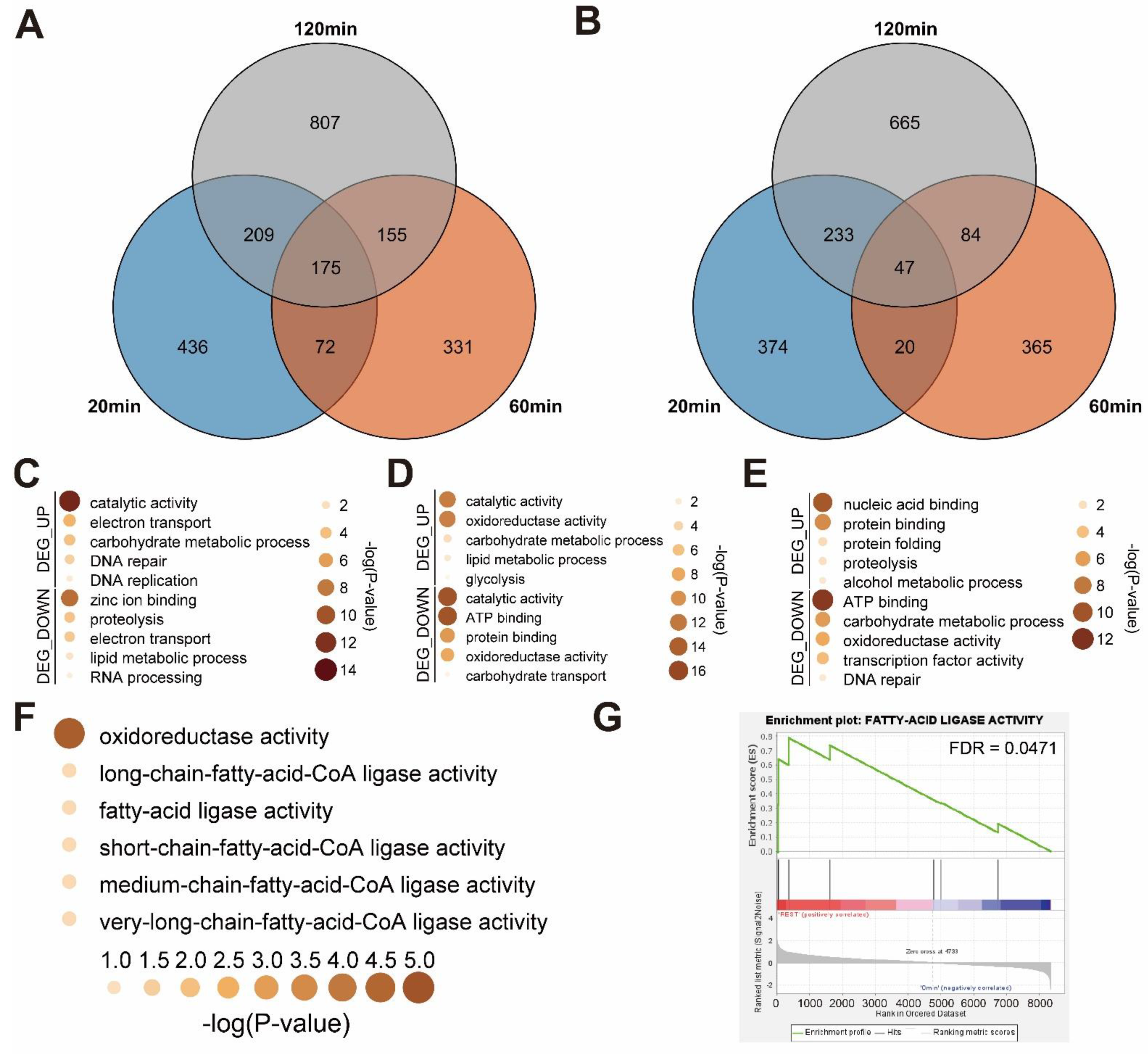
3.2. Time-Course Transcriptome Analysis of Mechanical Stimulus Treated Button Mushroom
3.3. Functional Annotation of Mechanical Stimulus Treated Transcriptome in Time Course
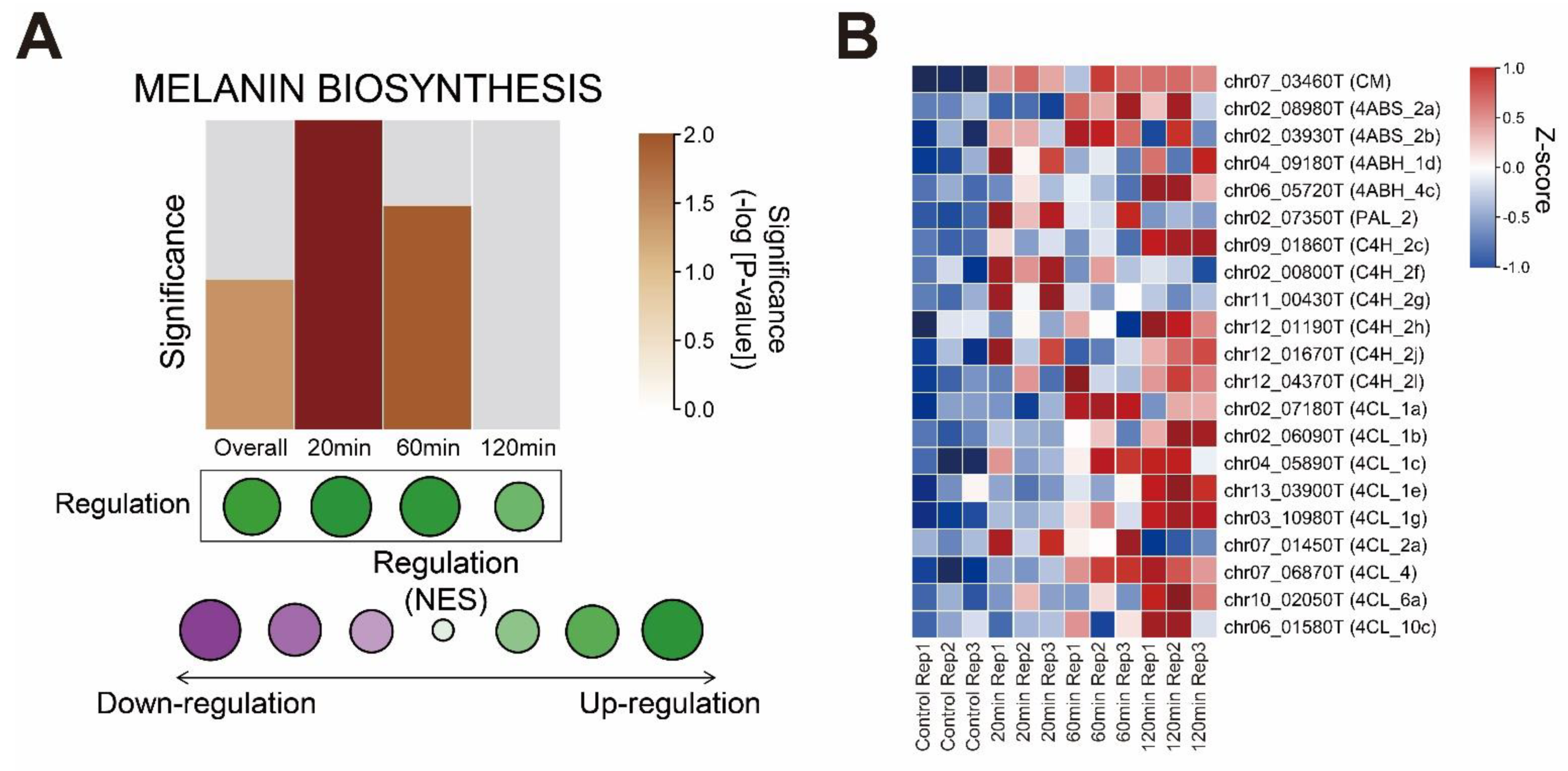
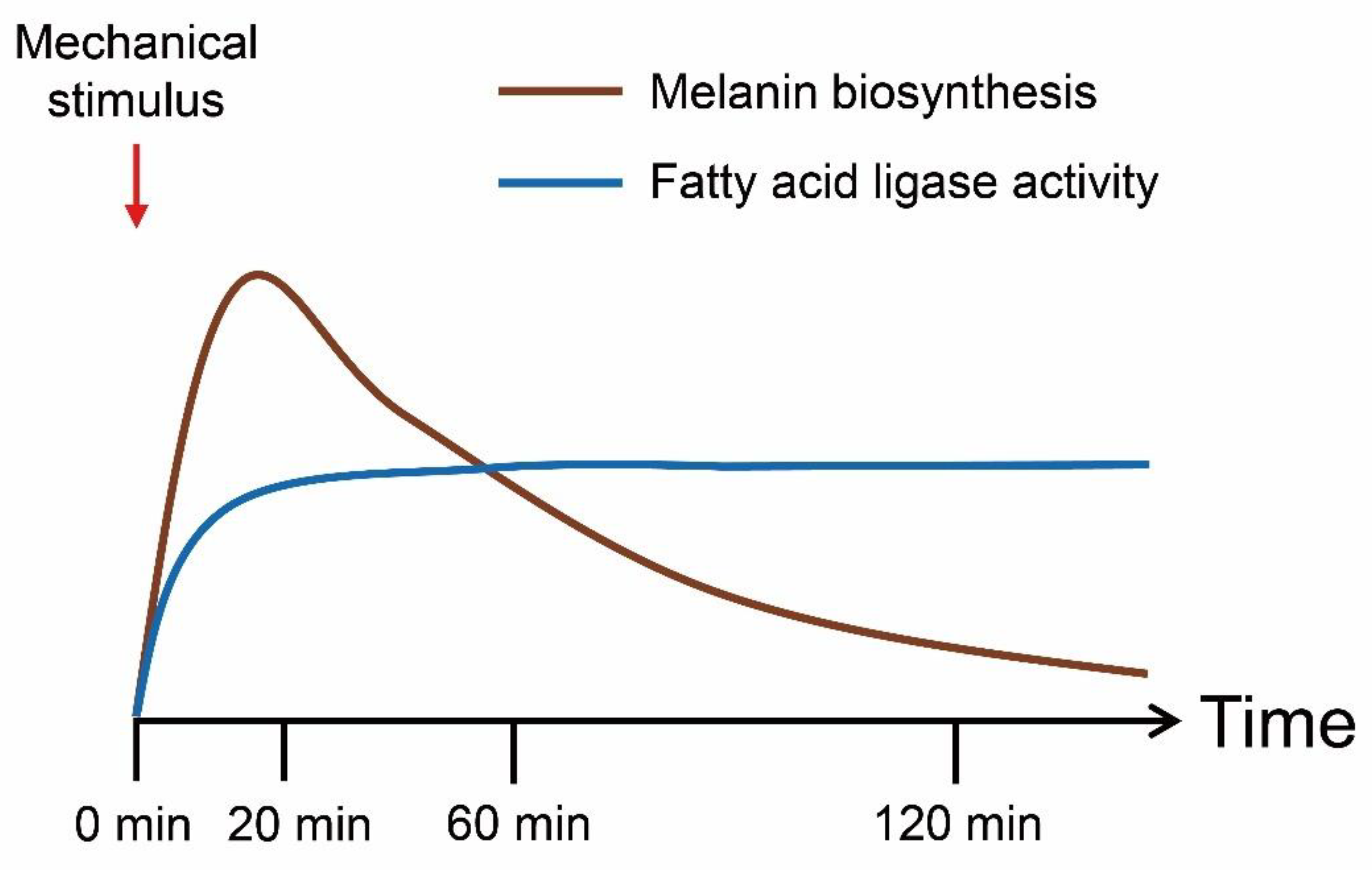
3.4. Melanin Biosynthesis Genes Were Moderately Up-Regulated in Response to Mechanical Stimulus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- An, H.; Jo, I.H.; Oh, Y.L.; Jang, K.Y.; Kong, W.S.; Sung, J.K.; So, Y.S.; Chung, J.W. Molecular Characterization of 170 New gDNA-SSR Markers for Genetic Diversity in Button Mushroom (Agaricus bisporus). Mycobiology 2019, 47, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Zhang, J.; Yan, R.; Shao, S.; Zhou, T.; Lei, J.; Wang, Z. Effects of UV-C treatment and cold storage on ergosterol and vitamin D2 contents in different parts of white and brown mushroom (Agaricus bisporus). Food Chem. 2016, 210, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Jovana Vunduk, I.D.; Petrović, P.; Tomašević, I.; Kozarski, M.; Despotović, S.; Nikšić, M.; Klaus, A. Challenging the difference between white and brown Agaricus bisporus mushrooms: Science behind consumers choice. Br. Food J. 2018, 120, 1381–1394. [Google Scholar] [CrossRef]
- Simon, A.; Gonzalez-Fandos, E.; Vazquez, M. Effect of washing with citric acid and packaging in modified atmosphere on the sensory and microbiological quality of sliced mushrooms (Agaricus bisporus L.). Food Control 2010, 21, 851–856. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, C.; Liu, Y.; Ding, Y.; Winters, E.; Li, W.; Cheng, F. Gibberellic acid maintains postharvest quality of Agaricus bisporus mushroom by enhancing antioxidative system and hydrogen sulfide synthesis. J. Food Biochem. 2021, 45, e13939. [Google Scholar] [CrossRef]
- Kwak, E.J.; Lim, S.I. Inhibition of browning by antibrowning agents and phenolic acids or cinnamic acid in the glucose-lysine model. J. Sci. Food Agric. 2005, 85, 1337–1342. [Google Scholar] [CrossRef]
- Gao, M.; Feng, L.; Jiang, T. Browning inhibition and quality preservation of button mushroom (Agaricus bisporus) by essential oils fumigation treatment. Food Chem. 2014, 149, 107–113. [Google Scholar] [CrossRef]
- Sonnenberg, A.S.; Gao, W.; Lavrijssen, B.; Hendrickx, P.; Sedaghat-Tellgerd, N.; Foulongne-Oriol, M.; Kong, W.S.; Schijlen, E.G.; Baars, J.J.; Visser, R.G. A detailed analysis of the recombination landscape of the button mushroom Agaricus bisporus var. bisporus. Fungal Genet. Biol. 2016, 93, 35–45. [Google Scholar] [CrossRef]
- Sun, L.; Fu, Y.; Yang, Y.; Wang, X.; Cui, W.; Li, D.; Yuan, X.; Zhang, Z.; Fu, Y.; Li, Y. Genomic Analyses Reveal Evidence of Independent Evolution, Demographic History, and Extreme Environment Adaptation of Tibetan Plateau Agaricus bisporus. Front. Microbiol. 2019, 10, 1786. [Google Scholar] [CrossRef]
- Sonnenberg, A.S.M.; Sedaghat-Telgerd, N.; Lavrijssen, B.; Ohm, R.A.; Hendrickx, P.M.; Scholtmeijer, K.; Baars, J.J.P.; van Peer, A. Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends. Sci. Rep. 2020, 10, 14653. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, J.T.; Moon, H.S.; Kim, Y.J.; Kim, S.H. Geographical origin discrimination of Agaricus bisporus produced by the complete medium: A pilot study in South Korea. Food Chem. 2022, 386, 132820. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.L.; Oh, M.J.; Im, J.H.; Jang, K.Y. Breeding a new white button mushroom cultivar ‘Hadam’ to produce mushrooms at high temperature. J. Mushrooms 2020, 18, 214–220. [Google Scholar]
- Weijn, A.; van den Berg-Somhorst, D.B.; Slootweg, J.C.; Vincken, J.P.; Gruppen, H.; Wichers, H.J.; Mes, J.J. Main phenolic compounds of the melanin biosynthesis pathway in bruising-tolerant and bruising-sensitive button mushroom (Agaricus bisporus) strains. J. Agric. Food Chem. 2013, 61, 8224–8231. [Google Scholar] [CrossRef] [PubMed]
- Weijn, A.; Tomassen, M.M.M.; Bastiaan-Net, S.; Wigham, M.L.I.; Boer, E.P.J.; Hendrix, E.A.H.J.; Baars, J.J.P.; Sonnenberg, A.S.M.; Wichers, H.J.; Mes, J.J. A new method to apply and quantify bruising sensitivity of button mushrooms. LWT Food Sci. Technol. 2012, 47, 308–314. [Google Scholar] [CrossRef]
- Arjun, A.D.; Chakraborty, S.K.; Mahanti, N.K.; Kotwaliwale, N. Non-destructive assessment of quality parameters of white button mushrooms (Agaricus bisporus) using image processing techniques. J. Food Sci. Technol. 2022, 59, 2047–2059. [Google Scholar] [CrossRef]
- Li, T.H.; Zhang, M. The Physiological and Quality Change of Mushroom Agaricus bisporus Stored in Modified Atmosphere Packaging with Various Sizes of Silicone Gum Film Window. Food Sci. Technol. Res. 2013, 19, 569–576. [Google Scholar] [CrossRef]
- O’Connor, E.; Doyle, S.; Amini, A.; Grogan, H.; Fitzpatrick, D.A. Transmission of mushroom virus X and the impact of virus infection on the transcriptomes and proteomes of different strains of Agaricus bisporus. Fungal Biol. 2021, 125, 704–717. [Google Scholar] [CrossRef]
- Fleming-Archibald, C.; Ruggiero, A.; Grogan, H.M. Brown mushroom symptom expression following infection of an Agaricus bisporus crop with MVX associated dsRNAs. Fungal Biol. 2015, 119, 1237–1245. [Google Scholar] [CrossRef]
- Hamidizade, M.; Taghavi, S.M.; Mafakheri, H.; Herschlag, R.A.; Martins, S.J.; Hockett, K.L.; Bull, C.T.; Osdaghi, E. First Report of Brown Spot on White Button Mushroom (Agaricus bisporus) Caused by Cedecea neteri in Iran. Plant Dis. 2022, 106, 1291. [Google Scholar] [CrossRef]
- Gea, F.; Navarro, M.; Santos, M.; Diánez, F.; Carrasco, J. Control of Fungal Diseases in Mushroom Crops while Dealing with Fungicide Resistance: A Review. Microorganisms 2021, 9, 585. [Google Scholar] [CrossRef]
- Karimirad, R.; Behnamian, M.; Dezhsetan, S.; Sonnenberg, A. Chitosan nanoparticles-loaded Citrus aurantium essential oil: A novel delivery system for preserving the postharvest quality of Agaricus bisporus. J. Sci. Food Agric. 2018, 98, 5112–5119. [Google Scholar] [CrossRef] [PubMed]
- Weijn, A.; Bastiaan-Net, S.; Wichers, H.J.; Mes, J.J. Melanin biosynthesis pathway in Agaricus bisporus mushrooms. Fungal Genet. Biol. 2013, 55, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Wakamatsu, K. Quantitative analysis of eumelanin and pheomelanin in humans, mice, and other animals: A comparative review. Pigment Cell Res. 2003, 16, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.X.; Chen, M.Y.; Lu, Y.P.; Guo, Z.J.; Zeng, Z.H.; Liao, J.H.; Zeng, H. Metabolomics and transcriptomics unravel the mechanism of browning resistance in Agaricus bisporus. PLoS ONE 2022, 17, e0255765. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, S.; Li, A.; Ruan, J. SMARTdenovo: A de novo assembler using long noisy reads. Gigabyte 2021, 2021, 2020090207. [Google Scholar] [CrossRef]
- Kajitani, R.; Yoshimura, D.; Okuno, M.; Minakuchi, Y.; Kagoshima, H.; Fujiyama, A.; Kubokawa, K.; Kohara, Y.; Toyoda, A.; Itoh, T. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 2019, 10, 1702. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Handsaker, R.E.; Pulit, S.L.; Nizzari, M.M.; O’Donnell, C.J.; de Bakker, P.I. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008, 24, 2938–2939. [Google Scholar] [CrossRef] [PubMed]
- Nachtweide, S.; Stanke, M. Multi-Genome Annotation with AUGUSTUS. Methods Mol. Biol. 2019, 1962, 139–160. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Baars, J.J.; Dolstra, O.; Visser, R.G.; Sonnenberg, A.S. Genetic Variation and Combining Ability Analysis of Bruising Sensitivity in Agaricus bisporus. PLoS ONE 2013, 8, e76826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, W.; Weijn, A.; Baars, J.J.; Mes, J.J.; Visser, R.G.; Sonnenberg, A.S. Quantitative trait locus mapping for bruising sensitivity and cap color of Agaricus bisporus (button mushrooms). Fungal Genet. Biol. 2015, 77, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.P.; Kim, J.; Lee, J.; Yoo, S.I.; Bae, W.; Geem, K.R.; Yu, J.; Jang, I.; Jo, I.H.; Cho, H.; et al. Gibberellin Signaling Promotes the Secondary Growth of Storage Roots in Panax ginseng. Int. J. Mol. Sci. 2021, 22, 8694. [Google Scholar] [CrossRef]
- Maulucci, G.; Cohen, O.; Daniel, B.; Sansone, A.; Petropoulou, P.I.; Filou, S.; Spyridonidis, A.; Pani, G.; De Spirito, M.; Chatgilialoglu, C.; et al. Fatty acid-related modulations of membrane fluidity in cells: Detection and implications. Free Radic. Res. 2016, 50, S40–S50. [Google Scholar] [CrossRef]
- Yan, J.J.; Tong, Z.J.; Liu, Y.Y.; Lin, Z.Y.; Long, Y.; Han, X.; Xu, W.N.; Huang, Q.H.; Tao, Y.X.; Xie, B.G. The NADPH oxidase in Volvariella volvacea and its differential expression in response to mycelial ageing and mechanical injury. Braz. J. Microbiol. 2020, 51, 87–94. [Google Scholar] [CrossRef]
- Hernandez-Onate, M.A.; Esquivel-Naranjo, E.U.; Mendoza-Mendoza, A.; Stewart, A.; Herrera-Estrella, A.H. An injury-response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc. Natl. Acad. Sci. USA 2012, 109, 14918–14923. [Google Scholar] [CrossRef]
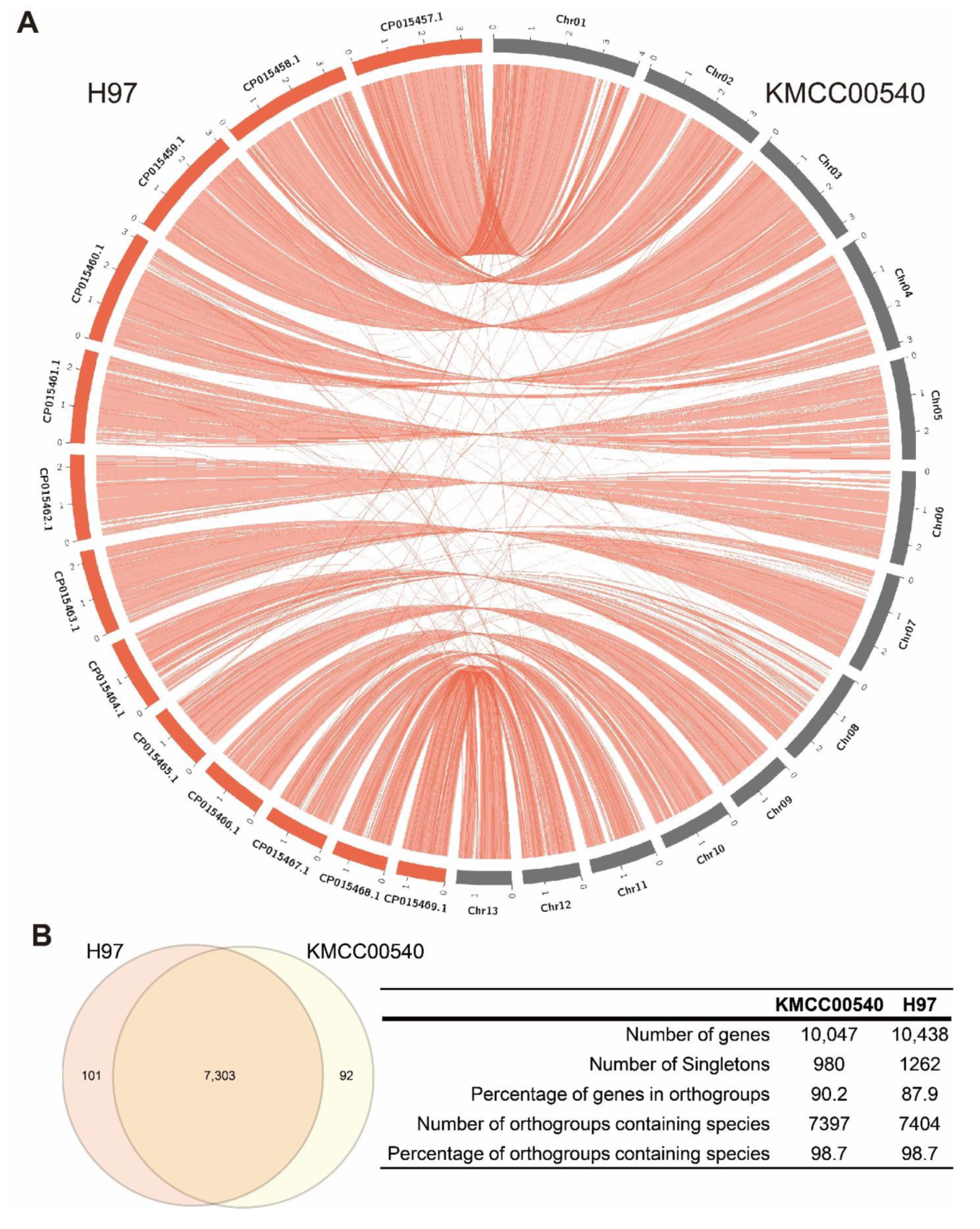

| Strain | KMCC00540 | H97 | H39 | AB58 | H119 | ARP23 | JB137-s8 |
|---|---|---|---|---|---|---|---|
| Assembly level | Chromosome | Chromosome | Chromosome | Chromosome | Chromosome | Scaffold | Contig |
| Number of contigs (≥0 bp) | 14 | 13 | 16 | 18 | 16 | 169 | 2016 |
| Number of contigs (≥10,000 bp) | 14 | 13 | 16 | 18 | 16 | 167 | 121 |
| Number of contigs (≥50,000 bp) | 14 | 13 | 16 | 17 | 16 | 121 | 52 |
| Total length (≥0 bp) | 33,281,038 | 30,417,844 | 30,702,502 | 30,069,126 | 30,702,502 | 33,487,476 | 32,614,392 |
| Number of contigs | 14 | 13 | 16 | 18 | 16 | 169 | 2015 |
| Largest contig (bp) | 3,927,320 | 3,550,205 | 3,464,045 | 3,600,717 | 3,464,045 | 1,506,893 | 2,973,556 |
| Total length (bp) | 33,281,038 | 30,417,844 | 30,702,502 | 30,069,126 | 30,702,502 | 33,487,476 | 32,613,893 |
| GC (%) | 46.45 | 46.5 | 46.64 | 46.53 | 46.64 | 46.34 | 46.59 |
| N50 | 2,750,289 | 2,550,681 | 1,931,181 | 2,300,414 | 1,931,181 | 350,711 | 1,225,131 |
| N90 | 1,704,996 | 1,688,379 | 1,323,473 | 1,336,791 | 1,323,473 | 91,735 | 19,667 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, I.-H.; Kim, J.; An, H.; Lee, H.-Y.; So, Y.-S.; Ryu, H.; Sung, G.-H.; Shim, D.; Chung, J.-W. Pseudo-Chromosomal Genome Assembly in Combination with Comprehensive Transcriptome Analysis in Agaricus bisporus Strain KMCC00540 Reveals Mechanical Stimulus Responsive Genes Associated with Browning Effect. J. Fungi 2022, 8, 886. https://doi.org/10.3390/jof8080886
Jo I-H, Kim J, An H, Lee H-Y, So Y-S, Ryu H, Sung G-H, Shim D, Chung J-W. Pseudo-Chromosomal Genome Assembly in Combination with Comprehensive Transcriptome Analysis in Agaricus bisporus Strain KMCC00540 Reveals Mechanical Stimulus Responsive Genes Associated with Browning Effect. Journal of Fungi. 2022; 8(8):886. https://doi.org/10.3390/jof8080886
Chicago/Turabian StyleJo, Ick-Hyun, Jaewook Kim, Hyejin An, Hwa-Yong Lee, Yoon-Sup So, Hojin Ryu, Gi-Ho Sung, Donghwan Shim, and Jong-Wook Chung. 2022. "Pseudo-Chromosomal Genome Assembly in Combination with Comprehensive Transcriptome Analysis in Agaricus bisporus Strain KMCC00540 Reveals Mechanical Stimulus Responsive Genes Associated with Browning Effect" Journal of Fungi 8, no. 8: 886. https://doi.org/10.3390/jof8080886
APA StyleJo, I.-H., Kim, J., An, H., Lee, H.-Y., So, Y.-S., Ryu, H., Sung, G.-H., Shim, D., & Chung, J.-W. (2022). Pseudo-Chromosomal Genome Assembly in Combination with Comprehensive Transcriptome Analysis in Agaricus bisporus Strain KMCC00540 Reveals Mechanical Stimulus Responsive Genes Associated with Browning Effect. Journal of Fungi, 8(8), 886. https://doi.org/10.3390/jof8080886







