Transcriptomics Integrated with Metabolomics: Assessing the Central Metabolism of Different Cells after Cell Differentiation in Aureobasidium pullulans NG
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Cultivation Conditions
2.2. Sample Collection and Observation
2.3. Electron Microscopic Observation of SCs and YL
2.4. Treatment of the Two Types of Cells under Different Conditions
2.5. Determination of Intracellular Reactive Oxygen Species
2.6. Determination of Intracellular Superoxide Dismutase, Catalase and Total Antioxidant Capacity
2.7. Transcriptome Analysis
2.8. Metabolome Analysis
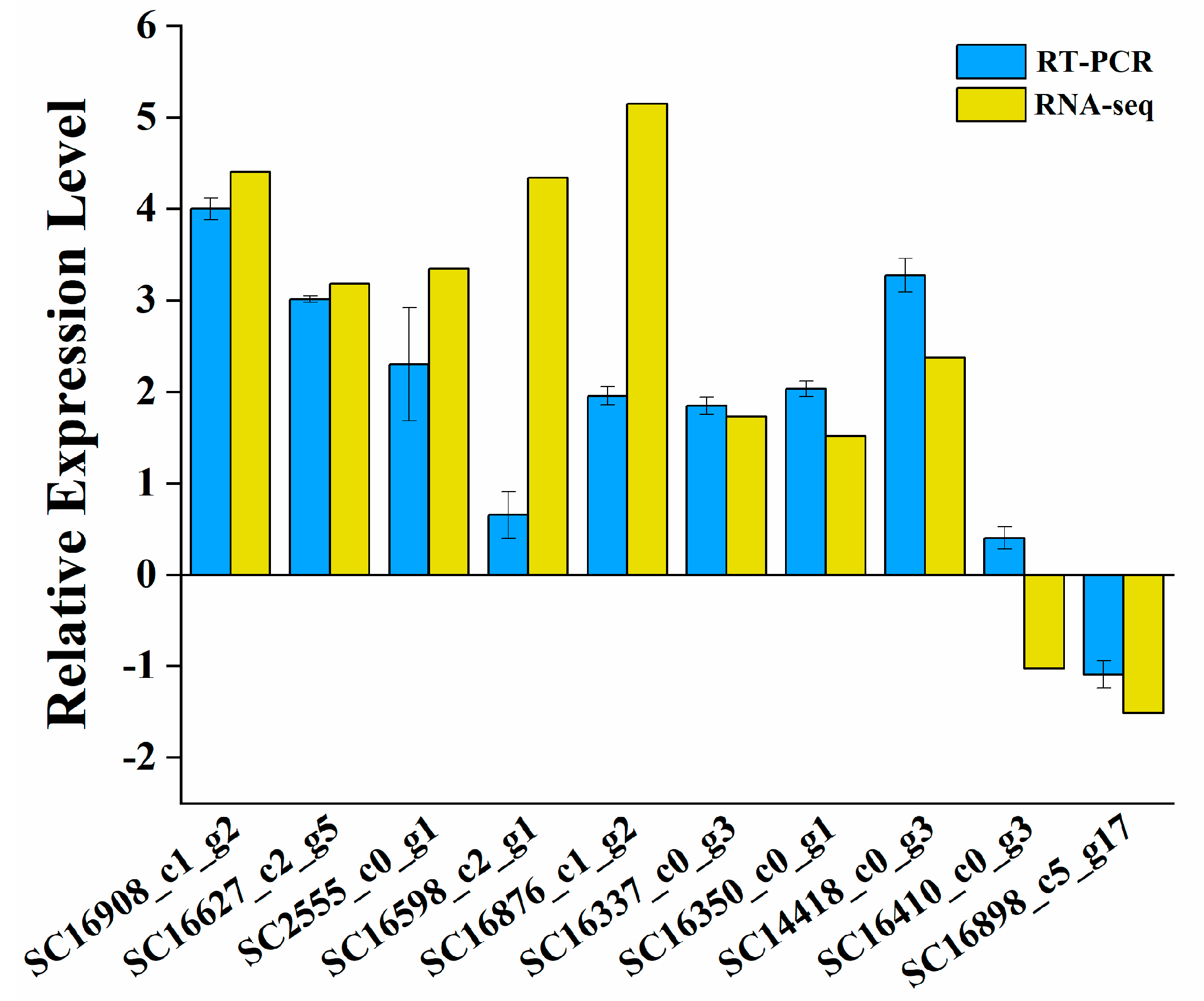
2.9. Real-Time Quantitative PCR Confirmation of the RNA-Seq Data
2.10. Statistical Analysis
3. Results
3.1. Determination of the Antioxidant Capacity of SCs and YL
3.2. Survival of Two Types of Cells in Response to Different External Stress Conditions
3.3. Transcriptome Analysis
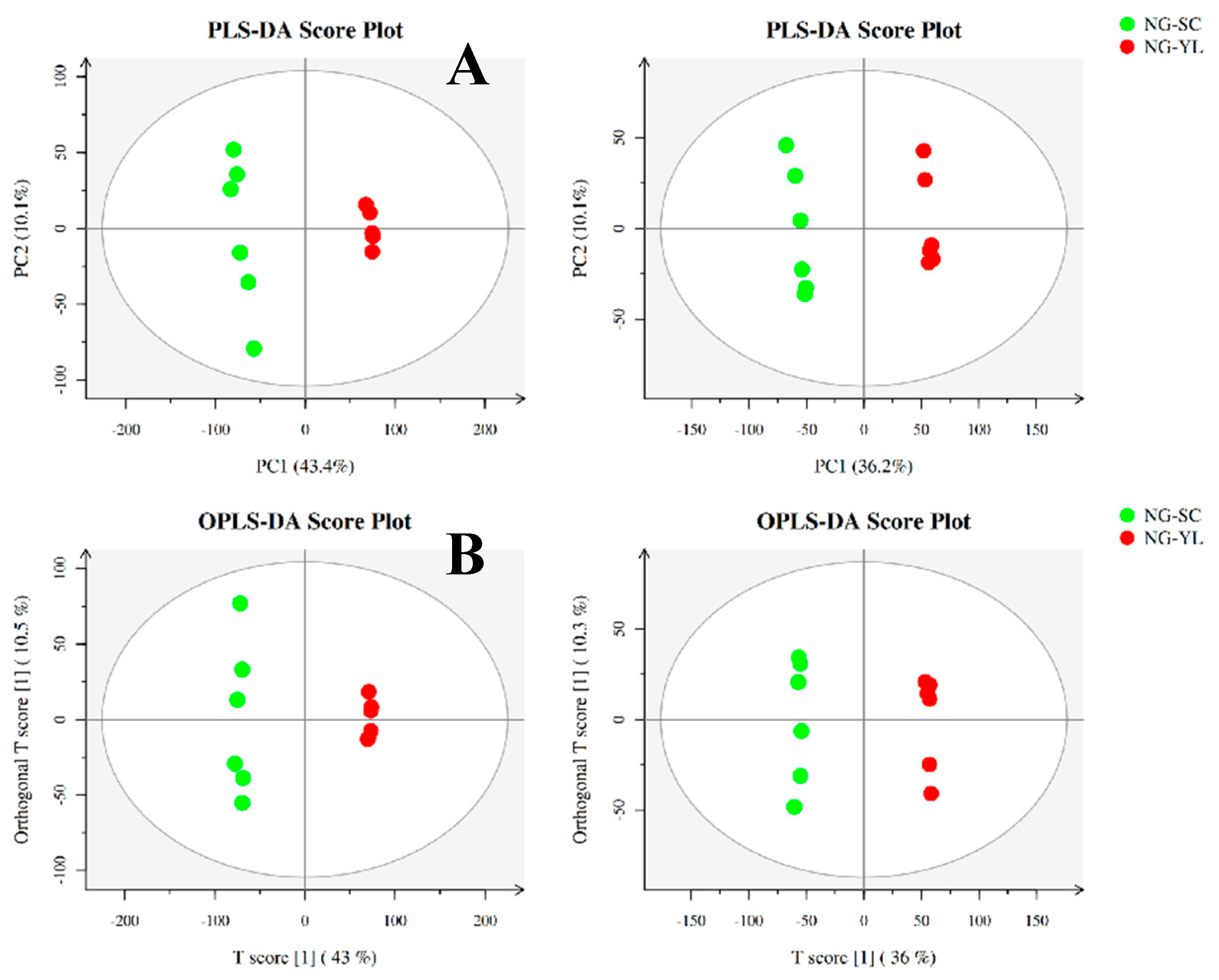
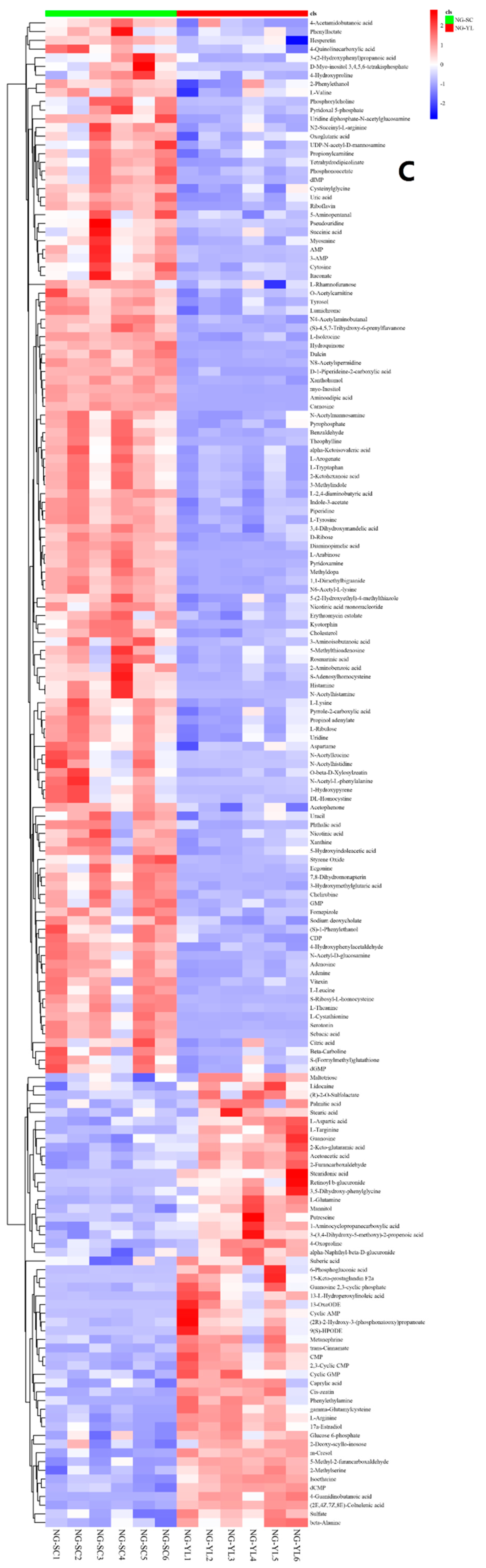
3.4. Metabolome Analysis
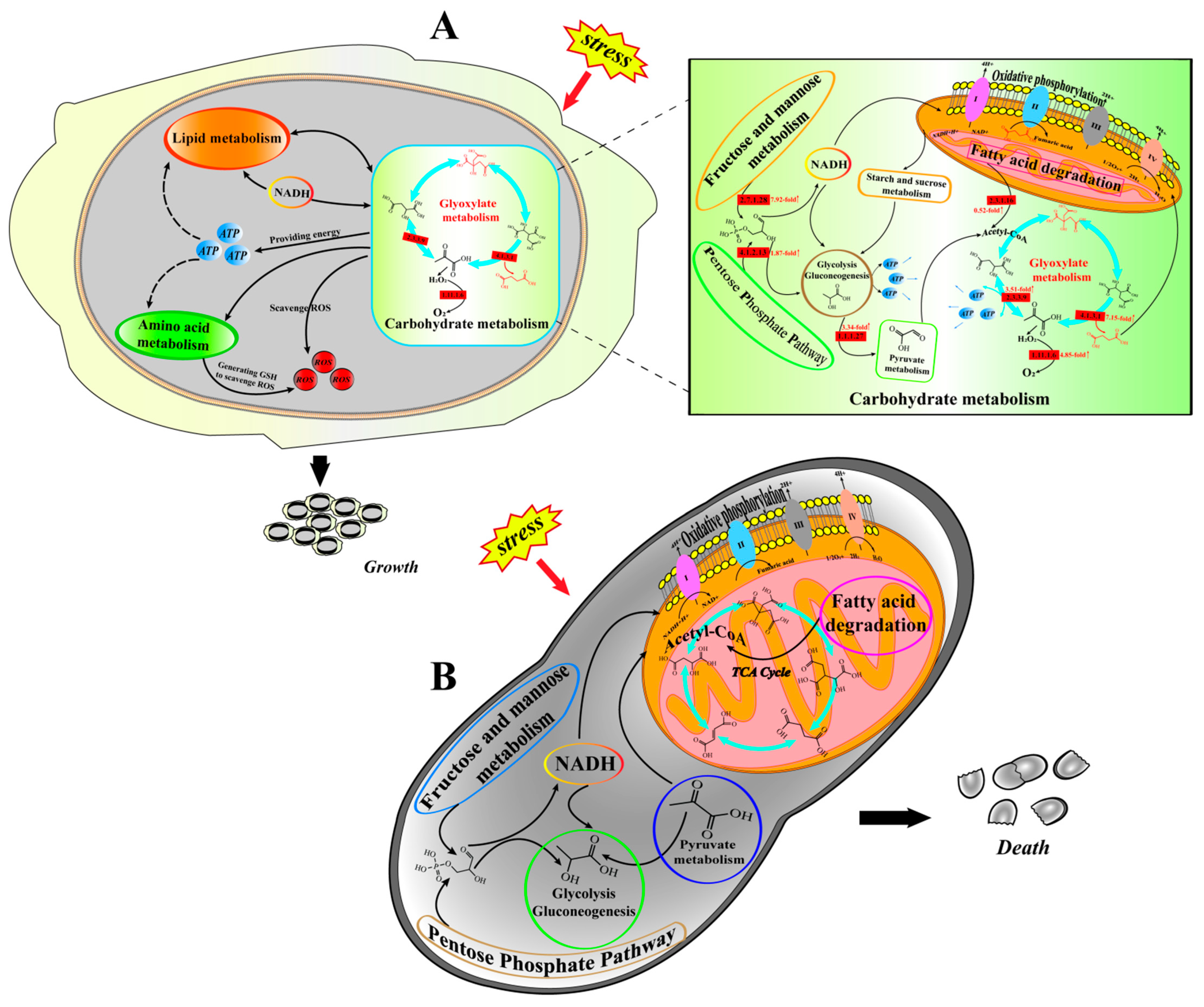
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bermejo, J.M.; Dominguez, J.B.; Goñi, F.M.; Uruburu, F. Influence of carbon and nitrogen sources on the transition from yeast-like cells to chlamydospores in Aureobasidium pullulans. Antonie Van Leeuwenhoek 1981, 47, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Gunde-Cimerman, N. Overview of Oxidative Stress Response Genes in Selected Halophilic Fungi. Genes 2018, 9, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, M.J. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 2009, 7, 237–245. [Google Scholar] [CrossRef]
- Deavall, D.G.; Martin, E.A.; Horner, J.M.; Roberts, R.; Kang, Y.J. Drug-Induced Oxidative Stress and Toxicity. J. Toxicol. 2012, 2012, 645460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjum, N.A.; Sofo, A.; Scopa, A.; Roychoudhury, A.; Gill, S.S.; Iqbal, M.; Lukatkin, A.S.; Pereira, E.; Duarte, A.C.; Ahmad, I. Lipids and proteins—Major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. 2015, 22, 4099–4121. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, G.; Nam, S.; Yin, J.; Yoon, J. Visualization of Endogenous and Exogenous Hydrogen Peroxide Using A Lysosome-Targetable Fluorescent Probe. Sci. Rep. 2015, 5, 8488. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.; Qian, P.; Xin, W.; Li, H.; Burritt, D.J.; Fujita, M.; Tran, L.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef] [Green Version]
- Pandey, P.; Zaman, K.; Prokai, L.; Shulaev, V. Comparative Proteomics Analysis Reveals Unique Early Signaling Response of Saccharomyces cerevisiae to Oxidants with Different Mechanism of Action. Int. J. Mol. Sci. 2021, 22, 167. [Google Scholar] [CrossRef]
- Piccirillo, S.; Morales, R.; White, M.G.; Smith, K.; Kapros, T.; Honigberg, S.M. Cell Differentiation and Spatial Organization in Yeast Colonies: Role of Cell-Wall Integrity Pathway. Genetics 2015, 201, 1427–1438. [Google Scholar] [CrossRef] [Green Version]
- Lorna, M.Y.; Mitchison-Field, J.M.V.B.; Vogt, S.V.D.J.; Christine, M.; Field, A.S.G. Unconventional Cell Division Cycles from Marine-Derived Yeasts. Curr. Biol. 2019, 29, 3439–3456. [Google Scholar]
- Opalek, M.; Wloch-Salamon, D. Aspects of Multicellularity in Saccharomyces cerevisiae Yeast: A Review of Evolutionary and Physiological Mechanisms. Genes 2020, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Váchová, Z.P.L. Yeast cell differentiation: Lessons from pathogenic and non-pathogenic yeasts. Semin. Cell Dev. Biol. 2016, 57, 110–119. [Google Scholar]
- Dolan, S.; Welch, M. The Glyoxylate Shunt, 60 Years on. Annu. Rev. Microbiol. 2018, 72, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, M.; Nathan, C.; Rhee, K.Y. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 2014, 5, 4306. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef]
- Xiang Zou, C.C.J.F. Biosynthesis of polymalic acid in fermentation: Advances and prospects for industrial application. Crit. Rev. Biotechnol. 2019, 39, 408–421. [Google Scholar]
- Zalar, P.; Novak, M.; de Hoog, G.S.; Gunde-Cimerman, N. Dishwashers—A man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011, 115, 997–1007. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Zalar, P.; de Hoog, S.; Plemenitaš, A. Hypersaline waters in salterns—natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar]
- Botić, T.; Kralj-Kunčič, M.; Sepčić, K.; Batista, U.; Zalar, P.; Knez, Ž.; Gunde-Cimerman, N. Biological activities of organic extracts of four Aureobasidium pullulans varieties isolated from extreme marine and terrestrial habitats. Nat. Prod. Res. 2014, 28, 874–882. [Google Scholar] [CrossRef]
- Mouaheb, N.; Thomas, D.; Verdoucq, L.; Monfort, P.; Meyer, Y. In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 3312–3317. [Google Scholar] [CrossRef] [Green Version]
- Ramos, S.; Acha, I.G. A vegetative cycle of Pullularia pullulans. Trans. Br. Mycol. Soc. 1975, 64, 129–135. [Google Scholar] [CrossRef]
- Zeng, W.; Zhang, B.; Jiang, L.; Liu, Y.; Ding, S.; Chen, G.; Liang, Z. Poly(malic acid) production from liquefied corn starch by simultaneous saccharification and fermentation with a novel isolated Aureobasidium pullulans GXL-1 strain and its techno-economic analysis. Bioresour. Technol. 2020, 304, 122990. [Google Scholar] [CrossRef] [PubMed]
- No, H.; Kim, J.; Seo, C.; Lee, D.E.; Kim, J.H.; Kuge, T.; Mori, T.; Kimoto, H.; Kim, J. Anti-inflammatory effects of β-1,3-1,6-glucan derived from black yeast Aureobasidium pullulans in RAW264.7 cells. Int. J. Biol. Macromol. 2021, 193, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, N.; Peng, Q.; Yin, T.; Guan, F.; Wang, G.; Li, Y. Production of pigment-free pullulan by swollen cell in Aureobasidium pullulans NG which cell differentiation was affected by pH and nutrition. Appl. Microbiol. Biot. 2009, 84, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Esaki, M.; Ogura, T.; Arai, S.; Yamamoto, Y.; Tanaka, N. Whole-cell imaging of the budding yeast Saccharomyces cerevisiae by high-voltage scanning transmission electron tomography. Ultramicroscopy 2014, 146, 39–45. [Google Scholar] [CrossRef]
- Sangster, T.; Major, H.; Plumb, R.; Wilson, A.J.; Wilson, I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst 2006, 131, 1075–1078. [Google Scholar] [CrossRef]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef]
- Li, C.; Zhao, D.; Yan, J.; Zhang, N.; Li, B. Metabolomics integrated with transcriptomics: Assessing the central metabolism of marine red yeast Sporobolomyces pararoseus under salinity stress. Arch. Microbiol. 2021, 203, 889–899. [Google Scholar] [CrossRef]
- Ahmadi, F.I.; Karimi, K.; Struik, P.C. Effect of exogenous application of methyl jasmonate on physiological and biochemical characteristics of Brassica napus L. cv. Talaye under salinity stress. S. Afr. J. Bot. 2018, 115, 5–11. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, Z.; Wang, L.; Li, M.; Wang, W.; Yi, Q.; Huang, S.; Song, L. Dopamine and Serotonin Modulate Free Amino Acids Production and Na+/K+ Pump Activity in Chinese Mitten Crab Eriocheir sinensis Under Acute Salinity Stress. Front. Physiol. 2018, 9, 1080. [Google Scholar] [CrossRef]
- Quintana-Cabrera, R.; Bolaños, J.P. Glutathione and γ-glutamylcysteine in the antioxidant and survival functions of mitochondria. Biochem. Soc. Trans. 2013, 41, 106–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iskusnykh, I.Y.; Zakharova, A.A.; Pathak, D. Glutathione in Brain Disorders and Aging. Molecules 2022, 27, 324. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Liu, T.; Zhang, W.; Li, S.; Zhu, M.; Li, H.; Kong, Y.; Xu, L. Disclosure of the Molecular Mechanism of Wheat Leaf Spot Disease Caused by Bipolaris sorokiniana through Comparative Transcriptome and Metabolomics Analysis. Int. J. Mol. Sci. 2019, 20, 6090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Wang, Q.; Ma, X.; Lang, D.; Guo, Z.; Zhang, X. Physiological Biochemistry-Combined Transcriptomic Analysis Reveals Mechanism of Bacillus cereus G2 Improved Salt-Stress Tolerance of Glycyrrhiza uralensis Fisch. Seedlings by Balancing Carbohydrate Metabolism. Front. Plant Sci. 2022, 12, 712363. [Google Scholar] [CrossRef]
- Li, L.; Wang, F.; Yan, P.; Jing, W.; Zhang, C.; Kudla, J.; Zhang, W. A phosphoinositide-specific phospholipase C pathway elicits stress-induced Ca2+ signals and confers salt tolerance to rice. New Phytol. 2017, 214, 1172–1187. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.; Marinho, H.S.; Cyrne, L. Regulation of the inositol transporter Itr1p by hydrogen peroxide in Saccharomyces cerevisiae. Arch. Microbiol. 2019, 201, 123–134. [Google Scholar] [CrossRef]
- Ahn, S.; Jung, J.; Jang, I.; Madsen, E.L.; Park, W. Role of Glyoxylate Shunt in Oxidative Stress Response. J. Biol. Chem. 2016, 291, 11928–11938. [Google Scholar] [CrossRef] [Green Version]
- Tahjib-Ul-Arif, M.; Zahan, M.I.; Karim, M.M.; Imran, S.; Hunter, C.T.; Islam, M.S.; Mia, M.A.; Hannan, M.A.; Rhaman, M.S.; Hossain, M.A.; et al. Citric Acid-Mediated Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 7235. [Google Scholar] [CrossRef]
- Wu, X.; Dai, H.; Yu, S.; Zhao, Y.; Long, Y.; Li, W.; Tu, J. Citrate regulates extracellular matrix mineralization during osteoblast differentiation in vitro. J. Inorg. Biochem. 2021, 214, 111269. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Liao, W.; Peng, W.; Qin, J.; Chen, D.; Zheng, L.; Yan, W.; Li, L.; Guo, Z.; et al. Citrate-Stabilized Gold Nanorods-Directed Osteogenic Differentiation of Multiple Cells. Int. J. Nanomed. 2021, 16, 2789. [Google Scholar] [CrossRef]
- Fan, J.; Zheng, L. Acclimation to NaCl and light stress of heterotrophic Chlamydomonas reinhardtii for lipid accumulation. J. Biosci. Bioeng. 2017, 124, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Park, J.C.; Kim, D.; Kang, S.; Shin, K.; Park, H.G.; Han, J.; Lee, J. Interrelationship of salinity shift with oxidative stress and lipid metabolism in the monogonont rotifer Brachionus koreanus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 214, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ge, H.; Liu, T.; Tian, X.; Wang, Z.; Guo, M.; Chu, J.; Zhuang, Y. Salt stress induced lipid accumulation in heterotrophic culture cells of Chlorella protothecoides: Mechanisms based on the multi-level analysis of oxidative response, key enzyme activity and biochemical alteration. J. Biotechnol. 2016, 228, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.; Himbergen, J.V.; Musgrave, A.; Munnik, T. Acclimation to salt modifies the activation of several osmotic stress-activated lipid signalling pathways in Chlamydomonas. Phytochemistry 2017, 135, 64–72. [Google Scholar] [CrossRef]


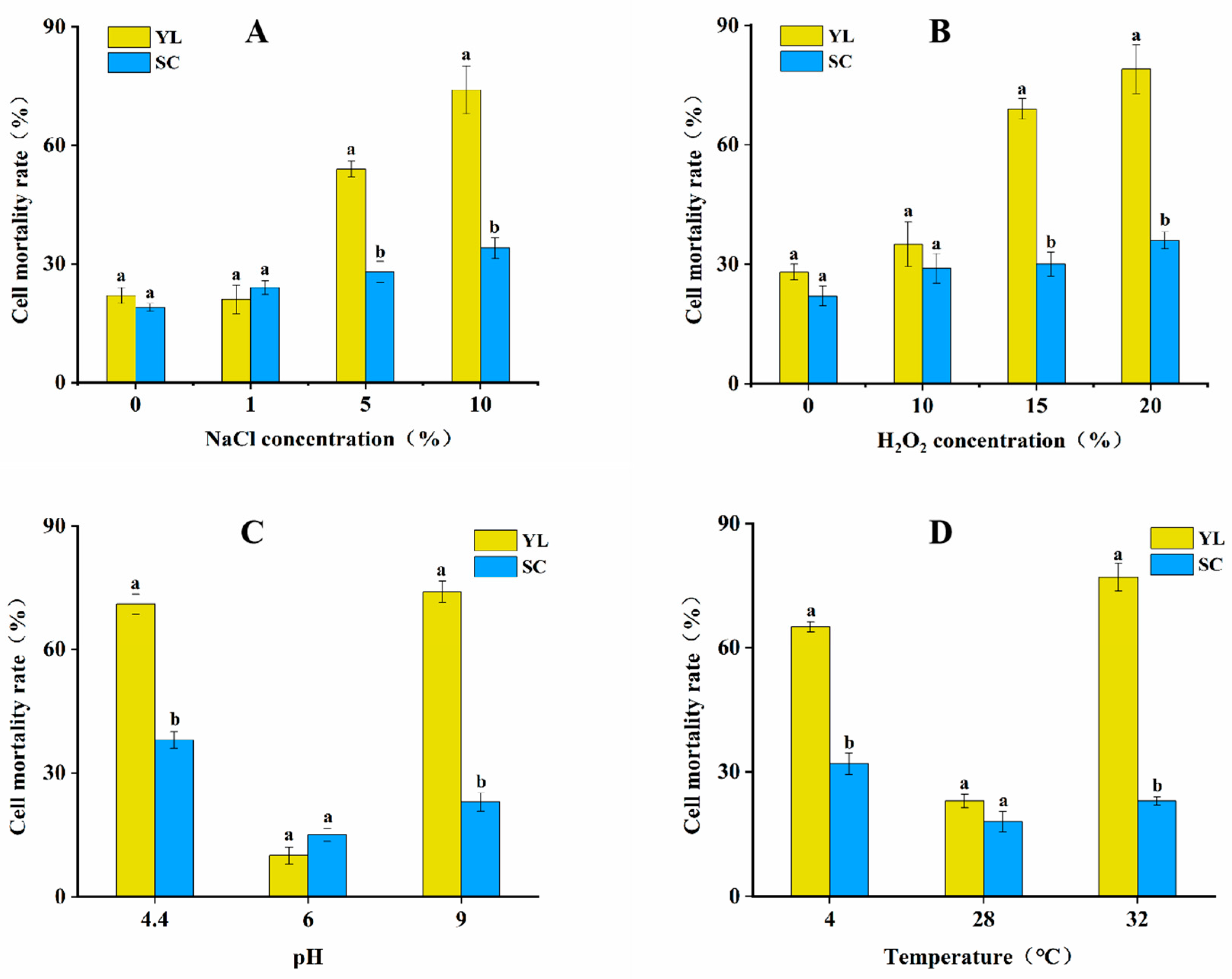

| Catalase (CAT) Activity (U/mg) | Superoxide Dismutase (SOD) Activity (U/mg) | SOD Inhibition Rate (%) | Total Antioxidant Capacity (T−AOC) (U/mg) | |
|---|---|---|---|---|
| Yeast cells | 2.87 ± 0.74 b | 63.03 ± 1.60 b | 60 | 4.067 ± 0.011 b |
| Swollen cells | 7.64 ± 2.44 a | 103.54 ± 7.71 a | 69 | 7.949 ± 0.017 a |
| Classification | ID | Description | Up/Downregulated |
|---|---|---|---|
| UniGene | SC16908_c1_g2 | Catalase domain-containing protein | Up |
| SC16627_c2_g5 | CAT2 catalase | Up | |
| SC2555_c0_g1 | L-lactate dehydrogenase | Up | |
| SC16598_c2_g1 | Acyl-CoA oxidase | Up | |
| SC16876_c1_g2 | Alcohol dehydrogenase | Up | |
| SC16337_c0_g3 | Glyceraldehyde-3-phosphate Dehydrogenase | Up | |
| SC16350_c0_g1 | Up | ||
| SC14418_c0_g3 | Alpha-amylase | Up | |
| SC16410_c0_g3 | Citrate synthase | Down | |
| SC16898_c5_g17 | Aconitate hydratase | Down | |
| Metabolites | com00158 | Citrate | Up |
| com00042 | Succinate | Up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, N.; Zhang, N.; Ma, X.; Wang, Y.; Zhang, Y.; Wang, D.; Pu, F.; Li, B. Transcriptomics Integrated with Metabolomics: Assessing the Central Metabolism of Different Cells after Cell Differentiation in Aureobasidium pullulans NG. J. Fungi 2022, 8, 882. https://doi.org/10.3390/jof8080882
Zeng N, Zhang N, Ma X, Wang Y, Zhang Y, Wang D, Pu F, Li B. Transcriptomics Integrated with Metabolomics: Assessing the Central Metabolism of Different Cells after Cell Differentiation in Aureobasidium pullulans NG. Journal of Fungi. 2022; 8(8):882. https://doi.org/10.3390/jof8080882
Chicago/Turabian StyleZeng, Nan, Ning Zhang, Xin Ma, Yunjiao Wang, Yating Zhang, Dandan Wang, Fangxiong Pu, and Bingxue Li. 2022. "Transcriptomics Integrated with Metabolomics: Assessing the Central Metabolism of Different Cells after Cell Differentiation in Aureobasidium pullulans NG" Journal of Fungi 8, no. 8: 882. https://doi.org/10.3390/jof8080882
APA StyleZeng, N., Zhang, N., Ma, X., Wang, Y., Zhang, Y., Wang, D., Pu, F., & Li, B. (2022). Transcriptomics Integrated with Metabolomics: Assessing the Central Metabolism of Different Cells after Cell Differentiation in Aureobasidium pullulans NG. Journal of Fungi, 8(8), 882. https://doi.org/10.3390/jof8080882






