Yeast Diversity in the Qaidam Basin Desert in China with the Description of Five New Yeast Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Soil Chemical Analyses
2.3. Yeast Isolation
2.4. Molecular Phylogenetic Analysis
2.5. Phenotypical Characterization
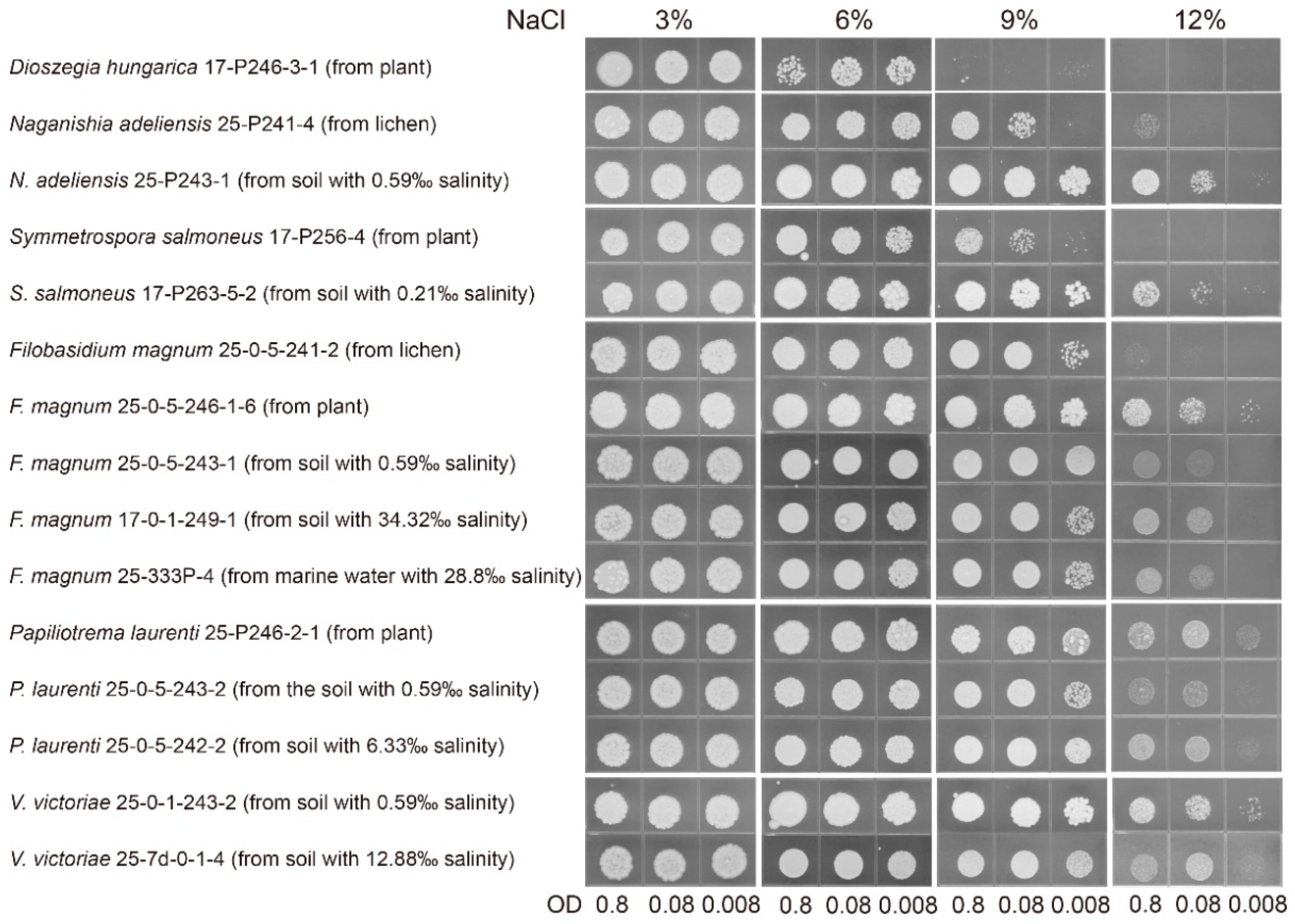
2.6. Salinity Tolerance Analysis
2.7. Statistical Analysis
3. Results
3.1. Yeast Diversity
3.2. Salt-Tolerant Ability
3.3. Phylogeny and Taxonomy of Novel Yeast Species
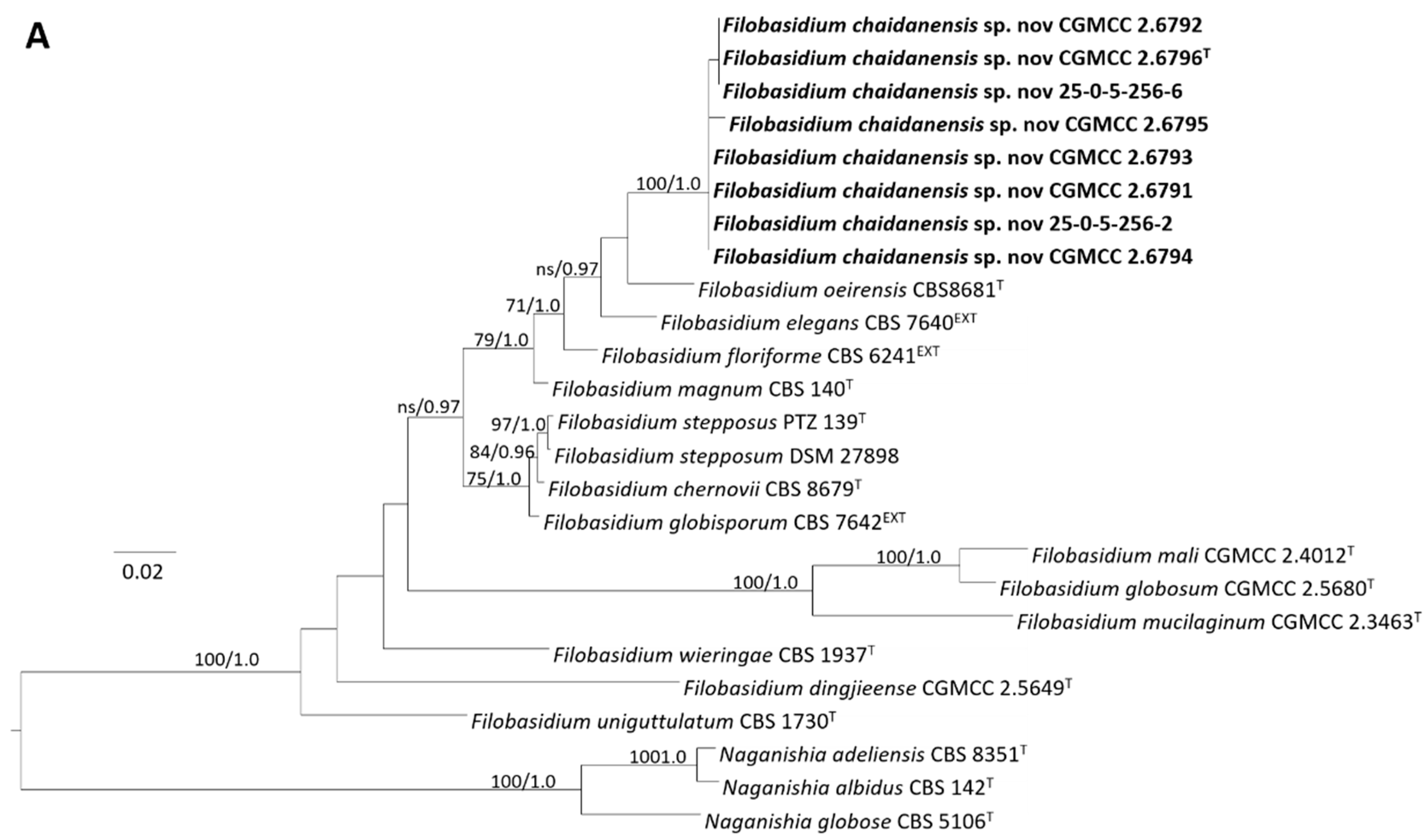
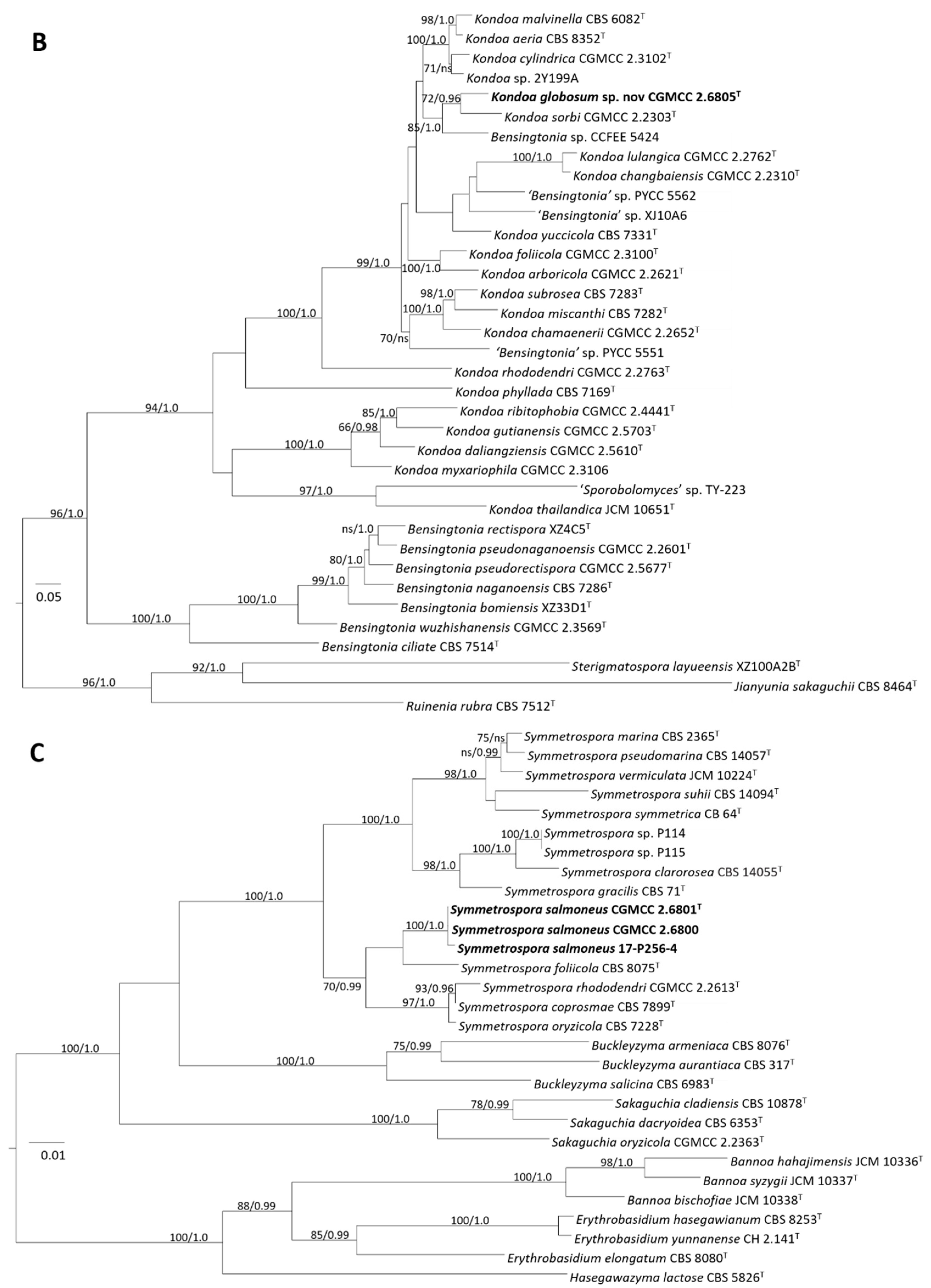
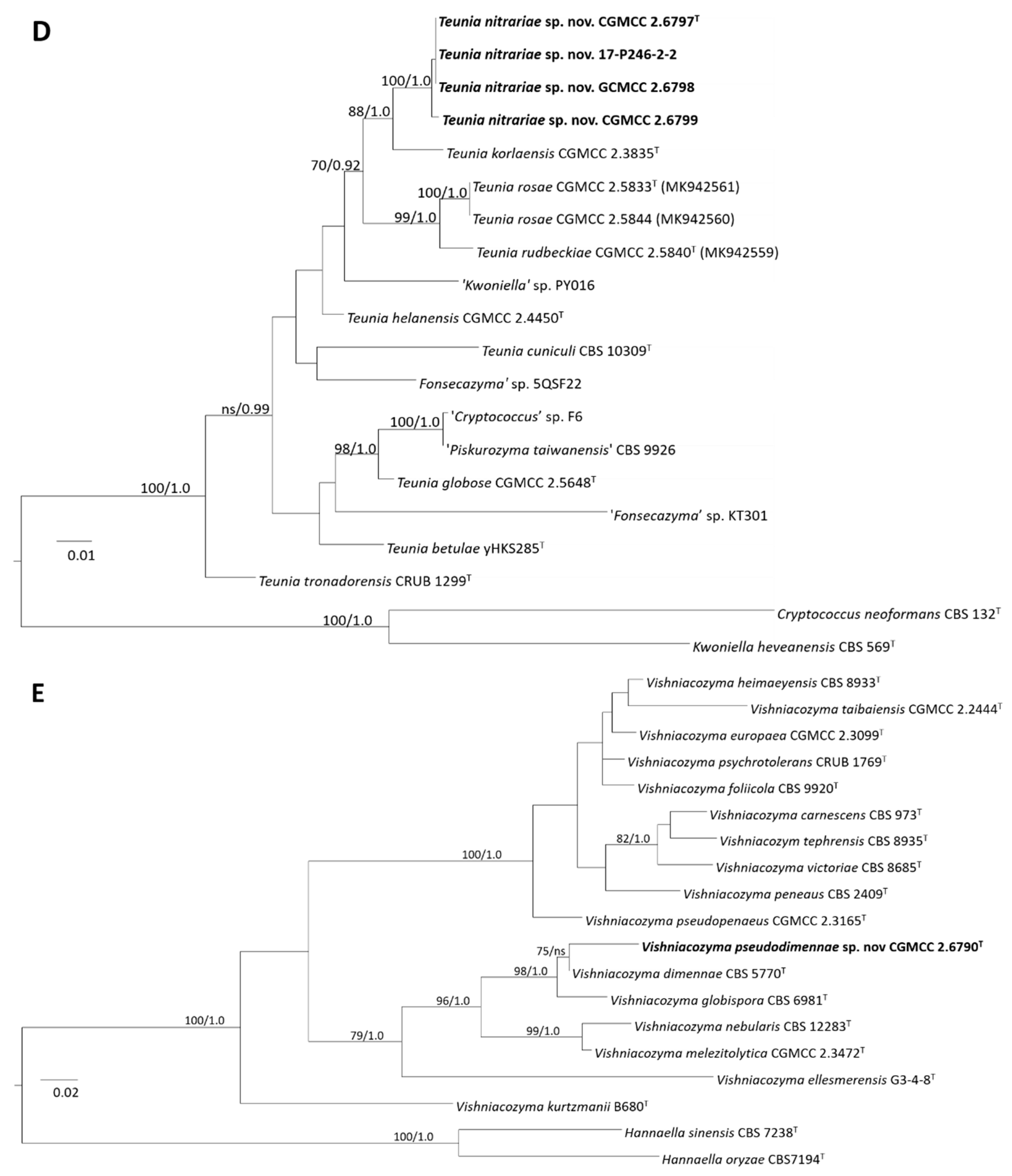
3.3.1. Phylogeny
3.3.2. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farr, T.G. Terrestrial analogs to Mars: The NRC community decadal report. Planet Space Sci. 2004, 52, 3–10. [Google Scholar] [CrossRef]
- Léveillé, R.; Mars, A.S. Encyclopedia of Astrobiology; Gargaud, M., Irvine, W.M., Amils, R., Cleaves, H.J., Pinti, D.L., Quintanilla, J.C., Rouan, D., Spohn, T., Tirard, S., Viso, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; p. 977. [Google Scholar]
- McKay, C.P.; Mancinelli, R.L.; Stoker, C.R.; Wharton, R.A., Jr. The possibility of life on Mars during a water-rich past. In Mars; Kieffer, H.H., Jakosky, B.M., Snyder, C.W., Matthews, M.S., Eds.; The University of Arizona Press: Tucson, AZ, USA, 1992; pp. 1234–1245. [Google Scholar]
- Brack, A. Life on Mars: A clue to life on Earth? Chem. Biol. 1997, 4, 9–12. [Google Scholar] [CrossRef]
- Fairén, A.; Davila, A.; Darlene, L.; Nathan, B.; Rosalba, B.; Jhony, Z.; Uceda, E.; Carol, S.; Jacek, W.; Dohm, J.; et al. Astrobiology through the ages of Mars: The study of terrestrial analogues to understand the habitability of Mars. Astrobiology 2010, 10, 821–843. [Google Scholar] [CrossRef] [PubMed]
- Cockell, C.S. Trajectories of Martian habitability. Astrobiology 2014, 14, 182–203. [Google Scholar] [CrossRef]
- Grotzinger, J.P.; Sumner, D.Y.; Kah, L.C.; Stack, K.; Gupta, S.; Edgar, L.; Rubin, D.; Lewis, K.; Schieber, J.; Mangold, N.; et al. A habitable fluvio-lacustrine environment at Yellowknife Bay, Gale Crater, Mars. Science 2014, 343, 1242777. [Google Scholar] [CrossRef]
- Westall, F.; Foucher, F.; Bost, N.; Bertrand, M.; Loizeau, D.; Vago, J.L.; Kminek, G.; Gaboyer, F.; Campbell, K.A.; Bréhéret, J.G.; et al. Biosignatures on Mars: What, Where, and How? Implications for the Search for Martian life. Astrobiology 2015, 15, 998–1029. [Google Scholar] [CrossRef]
- Mormile, M.R.; Hong, B.Y.; Benison, K.C. Molecular analysis of the microbial communities of Mars analog lakes in Western Australia. Astrobiology 2009, 9, 919–930. [Google Scholar] [CrossRef]
- Foing, B.H.; Stoker, C.; Zavaleta, J.; Ehrenfreund, P.; Thiel, C.; Sarrazin, P.; Blake, D.; Page, J.; Pletser, V.; Hendrikse, J.; et al. Field astrobiology research in Moon-Mars analogue environments: Instruments and methods. Int. J. Astrobiol. 2011, 10, 141–160. [Google Scholar] [CrossRef]
- Amils, R.; Fernández-Remolar, D.; The Ipbsl Team. Río Tinto: A Geochemical and Mineralogical Terrestrial Analogue of Mars. Life 2014, 4, 511–534. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, J.; Dang, Y.; Cheng, Z.; Huang, T.; Zhao, J.; Xu, Y.; Huang, J.; Xiao, Z.; Komatsu, G. A new terrestrial analogue site for Mars research: The Qaidam Basin, Tibetan Plateau (NW China). Earth-Sci. Rev. 2017, 164, 84–101. [Google Scholar] [CrossRef]
- Bull, A.T.; Andrews, B.A.; Dorador, C.; Goodfellow, M. Introducing the Atacama Desert. Antonie Leeuwenhoek 2018, 111, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Cassaro, A.; Pacelli, C.; Aureli, L.; Catanzaro, I.; Leo, P.; Onofri, S. Antarctica as a reservoir of planetary analogue environments. Extremophiles 2021, 25, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Azua-Bustos, A.; González-Silva, C.; Fairén, A.G. The Atacama Desert in Northern Chile as an Analog Model of Mars. Front. Astron. Space Sci. 2022, 8, 810426. [Google Scholar] [CrossRef]
- Hallsworth, J.E.; Mancinelli, R.L.; Conley, C.A.; Dallas, T.D.; Rinaldi, T.; Davila, A.F.; Benison, K.C.; Rapoport, A.; Cavalazzi, B.; Selbmann, L.; et al. Astrobiology of life on Earth. Environ. Microbiol. 2021, 23, 3335–3344. [Google Scholar] [CrossRef]
- Thiel, C.; Ehrenfreund, P.; Foing, B.; Pletser, V.; Ullrich, O. PCR-based analysis of microbial communities during the EuroGeoMars campaign at Mars Desert Research Station, Utah. Int. J. Astrobiol. 2011, 10, 177–190. [Google Scholar] [CrossRef]
- Dorador, C.; Fink, P.; Hengst, M.; Icaza, G.; Villalobos, A.S.; Vejar, D.; Meneses, D.; Zadjelovic, V.; Burmann, L.; Moelzner, J.; et al. Microbial community composition and trophic role along a marked salinity gradient in Laguna Puilar, Salar de Atacama, Chile. Antonie Van Leeuwenhoek 2018, 111, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Santiago, I.F.; Gonçalves, V.N.; Gómez-Silva, B.; Galetovic, A.; Rosa, L.H. Fungal diversity in the Atacama Desert. Antonie Leeuwenhoek 2018, 111, 1345–1360. [Google Scholar] [CrossRef]
- Aszalós, J.M.; Szabó, A.; Megyes, M.; Anda, D.; Nagy, B.; Borsodi, A.K. Bacterial Diversity of a High-Altitude Permafrost Thaw Pond Located on Ojos del Salado (Dry Andes, Altiplano-Atacama Region). Astrobiology 2020, 20, 754–765. [Google Scholar] [CrossRef]
- Bashir, A.K.; Wink, L.; Duller, S.; Schwendner, P.; Cockell, C.; Rettberg, P.; Mahnert, A.; Beblo-Vranesevic, K.; Bohmeier, M.; Rabbow, E.; et al. Taxonomic and functional analyses of intact microbial communities thriving in extreme, astrobiology-relevant, anoxic sites. Microbiome 2021, 9, 50. [Google Scholar] [CrossRef]
- O’Connor, B.R.W.; Fernández-Martínez, M.Á.; Léveillé, R.J.; Whyte, L.G. Taxonomic Characterization and Microbial Activity Determination of Cold-Adapted Microbial Communities in Lava Tube Ice Caves from Lava Beds National Monument, a High-Fidelity Mars Analogue Environment. Astrobiology 2021, 21, 613–627. [Google Scholar] [CrossRef]
- Wu, D.; Senbayram, M.; Moradi, G.; Mörchen, R.; Knief, C.; Klumpp, E.; Jones, D.L.; Well, R.; Chen, R.; Bol, R. Microbial potential for denitrification in the hyperarid Atacama Desert soils. Soil Biol. Biochem. 2021, 157, 108248. [Google Scholar] [CrossRef]
- Azua-Bustos, A.; Caro-Lara, L.; Vicuña, R. Discovery and microbial content of the driest site of the hyperarid Atacama Desert, Chile. Environ. Microbiol. Rep. 2015, 7, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Hynek, B.M.; Rogers, K.L.; Antunovich, M.; Avard, G.; Alvarado, G.E. Lack of Microbial Diversity in an Extreme Mars Analog Setting: Poás Volcano, Costa Rica. Astrobiology 2018, 18, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Fonseca, Á.; Sampaio, J.P.; Bandoni, R.J.; Fell, J.W.; Kwon-Chung, K.J. Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In The Yeasts: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1339–1372. [Google Scholar]
- Groenewald, M.; Boundy-Mills, K.; Čadež, N.; Endoh, R.; Jindamorakot, S.; Pohl-Albertyn, C.; Rosa, C.A.; Turchetti, B.; Yurkov, A. Census of yeasts isolated from natural ecosystem and conserved in worldwide collections. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 455–476. [Google Scholar]
- Gadanho, M.; Libkind, D.; Sampaio, J.P. Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb. Ecol. 2006, 52, 552–563. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Rodrigues, F.; Duarte, R.T.; Araujo, G.G.; Santiago, I.F.; Paulino-Lima, I.G.; Rosa, C.A.; Kato, M.J.; Pellizari, V.H.; Galante, D. UV-resistant yeasts isolated from a high-altitude volcanic area on the Atacama Desert as eukaryotic models for astrobiology. Microbiologyopen 2015, 4, 574–588. [Google Scholar] [CrossRef]
- Mokhtarnejad, L.; Arzanlou, M.; Babai-Ahari, A.; Di Mauro, S.; Onofri, A.; Buzzini, P.; Turchetti, B. Characterization of basidiomycetous yeasts in hypersaline soils of the Urmia Lake National Park, Iran. Extremophiles 2016, 20, 915–928. [Google Scholar] [CrossRef]
- Buzzini, P.; Turk, M.; Perini, L.; Turchetti, B.; Gunde-Cimerman, N. Yeasts in polar and subpolar habitats. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 331–365. [Google Scholar]
- Mitsuya, D.; Hayashi, T.; Wang, Y.; Tanaka, M.; Okai, M.; Ishida, M.; Urano, N. Isolation of aquatic yeasts with the ability to neutralize acidic media, from an extremely acidic river near Japan’s Kusatsu-Shirane Volcano. J. Biosci. Bioeng. 2017, 124, 43–46. [Google Scholar] [CrossRef]
- Sannino, C.; Tasselli, G.; Filippucci, S.; Turchetti, B.; Buzzini, P. Yeasts in nonpolar cold habitats. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 366–396. [Google Scholar]
- Luo, B.; Sun, H.; Zhang, Y.; Gu, Y.; Yan, W.; Zhang, R.; Ni, Y. Habitat-specificity and diversity of culturable cold-adapted yeasts of a cold-based glacier in the Tianshan Mountains, northwestern China. Appl. Microbiol. Biotechnol. 2019, 103, 2311–2327. [Google Scholar] [CrossRef]
- Anglés, A.; Li, Y. The western Qaidam Basin as a potential Martian environmental analogue: An overview. J. Geophys. Res.-Planets 2017, 122, 856–888. [Google Scholar] [CrossRef]
- Ding, Z.J.; Zhao, J.N.; Wang, J.; Lai, Z.P. Yardangs on Earth and implications to Mars: A review. Geomorphology 2020, 364, 107230. [Google Scholar] [CrossRef]
- Chen, L.; Xu, Y.; Li, B. Comparative study of the geomorphological characteristics of valley networks between Mars and the Qaidam Basin. Remote Sens. 2021, 13, 4471. [Google Scholar] [CrossRef]
- Liu, W.; Li, J.; Gu, W.; Santos, L.F.E.D.; Boman, J.; Zhang, X.; Tang, M.; Wang, S.; Kong, X. Chemical and hygroscopic characterization of surface salts in the Qaidam Basin: Implications for climate impacts on Planet Earth and Mars. ACS Earth Space Chem. 2021, 5, 651–662. [Google Scholar] [CrossRef]
- Zhong, Z.P.; Liu, Y.; Miao, L.L.; Wang, F.; Chu, L.M.; Wang, J.L.; Liu, Z.P. Prokaryotic Community Structure Driven by Salinity and Ionic Concentrations in Plateau Lakes of the Tibetan Plateau. Appl. Environ. Microbiol. 2016, 82, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Xiao, L.; Wang, H.; Yang, H.; Li, J.; Huang, T.; Xu, Y.; Ma, N. Bacterial and archaeal lipids recovered from subsurface evaporites of Dalangtan Playa on the Tibetan Plateau and their astrobiological implications. Astrobiology 2017, 17, 1112–1122. [Google Scholar] [CrossRef]
- Zhang, W.; Bahadur, A.; Zhang, G.; Zhang, B.; Wu, X.; Chen, T.; Liu, G. Diverse bacterial communities from Qaidam Basin of the Qinghai-Tibet Plateau: Insights into variations in bacterial diversity across different regions. Front. Microbiol. 2020, 11, 554105. [Google Scholar] [CrossRef]
- Makimura, K.; Murayama, S.Y.; Yamaguchi, H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 1994, 40, 358–364. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, Q.M.; Theelen, B.; Groenewald, M.; Bai, F.Y.; Boekhout, T. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud. Mycol. 2015, 81, 1–26. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Effificient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts, A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 87–110. [Google Scholar]
- Do Carmo-Sousa, L.; Phaff, H.J. An improved method for the detection of spore discharge in the Sporobolomycetaceae. J. Bacteriol. 1962, 83, 434–435. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 15 February 2022).
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vires, M.; Verkleij, G.J.M.; Crous, P.W.; Boekhout, T.; et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Selbmann, L.; Zucconi, L.; Onofri, S.; Cecchini, C.; Isola, D.; Turchetti, B.; Buzzini, P. Taxonomic and phenotypic characterization of yeasts isolated from worldwide cold rock-associated habitats. Fungal Biol. 2014, 118, 61–71. [Google Scholar] [CrossRef]
- Péter, G.; Takashima, M.; Čadež, N. Yeast habitats: Different but global. In Yeasts in Natural Ecosystems: Ecology; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 38–71. [Google Scholar]
- Lisichkina, G.A.; Babjeva, I.P.; Sorokin, D.Y. Alkalitolerant yeasts from natural biotopes. Microbiology 2003, 72, 618–620. [Google Scholar] [CrossRef]
- Su, J.; Ma, X.R.; Abduwaly, A.; Suo, F.; Muhtar, A.; Erkin, R. Isolation and identification of a new alkalitolerant yeast strain Trichosporon asahii var. asahii XJU-1 from spontaneous fermentation millet juice. Biotechnology 2012, 22, 55–60. [Google Scholar]
- Turchetti, B.; Buzzini, P.; Goretti, M.; Branda, E.; Diolaiuti, G.; D’Agata, C.; Smiraglia, C.; Vaughan-Martini, A. Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol. Ecol. 2008, 63, 73–83. [Google Scholar] [CrossRef]
- Buzzini, P.; Turchetti, B.; Yurkov, A. Extremophilic yeasts: The toughest yeasts around? Yeast 2018, 35, 487–497. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Assessment of yeast diversity in soils under different management regimes. Fungal Ecol. 2012, 5, 24–35. [Google Scholar] [CrossRef]
- Vishniac, H.S. A multivariate analysis of soil yeasts isolated from a latitudinal gradient. Microb. Ecol. 2006, 52, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Mestre, M.C.; Fontenla, S. Yeast communities associated with ectomycorrehizal fungi indifferent Nothofagus forests of northwestern Patagonia. Symbiosis 2021, 84, 179–193. [Google Scholar] [CrossRef]
- Butinar, L.; Spencer-Martins, I.; Gunde-Cimerman, N. Yeasts in high Arctic glaciers: The discovery of a new habitat for eukaryotic microorganisms. Antonie Leeuwenhoek 2007, 91, 277–289. [Google Scholar] [CrossRef]
- Abughosh, S.; Droby, S.; Korine, C. Seasonal and plant-dependent variations in diversity, abundance and stress tolerance of epiphytic yeasts in desert habitats. Environ. Microbiol. Rep. 2014, 6, 373–382. [Google Scholar] [CrossRef]
- Deng, Z.J.; Wang, W.F.; Tan, H.M.; Cao, L.X. Characterization of heavy metal-resistant endophytic yeast Cryptococcus sp. CBSB78 from rapes (Brassica chinensis) and its potential in promoting the growth of Brassica spp. in metal-contaminated soils. Water Air Soil Pollut. 2012, 223, 5321–5329. [Google Scholar] [CrossRef]
- Loureiro, S.T.A.; de Queiroz Cavalcanti, M.A.; Neves, R.P.; de Oliveira Passavante, J.Z. Yeasts isolated from sand and sea water in beaches of Olinda, Pernambuco state, Brazil. Braz. J. Microbiol. 2005, 36, 333–337. [Google Scholar] [CrossRef]
- Vogel, C.; Rogerson, A.; Schatz, S.; Laubach, H.; Tallman, A.; Fell, J. Prevalence of yeasts in beach sand at three bathing beaches in South Florida. Water Res. 2007, 41, 1915–1920. [Google Scholar] [CrossRef]
- Raspor, P.; Zupan, J. Yeasts in extreme environments. In Biodiversity and Ecophysiology of Yeasts; Rosa, C.A., Péter, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 371–417. [Google Scholar]
- Gunde-Cimerman, N.; Plemenitaš, A. Ecology and molecular adaptions of the halophilic black yeast Hortaea werneckii. Rev. Environ. Sci. Biotechnol. 2006, 5, 323–331. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Buzzini, P. Change in lipids composition and fluidity of yeast plasma membrane as response to cold. In Cold-Adapted Yeasts; Buzzini, P., Margesin, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 225–242. [Google Scholar]
- Buzzini, P.; Branda, E.; Goretti, M.; Turchetti, B. Psychrophilic yeasts from worldwide glacial habitats: Diversity, adaptation strategies and biotechnological potential. FEMS Microbiol. Ecol. 2012, 82, 217–241. [Google Scholar] [CrossRef]
- Nguyen, V.A.T.; Senoo, K.; Mishima, T.; Hisamatsu, M. Multiple tolerance of Rhodotorula glutinis R-1 to acid, aluminum ion and manganese ion, and its unusual ability of neutralizing acidic medium. J. Biosci. Bioeng. 2001, 92, 366–371. [Google Scholar] [CrossRef]
- Aksenov, S.I.; Babyeva, I.P.; Golubev, V. On the mechanism of adaptation of micro-organisms to conditions of extreme low humidity. Life Sci. Space Res. 1973, 11, 55–61. [Google Scholar] [PubMed]
- Petrovič, U.; Gunde-Cimerman, N.; Plemenitaš, A. Cellular responses to environmental salinity in the halophilic black yeast Hortaea werneckii. Mol. Microbiol. 2002, 45, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Hong, C.P.; Thak, E.J.; Park, S.G.; Lee, C.H.; Lim, J.Y.; Seo, J.A.; Kang, H.A. Integrated genomic and transcriptomic analysis reveals unique mechanisms for high osmotolerance and halotolerance in Hyphopichia yeast. Environ. Microbiol. 2021, 23, 3499–3522. [Google Scholar] [CrossRef] [PubMed]
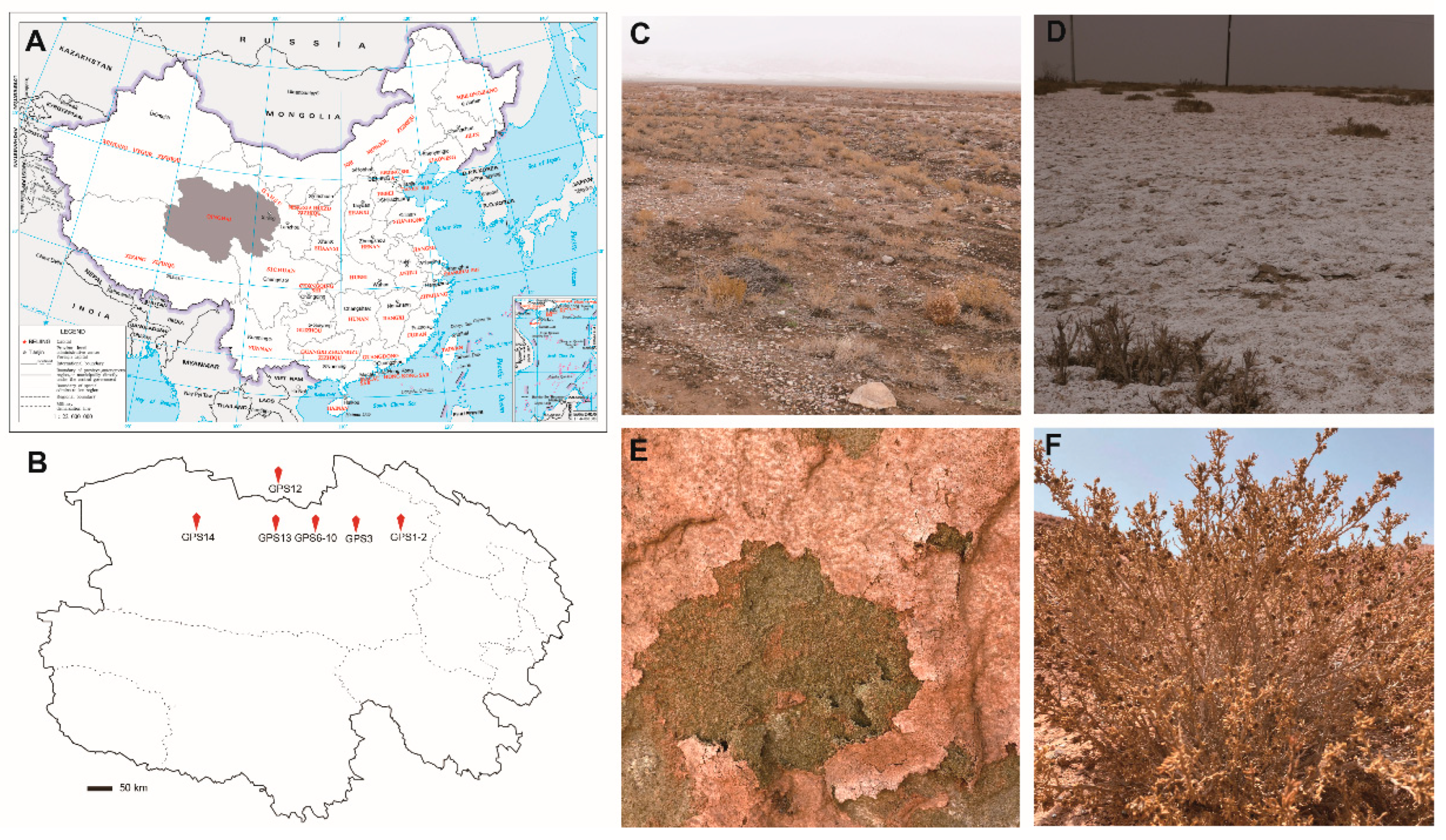


| Sampling Sites | Coordinates | Altitude (m) | Nature of Sample | Sample Type | Sample No. | Salinity (‰) | pH | Presence of Yeasts |
|---|---|---|---|---|---|---|---|---|
| GPS1 | 97°13′00″ E 37°20′27″ N | 2922 | Gobi desert | Soil | 248 | 1.15 ± 0.00 | 7.93 ± 0.01 | No |
| GPS2 | 97°08′44″ E 37°23′02″ N | 3041.5 | Gobi desert, colonized by Nitraria tangutorum and lichen | Soil | 245 | 0.16 ± 0.00 | 7.86 ± 0.01 | No |
| Vegetation soil | 247 | 0.25 ± 0.00 | 8.10 ± 0.01 | Yes | ||||
| Plant | 246 | / | / | Yes | ||||
| Lichen(Endocarpon) | 241 | / | / | Yes | ||||
| GPS3 | 96°51′15″ E 37°15′49″ N | 2813.6 | Saline-alkali land | Soil | 249 | 34.32 ± 1.13 | 8.22 ± 0.00 | Yes |
| Saline-alkali soil | 250 | 50.36 ± 0.39 | 8.03 ± 0.01 | No | ||||
| Plant | 251 | / | / | Yes | ||||
| GPS6 | 95°10′49″ E 37°34′51″ N | 3499 | Gobi desert | Plant | 255 | / | / | Yes |
| GPS7 | 95°09′41″ E 37°33′19″ N | 3370 | Yardang/Danxia landforms | Soil | 254 | 1.85 ± 0.01 | 8.85 ± 0.01 | Yes |
| Salt crystal | 252 | 58.51 ± 0.55 | 7.75 ± 0.01 | No | ||||
| GPS8 | 95°08′54″ E 37°31′09″ N | 3261 | Yardang/Danxia landforms | Red soil | 263-8 | 1.23 ± 0.00 | 8.37 ± 0.01 | Yes |
| Sandy soil | 263-5 | 0.21 ± 0.01 | 7.95 ± 0.01 | Yes | ||||
| Green sandy soil | 263-1 | 4.17 ± 0.02 | 7.82 ± 0.01 | Yes | ||||
| Green soil crust | 263-4 | 8.69 ± 0.02 | 7.54 ± 0.01 | No | ||||
| Yellow soil crust | 263-10 | 16.78 ± 0.00 | 7.88 ± 0.00 | No | ||||
| Plant | 263 | / | / | Yes | ||||
| GPS9 | 95°11′21″ E 37°32′25″ N | 3354 | Yardang/Danxia landforms | Soil | 261 | 1.84 ± 0.01 | 8.03 ± 0.00 | No |
| Rhizosphere soil | 260 | 0.69 ± 0.00 | 8.34 ± 0.02 | Yes | ||||
| Plant | 259 | / | / | No | ||||
| GPS10 | 95°12′20″ E 37°31′54″ N | 3424 | Yardang/Danxia landforms | Soil | 258 | 0.26 ± 0.00 | 9.74 ± 0.00 | No |
| Rhizosphere soil | 257 | 1.32 ± 0.01 | 9.01 ± 0.00 | No | ||||
| Plant | 256 | / | / | Yes | ||||
| GPS12 | 94°39′13″ E 38°1′36″ N | 2921 | Gobi desert, colonized by Nitraria tangutorum | Soil | 243 | 0.59 ± 0.00 | 7.99 ± 0.01 | Yes |
| Plant | 244 | / | / | No | ||||
| GPS13 | 94°17′23″ E 37°57′35″ N | 2754 | Yardang landforms | Saline-alkali soil | 264 | 12.88 ± 0.01 | 7.65 ± 0.01 | Yes |
| GPS14 | 93°46′19″ E 37°37′27″ N | 3198 | Sandy area and Saline-alkali land | Soil | 242 | 6.33 ± 0.02 | 7.87 ± 0.01 | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.-Y.; Zhu, H.-Y.; Song, L.; Zhang, R.-P.; Li, A.-H.; Niu, Q.-H.; Liu, X.-Z.; Bai, F.-Y. Yeast Diversity in the Qaidam Basin Desert in China with the Description of Five New Yeast Species. J. Fungi 2022, 8, 858. https://doi.org/10.3390/jof8080858
Wei X-Y, Zhu H-Y, Song L, Zhang R-P, Li A-H, Niu Q-H, Liu X-Z, Bai F-Y. Yeast Diversity in the Qaidam Basin Desert in China with the Description of Five New Yeast Species. Journal of Fungi. 2022; 8(8):858. https://doi.org/10.3390/jof8080858
Chicago/Turabian StyleWei, Xu-Yang, Hai-Yan Zhu, Liang Song, Ri-Peng Zhang, Ai-Hua Li, Qiu-Hong Niu, Xin-Zhan Liu, and Feng-Yan Bai. 2022. "Yeast Diversity in the Qaidam Basin Desert in China with the Description of Five New Yeast Species" Journal of Fungi 8, no. 8: 858. https://doi.org/10.3390/jof8080858
APA StyleWei, X.-Y., Zhu, H.-Y., Song, L., Zhang, R.-P., Li, A.-H., Niu, Q.-H., Liu, X.-Z., & Bai, F.-Y. (2022). Yeast Diversity in the Qaidam Basin Desert in China with the Description of Five New Yeast Species. Journal of Fungi, 8(8), 858. https://doi.org/10.3390/jof8080858






