Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Plot Setup and Soil Sampling
2.3. Determination of Soil Physicochemical Properties
2.4. DNA Extraction, MiSeq Sequencing, and Bioinformatics
2.5. Statistical Analysis
3. Results
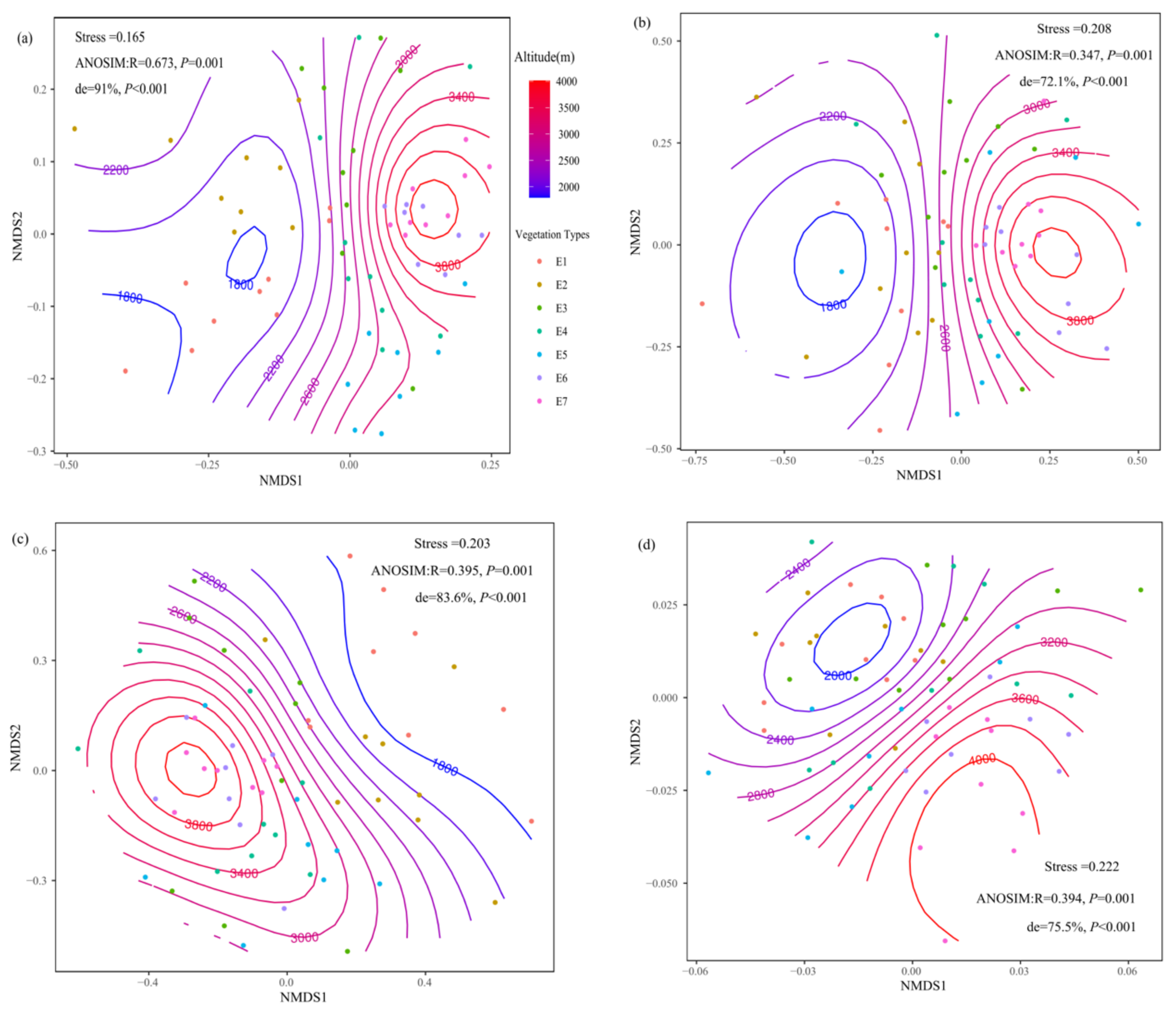
3.1. Soil Fungal Community Composition
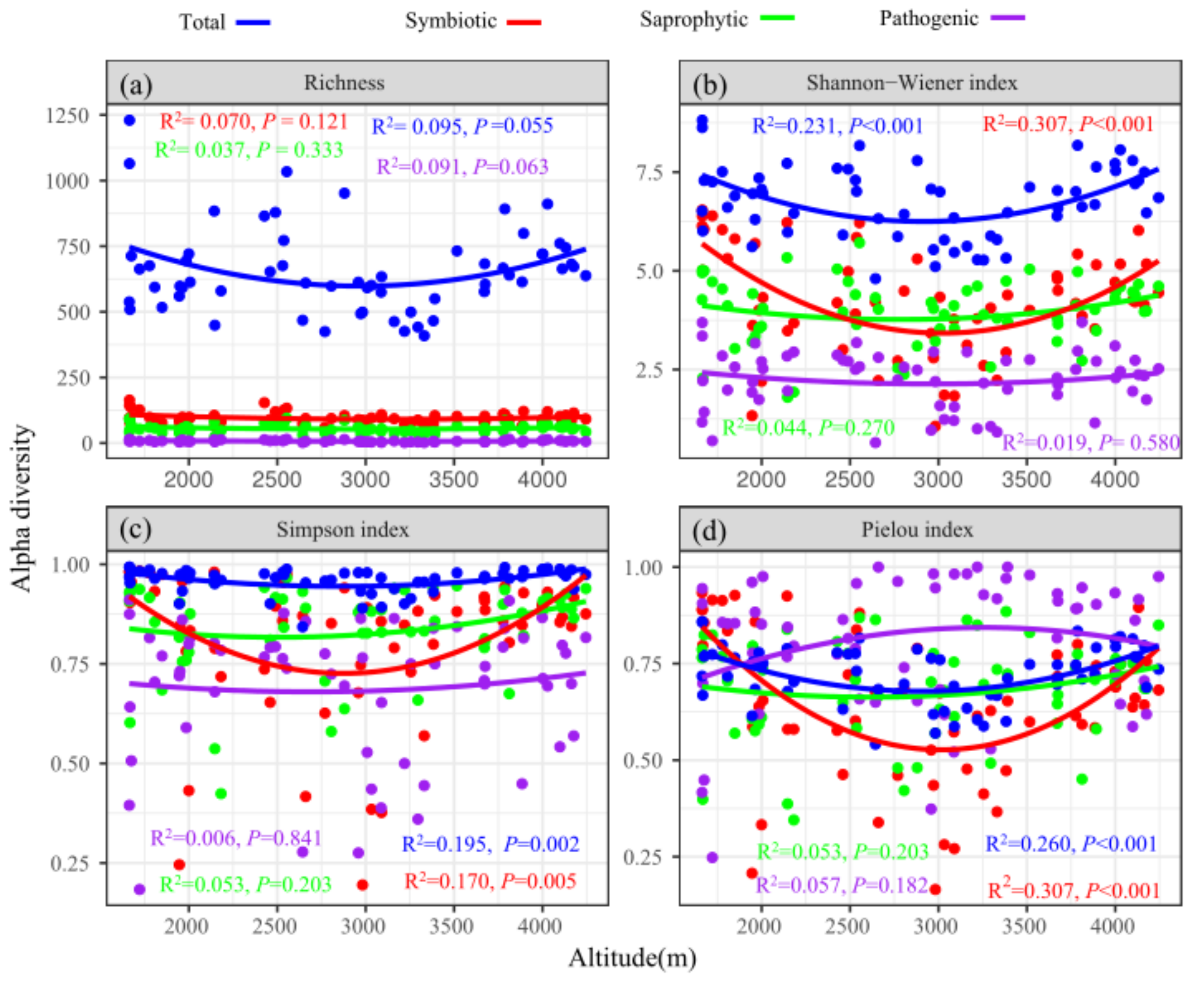
3.2. Soil Fungal Diversity
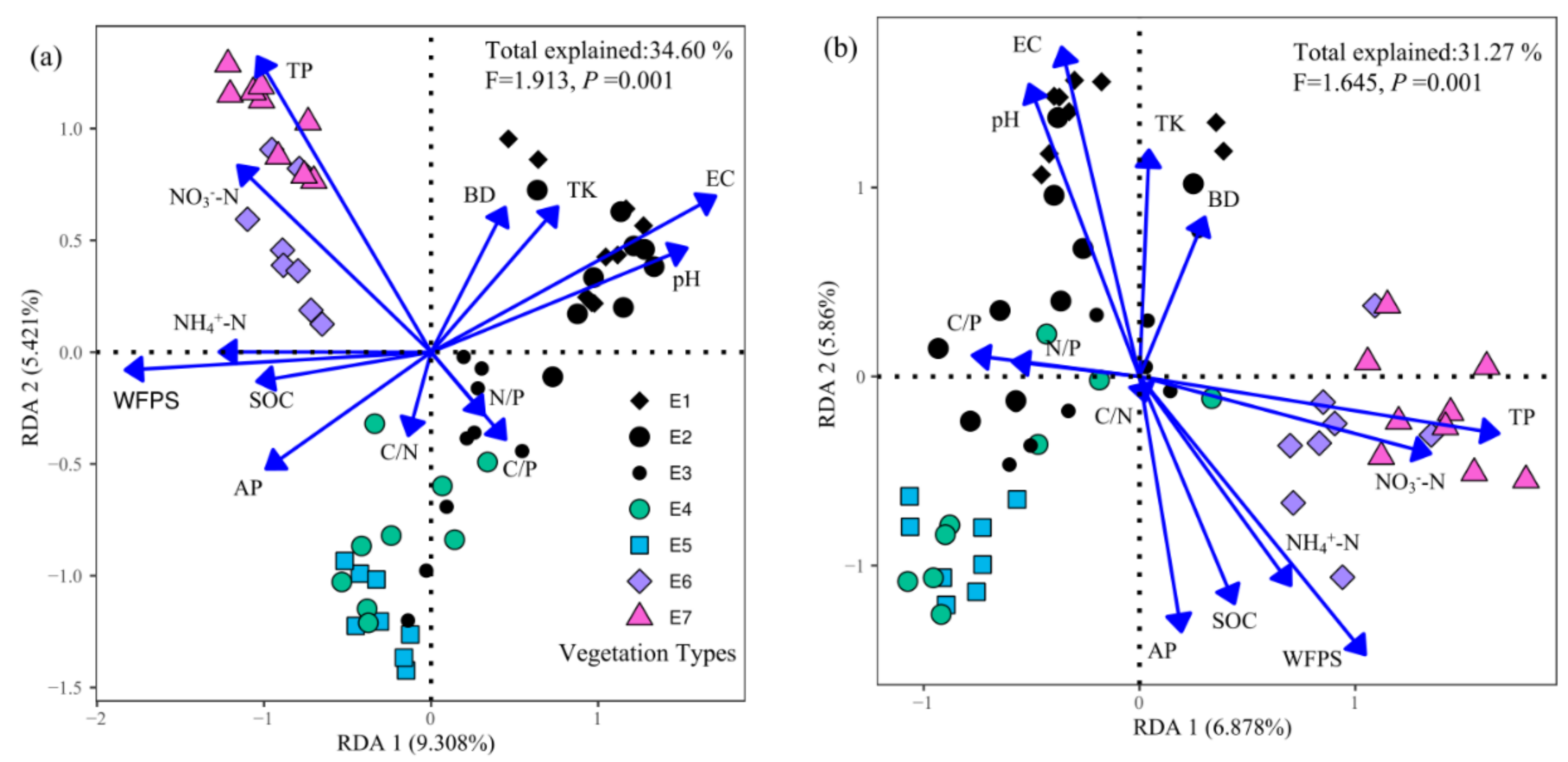
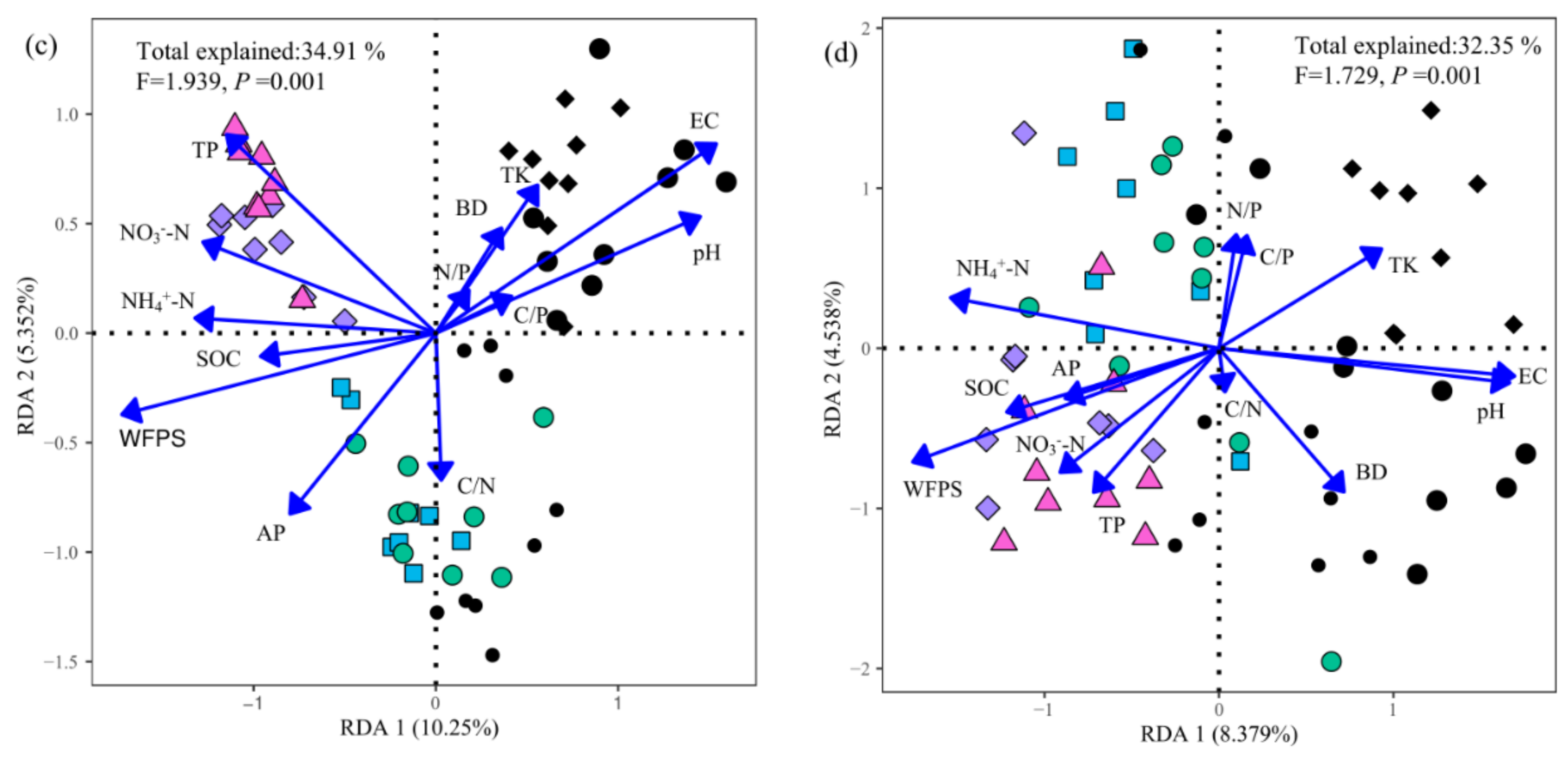
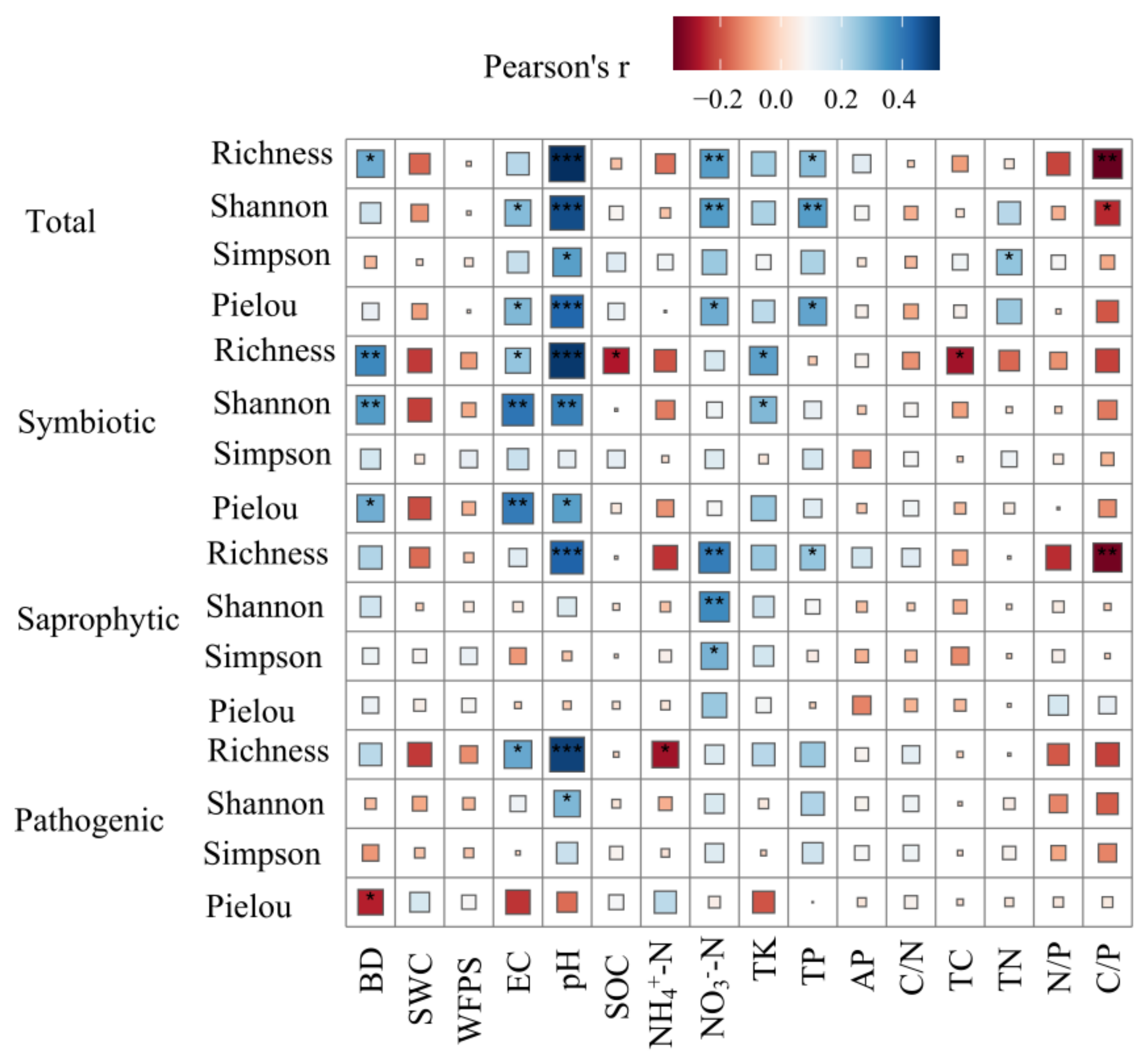
3.3. Drivers of the Soil Fungal Community
4. Discussion
4.1. Soil Fungal Community Composition
4.2. Soil Fungal Diversity
4.3. Drivers of the Soil Fungal Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal, R.L.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Rewald, B.; Matthews, B.; Sanden, H.; Rosinger, C.; Katzensteiner, K.; Gorfer, M.; Berger, H.; Tallian, C.; Berger, T.W.; et al. Soil fertility relates to fungal-mediated decomposition and organic matter turnover in a temperate mountain forest. New Phytol. 2021, 231, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef] [PubMed]
- Steidinger, B.S.; Bhatnagar, J.M.; Vilgalys, R.; Taylor, J.W.; Qin, C.; Zhu, K.; Bruns, T.D.; Peay, K.G. Ectomycorrhizal fungal diversity predicted to substantially decline due to climate changes in North American Pinaceae forests. J. Biogeogr. 2020, 47, 772–782. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Guerra, C.A.; Cano-Díaz, C.; Egidi, E.; Wang, J.; Eisenhauer, N.; Singh, B.K.; Maestre, F.T. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Chang. 2020, 10, 550–554. [Google Scholar] [CrossRef]
- Korner, C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.J.; Green, J.L. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. 1), 11505–11511. [Google Scholar] [CrossRef]
- Li, J.; Shen, Z.; Li, C.; Kou, Y.; Wang, Y.; Tu, B.; Zhang, S.; Li, X. Stair-step pattern of soil bacterial diversity mainly driven by pH and vegetation types along the elevational gradients of Gongga Mountain, China. Front. Microbiol. 2018, 9, 569. [Google Scholar] [CrossRef]
- Duan, Y.; Lian, J.; Wang, L.; Wang, X.; Luo, Y.; Wang, W.; Wu, F.; Zhao, J.; Ding, Y.; Ma, J.; et al. Variation in soil microbial communities along an elevational gradient in alpine meadows of the Qilian Mountains, China. Front. Microbiol. 2021, 12, 684386. [Google Scholar] [CrossRef]
- Marin, C.; Kohout, P. Response of soil fungal ecological guilds to global changes. New Phytol. 2021, 229, 656–658. [Google Scholar] [CrossRef]
- Peay, K.G.; Baraloto, C.; Fine, P.V. Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J. 2013, 7, 1852–1861. [Google Scholar] [CrossRef]
- Geml, J.; Arnold, A.E.; Semenova-Nelsen, T.A.; Nouhra, E.R.; Drechsler-Santos, E.R.; Góes-Neto, A.; Morgado, L.N.; Ódor, P.; Hegyi, B.; Oriol, G.; et al. Community dynamics of soil-borne fungal communities along elevation gradients in neotropical and palaeotropical forests. Mol. Ecol. 2022, 31, 2044–2060. [Google Scholar] [CrossRef]
- Zhou, Y.; Jia, X.; Han, L.; Liu, Z.; Kang, S.; Zhao, Y. Fungal community diversity in soils along an elevation gradient in a Quercus aliena var. acuteserrata forest in Qinling Mountains, China. Appl. Soil Ecol. 2021, 167, 104104. [Google Scholar] [CrossRef]
- Odriozola, I.; Navrátilová, D.; Tláskalová, P.; Klinerová, T.; Ervenková, Z.; Kohout, P.; Větrovský, T.; Ížková, P.; Starý, M.; Baldrian, P. Predictors of soil fungal biomass and community composition in temperate mountainous forests in Central Europe. Soil Biol. Biochem. 2021, 161, 108366. [Google Scholar] [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; Debruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial Taxa distribution is associated with ecological trophic Cascades along an elevation gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef]
- Meng, H.; Li, K.; Nie, M.; Wan, J.; Quan, Z.; Fang, C.; Chen, J.; Gu, J.; Li, B. Responses of bacterial and fungal communities to an elevation gradient in a subtropical montane forest of China. Appl. Microbiol. Biot. 2013, 97, 2219–2230. [Google Scholar] [CrossRef]
- Veach, A.M.; Stokes, C.E.; Knoepp, J.; Jumpponen, A.; Baird, R. Fungal communities and functional guilds shift along an elevational gradient in the southern Appalachian Mountains. Microb. Ecol. 2018, 76, 156–168. [Google Scholar] [CrossRef]
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Looby, C.I.; Martin, P.H. Diversity and function of soil microbes on montane gradients: The state of knowledge in a changing world. FEMS Microbiol. Ecol. 2020, 96, a122. [Google Scholar] [CrossRef]
- Hendershot, J.N.; Read, Q.D.; Henning, J.A.; Sanders, N.J.; Classen, A.T. Consistently inconsistent drivers of microbial diversity and abundance at macroecological scales. Ecology 2017, 98, 1757–1763. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, Y.; Jia, X.; Han, L.; Liu, L.; Ren, K.; Ye, X.; Qu, Z.; Pei, Y. Soil characteristics and microbial community structure on along elevation gradient in a Pinus armandii forest of the Qinling Mountains, China. For. Ecol. Manag. 2022, 503, 119793. [Google Scholar] [CrossRef]
- Ma, L.; Liu, L.; Lu, Y.; Chen, L.; Zhang, Z.; Zhang, H.; Wang, X.; Shu, L.; Yang, Q.; Song, Q.; et al. When microclimates meet soil microbes: Temperature controls soil microbial diversity along an elevational gradient in subtropical forests. Soil Biol. Biochem. 2022, 166, 108566. [Google Scholar] [CrossRef]
- Yang, T.; Tedersoo, L.; Fu, X.; Zhao, C.; Liu, X.; Gao, G.; Cheng, L.; Adams, J.M.; Chu, H. Saprotrophic fungal diversity predicts ectomycorrhizal fungal diversity along the timberline in the framework of island biogeography theory. ISME Commun. 2021, 1, 15. [Google Scholar] [CrossRef]
- Shen, C.; Gunina, A.; Luo, Y.; Wang, J.; He, J.Z.; Kuzyakov, Y.; Hemp, A.; Classen, A.T.; Ge, Y. Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 2020, 22, 3287–3301. [Google Scholar] [CrossRef]
- Shigyo, N.; Hirao, T. Saprotrophic and ectomycorrhizal fungi exhibit contrasting richness patterns along elevational gradients in cool-temperate montane forests. Fungal Ecol. 2021, 50, 101036. [Google Scholar] [CrossRef]
- Shen, C.; Liang, W.; Shi, Y.; Lin, X.; Zhang, H.; Wu, X.; Xie, G.; Chain, P.; Grogan, P.; Chu, H. Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 2014, 95, 3190–3202. [Google Scholar] [CrossRef]
- He, Z.; Chen, L.; Zhao, W.; Liu, J.; Song, M.; Kong, J.; Gao, Y. Co-ordinated elevational diversity patterns for soil bacteria, fungi, and plants in alkaline soils of arid northwestern China. Eur. J. Soil Sci. 2022, 73, e13209. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, Y.; Hu, H.; Zhang, L.; Li, J.; He, J. Soil pH determines the alpha diversity but not beta diversity of soil fungal community along altitude in a typical Tibetan forest ecosystem. J. Soil. Sediment. 2015, 15, 1224–1232. [Google Scholar] [CrossRef]
- Legendre, P. Interpreting the replacement and richness difference components of beta diversity. Glob. Ecol. Biogeogr. 2014, 23, 1324–1334. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Li, L.; Li, J.; Feng, Y.; Lu, Q. The patterns and drivers of bacterial and fungal β-diversity in a typical dryland ecosystem of northwest China. Front. Microbiol. 2017, 8, 2126. [Google Scholar] [CrossRef]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Xue, Y.; Deng, Y.; Li, H.; Zhou, J.; Li, D. Soil bacterial diversity patterns and drivers along an elevational gradient on Shennongjia Mountain, China. Microb. Biotechnol. 2015, 8, 739–746. [Google Scholar] [CrossRef]
- Burke, D.J.; Carrino-Kyker, S.R.; Burns, J.H. Is it climate or chemistry? Soil fungal communities respond to soil nutrients in a multi-year high-resolution analysis. Ecosphere 2019, 10, e2896. [Google Scholar] [CrossRef]
- Llado, S.; Lopez-Mondejar, R.; Baldrian, P. Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 2018, 102, 4331–4338. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, M.K.; Sanders, N.J.; Wardle, D.A. Community and ecosystem responses to elevational gradients: Processes, mechanisms, and insights for global change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 261–280. [Google Scholar] [CrossRef]
- Ji, L.; Yang, Y.; Yang, L. Seasonal variations in soil fungal communities and co-occurrence networks along an altitudinal gradient in the cold temperate zone of China: A case study on Oakley Mountain. Catena 2021, 204, 105448. [Google Scholar] [CrossRef]
- Lazzaro, A.; Hilfiker, D.; Zeyer, J. Structures of microbial communities in alpine soils: Seasonal and elevational effects. Front. Microbiol. 2015, 6, 1330. [Google Scholar] [CrossRef] [PubMed]
- Donhauser, J.; Frey, B. Alpine soil microbial ecology in a changing world. FEMS Microbiol. Ecol. 2018, 94, y99. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: What are the driving factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef]
- Liu, D.; Liu, G.; Chen, L.; Wang, J.; Zhang, L. Soil pH determines fungal diversity along an elevation gradient in Southwestern China. Sci. China Life Sci. 2018, 61, 718–726. [Google Scholar] [CrossRef]
- Zhang, P.; Luan, M.; Li, X.; Lian, Z.; Zhao, X. The distribution of soil fungal communities along an altitudinal gradient in an alpine meadow. Glob. Ecol. Conserv. 2021, 31, e1838. [Google Scholar] [CrossRef]
- Hussain, S.; Liu, H.; Liu, S.; Yin, Y.; Yuan, Z.; Zhao, Y.; Cao, H. Distribution and assembly processes of soil fungal communities along an altitudinal gradient in Tibetan Plateau. J. Fungi 2021, 7, 1082. [Google Scholar] [CrossRef]
- Sheng, Y.; Cong, W.; Yang, L.; Liu, Q.; Zhang, Y. Forest Soil fungal community elevational distribution pattern and their ecological assembly processes. Front. Microbiol. 2019, 10, 2226. [Google Scholar] [CrossRef]
- Cui, Y.; Bing, H.; Fang, L.; Wu, Y.; Yu, J.; Shen, G.; Jiang, M.; Wang, X.; Zhang, X. Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 2019, 338, 118–127. [Google Scholar] [CrossRef]
- Liang, Q.; Xu, X.; Mao, K.; Wang, M.; Wang, K.; Xi, Z.; Liu, J. Shifts in plant distributions in response to climate warming in a biodiversity hotspot, the Hengduan Mountains. J. Biogeogr. 2018, 45, 1334–1344. [Google Scholar] [CrossRef]
- Cao, R.; Yang, W.; Chang, C.; Wang, Z.; Wang, Q.; Li, H.; Tan, B. Differential seasonal changes in soil enzyme activity along an altitudinal gradient in an alpine-gorge region. Appl. Soil Ecol. 2021, 166, 104078. [Google Scholar] [CrossRef]
- Fuzhong, W.; Changhui, P.; Jianxiao, Z.; Jian, Z.; Bo, T.; Wanqin, Y. Impacts of freezing and thawing dynamics on foliar litter carbon release in alpine/subalpine forests along an altitudinal gradient in the eastern Tibetan Plateau. Biogeosciences 2014, 11, 6471–6481. [Google Scholar] [CrossRef]
- Gong, S.; Feng, B.; Jian, S.P.; Wang, G.S.; Ge, Z.W.; Yang, Z.L. Elevation matters more than season in shaping the heterogeneity of soil and root associated ectomycorrhizal fungal community. Microbiol. Spectr. 2022, 10, e195021. [Google Scholar] [CrossRef]
- Li, S.; Huang, X.; Shen, J.; Xu, F.; Su, J. Effects of plant diversity and soil properties on soil fungal community structure with secondary succession in the Pinus yunnanensis forest. Geoderma 2020, 379, 114646. [Google Scholar] [CrossRef]
- Linn, D.M.; Doran, J.W. Effect of water-filled pore space on carbon dioxide and nitrous oxide production in tilled and nontilled soils. Soil Sci. Soc. Am. J. 1984, 48, 1267–1272. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Furneaux, B.; Song, Z. FUNGuildR: Look up Guild Information for Fungi; R Package Version 0.2.0.9. Available online: https://github.com/brendanf/FUNGuildR/ (accessed on 10 March 2022).
- Taguchi, Y.H.; Oono, Y. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics 2005, 21, 730–740. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Package Version 2.6-2; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 18 April 2022).
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. Betapart: An R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Karney, C.; Williams, E.; Vennes, C. Geosphere: Spherical Trigonometry; R Package Version 1.5-14; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://cran.r-project.org/web/packages/geosphere/index.html (accessed on 18 April 2022).
- Goslee, S.; Urban, D. Ecodist: Dissimilarity-Based Functions for Ecological Analysis; R Package Version 2.0-9; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://cran.r-project.org/web/packages/ecodist/index.html (accessed on 18 May 2022).
- Chagnon, P.L.; Bradley, R.L.; Maherali, H.; Klironomos, J.N. A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant. Sci. 2013, 18, 484–491. [Google Scholar] [CrossRef]
- Looby, C.I.; Treseder, K.K. Shifts in soil fungi and extracellular enzyme activity with simulated climate change in a tropical montane cloud forest. Soil Biol. Biochem. 2018, 117, 87–96. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Nakano, T.; Hattori, M.; Nara, K. The mid-domain effect in ectomycorrhizal fungi: Range overlap along an elevation gradient on Mount Fuji, Japan. ISME J. 2014, 8, 1739–1746. [Google Scholar] [CrossRef]
- Peay, K.G.; von Sperber, C.; Cardarelli, E.; Toju, H.; Francis, C.A.; Chadwick, O.A.; Vitousek, P.M. Convergence and contrast in the community structure of Bacteria, Fungi and Archaea along a tropical elevation-climate gradient. FEMS Microbiol. Ecol. 2017, 93, fix045. [Google Scholar] [CrossRef]
- Park, K.H.; Yoo, S.; Park, M.S.; Kim, C.S.; Lim, Y.W. Different patterns of belowground fungal diversity along altitudinal gradients with respect to microhabitat and guild types. Environ. Microbiol. Rep. 2021, 13, 649–658. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, W.; Zhong, Z.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 2018, 610–611, 750–758. [Google Scholar] [CrossRef]
- Pellissier, L.; Pinto-Figueroa, E.; Niculita-Hirzel, H.; Moora, M.; Villard, L.; Goudet, J.; Guex, N.; Pagni, M.; Xenarios, I.; Sanders, I.; et al. Plant species distributions along environmental gradients: Do belowground interactions with fungi matter? Front. Plant. Sci. 2013, 4, 500. [Google Scholar] [CrossRef]
- Yamauchi, D.H.; Garcia, G.H.; Teixeira, M.M.; Rodrigues, G.; Ullmann, L.S.; Garcia, G.A.; Hebeler-Barbosa, F.; Bagagli, E. Soil mycobiome is shaped by vegetation and microhabitats: A regional-scale study in southeastern Brazil. J. Fungi 2021, 7, 587. [Google Scholar] [CrossRef]
- Yang, T.; Tedersoo, L.; Soltis, P.S.; Soltis, D.E.; Gilbert, J.A.; Sun, M.; Shi, Y.; Wang, H.; Li, Y.; Zhang, J.; et al. Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. ISME J. 2019, 13, 686–697. [Google Scholar] [CrossRef]
- Santonja, M.; Rancon, A.; Fromin, N.; Baldy, V.; Hättenschwiler, S.; Fernandez, C.; Montès, N.; Mirleau, P. Plant litter diversity increases microbial abundance, fungal diversity, and carbon and nitrogen cycling in a Mediterranean shrubland. Soil Biol. Biochem. 2017, 111, 124–134. [Google Scholar] [CrossRef]
- Hugoni, M.; Luis, P.; Guyonnet, J.; Haichar, F. Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 2018, 28, 451–463. [Google Scholar] [CrossRef]
- Boddy, L.; Hiscox, J.; Heitman, J.; Crous, P.W. Principles and mechanisms of colonization and competition by saprotrophic Fungi. Microbiol. Spectr. 2016, 4, 4–6. [Google Scholar] [CrossRef]
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, Å.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef]
- Guo, J.; Ling, N.; Chen, Z.; Xue, C.; Li, L.; Liu, L.; Gao, L.; Wang, M.; Ruan, J.; Guo, S.; et al. Soil fungal assemblage complexity is dependent on soil fertility and dominated by deterministic processes. New Phytol. 2020, 226, 232–243. [Google Scholar] [CrossRef]
- Soininen, J.; Heino, J.; Wang, J. A meta-analysis of nestedness and turnover components of beta diversity across organisms and ecosystems. Glob. Ecol. Biogeogr. 2018, 27, 96–109. [Google Scholar] [CrossRef]
- Nottingham, A.T.; Fierer, N.; Turner, B.L.; Whitaker, J.; Ostle, N.J.; Mcnamara, N.P.; Bardgett, R.D.; Leff, J.W.; Salinas, N.; Silman, M.R.; et al. Microbes follow Humboldt: Temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology 2018, 99, 2455–2466. [Google Scholar] [CrossRef]
- Ping, Y.; Han, D.; Wang, N.; Hu, Y.; Mu, L.; Feng, F. Vertical zonation of soil fungal community structure in a Korean pine forest on Changbai Mountain, China. World J. Microbiol. Biotechnol. 2016, 33, 12. [Google Scholar] [CrossRef]
- Roy, J.; Bonneville, J.M.; Saccone, P.; Ibanez, S.; Albert, C.H.; Boleda, M.; Gueguen, M.; Ohlmann, M.; Rioux, D.; Clement, J.C.; et al. Differences in the fungal communities nursed by two genetic groups of the alpine cushion plant, Silene acaulis. Ecol. Evol. 2018, 8, 11568–11581. [Google Scholar] [CrossRef]
- Bayranvand, M.; Akbarinia, M.; Salehi, J.G.; Gharechahi, J.; Kooch, Y.; Baldrian, P. Composition of soil bacterial and fungal communities in relation to vegetation composition and soil characteristics along an altitudinal gradient. FEMS Microbiol. Ecol. 2021, 97, fiaa201. [Google Scholar] [CrossRef]
- Jarvis, S.G.; Woodward, S.; Taylor, A.F.S. Strong altitudinal partitioning in the distributions of ectomycorrhizal fungi along a short (300 m) elevation gradient. New Phytol. 2015, 206, 1145–1155. [Google Scholar] [CrossRef]
- Adamo, I.; Castaño, C.; Bonet, J.A.; Colinas, C.; Martínez De Aragón, J.; Alday, J.G. Soil physico-chemical properties have a greater effect on soil fungi than host species in Mediterranean pure and mixed pine forests. Soil Biol. Biochem. 2021, 160, 108320. [Google Scholar] [CrossRef]
- Vašutová, M.; Edwards-Jonášová, M.; Baldrian, P.; Ermák, M.; Cudlín, P. Distinct environmental variables drive the community composition of mycorrhizal and saprotrophic fungi at the alpine treeline ecotone. Fungal Ecol. 2017, 27, 116–124. [Google Scholar] [CrossRef]
- Kivlin, S.N.; Hawkes, C.V. Tree species, spatial heterogeneity, and seasonality drive soil fungal abundance, richness, and composition in Neotropical rainforests. Environ. Microbiol. 2016, 18, 4662–4673. [Google Scholar] [CrossRef] [PubMed]
- Canini, F.; Zucconi, L.; Pacelli, C.; Selbmann, L.; Onofri, S.; Geml, J. Vegetation, pH and water content as main factors for shaping fungal richness, community composition and functional guilds distribution in soils of western Greenland. Front. Microbiol. 2019, 10, 2348. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Y.; Cui, X.; Yue, P.; Li, K.; Liu, X.; Tripathi, B.M.; Chu, H.; Lozupone, C. Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 2019, 4, e218–e225. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.; Liu, H.; Zhang, Y.; Yu, L. Soil pH is a key determinant of soil fungal community composition in the Ny-Ålesund Region, Svalbard (High Arctic). Front. Microbiol. 2016, 7, 227. [Google Scholar] [CrossRef]
- Schappe, T.; Albornoz, F.E.; Turner, B.L.; Jones, F.A. Co-occurring fungal functional groups respond differently to tree neighborhoods and soil properties across three tropical rainforests in Panama. Microb. Ecol. 2020, 79, 675–685. [Google Scholar] [CrossRef]
- Plett, J.M.; Martin, F. Blurred boundaries: Lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 2011, 27, 14–22. [Google Scholar] [CrossRef]
- Chen, W.; Wang, J.; Meng, Z.; Xu, R.; Chen, J.; Zhang, Y.; Hu, T. Fertility-related interplay between fungal guilds underlies plant richness-productivity relationships in natural grasslands. New Phytol. 2020, 226, 1129–1143. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Semchenko, M.; Leff, J.W.; Lozano, Y.M.; Saar, S.; Davison, J.; Wilkinson, A.; Jackson, B.G.; Pritchard, W.J.; De Long, J.R.; Oakley, S.; et al. Fungal diversity regulates plant-soil feedbacks in temperate grassland. Sci. Adv. 2018, 4, u4578. [Google Scholar] [CrossRef]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 2015, 79, 243–262. [Google Scholar] [CrossRef]
- Chiu, C.H.; Paszkowski, U. Mechanisms and impact of symbiotic phosphate acquisition. Cold Spring Harb. Perspect. Biol. 2019, 11, a034603. [Google Scholar] [CrossRef]
- Kanzaki, N.; Ekino, T.; Giblin-Davis, R.M. Feeding Dimorphism in a Mycophagous Nematode, Bursaphelenchus sinensis. Sci. Rep. 2019, 9, 13956. [Google Scholar] [CrossRef]
- Pimentel, C.S.; Firmino, P.N.; Ayres, M.P. Interactions between pinewood nematodes and the fungal community of pine trees. Fungal Ecol. 2021, 51, 101046. [Google Scholar] [CrossRef]
- Hodda, M. Phylum Nematoda: Feeding habits for all valid genera using a new, universal scheme encompassing the entire phylum, with descriptions of morphological characteristics of the stoma, a key, and discussion of the evidence for trophic relationships. Zootaxa 2022, 5114, 318–451. [Google Scholar] [CrossRef]
- Hernández-Cáceres, D.; Stokes, A.; Angeles-Alvarez, G.; Abadie, J.; Anthelme, F.; Bounous, M.; Freschet, G.T.; Roumet, C.; Weemstra, M.; Merino-Martín, L.; et al. Vegetation creates microenvironments that influence soil microbial activity and functional diversity along an elevation gradient. Soil Biol. Biochem. 2022, 165, 108485. [Google Scholar] [CrossRef]
- Rincon, A.; Santamaria-Perez, B.; Rabasa, S.G.; Coince, A.; Marcais, B.; Buee, M. Compartmentalized and contrasted response of ectomycorrhizal and soil fungal communities of Scots pine forests along elevation gradients in France and Spain. Environ. Microbiol. 2015, 17, 3009–3024. [Google Scholar] [CrossRef]
- Khalid, M.; Du, B.; Tan, H.; Liu, X.; Su, L.; Saeed-Ur-Rahman; Ali, M.; Liu, C.; Sun, N.; Hui, N. Phosphorus elevation erodes ectomycorrhizal community diversity and induces divergence of saprophytic community composition between vegetation types. Sci. Total Environ. 2021, 793, 148502. [Google Scholar] [CrossRef]
- Hou, W.; Lian, B.; Dong, H.; Jiang, H.; Wu, X. Distinguishing ectomycorrhizal and saprophytic fungi using carbon and nitrogen isotopic compositions. Geosci. Front. 2012, 3, 351–356. [Google Scholar] [CrossRef]
- Lin, L.; Jing, X.; Lucas-Borja, M.E.; Shen, C.; Wang, Y.; Feng, W. Rare taxa drive the response of soil fungal guilds to soil salinization in the Taklamakan desert. Front. Microbiol. 2022, 13, 862245. [Google Scholar] [CrossRef]
- Du, S.; Trivedi, P.; Wei, Z.; Feng, J.; Hu, H.W.; Bi, L.; Huang, Q.; Liu, Y.R. The proportion of soil-borne fungal pathogens increases with elevated organic carbon in agricultural soils. mSystems 2022, 7, e133721. [Google Scholar] [CrossRef]
- Makiola, A.; Holdaway, R.J.; Wood, J.R.; Orwin, K.H.; Glare, T.R.; Dickie, I.A. Environmental and plant community drivers of plant pathogen composition and richness. New Phytol. 2022, 233, 496–504. [Google Scholar] [CrossRef]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Galante, T.E.; Horton, T.R.; Swaney, D.P. 95% of basidiospores fall within 1 m of the cap: A field- and modeling-based study. Mycologia 2011, 103, 1175–1183. [Google Scholar] [CrossRef]
- García De León, D.; Moora, M.; Öpik, M.; Jairus, T.; Neuenkamp, L.; Vasar, M.; Bueno, C.G.; Gerz, M.; Davison, J.; Zobel, M. Dispersal of arbuscular mycorrhizal fungi and plants during succession. Acta Oecol. 2016, 77, 128–135. [Google Scholar] [CrossRef]
- Talbot, J.M.; Bruns, T.D.; Taylor, J.W.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Liao, H.L.; Smith, M.E.; et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. USA 2014, 111, 6341–6346. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Shan, X.; Yang, Y.; Wang, Y.; Druzhinina, I.S.; Pan, X.; Jin, W.; He, X.; Wang, X.; Zhang, X.; et al. Facultative symbiosis with a saprotrophic soil fungus promotes potassium uptake in American sweetgum trees. Plant Cell Environ. 2021, 44, 2793–2809. [Google Scholar] [CrossRef] [PubMed]


| Abbreviation | Altitude Range (m) | Vegetation Types | Dominant Plant Species |
|---|---|---|---|
| E7 | 3998–4245 | Alpine meadow | Carex atrofusca, Polygonum viviparum, Anemone rivularis |
| E6 | 3671–3893 | Alpine shrub meadow | Potentilla fruticose *, Sibiraea angustata, Berberis wilsoniae |
| E5 | 3161–3516 | Subalpine coniferous forest | Abies fargesii var. faxoniana# |
| E4 | 2805–3089 | Subalpine coniferous-broadleaved mixed forest | A. fargesii var. faxoniana #, Betula albosinensis # |
| E3 | 2427–2769 | Subalpine broadleaved mixed forest | B. albosinensis#, B. platyphylla#, Acer davidii |
| E2 | 1946–2182 | Valley-mountain ecotone Broadleaved mixed forest | Quercus baronii #, Cotinus coggygria *, Ostryopsis davidiana # |
| E1 | 1663–1848 | Dry valley shrub | Campylotropis macrocarpa *, Bauhinia brachycarpa *, Sophora davidii * |
| Soil Fungi | Bray–Curtis Index | Jaccard Index | ||||||
|---|---|---|---|---|---|---|---|---|
| Component | Mean | Slope (km−1) | R2 | Component | Mean | Slope (km−1) | R2 | |
| Total | β-diversity | 0.850 | 0.0535 | 0.22 *** | β-diversity | 0.750 | 0.0205 | 0.16 *** |
| Turnover | 0.794 | 0.0692 | 0.19 *** | Turnover | 0.706 | 0.026 | 0.15 *** | |
| Nestedness | 0.056 | −0.0157 | 0.02 *** | Nestedness | 0.044 | −0.0055 | <0.01 *** | |
| Symbiotic | β-diversity | 0.875 | 0.0205 | 0.02 *** | β-diversity | 0.818 | 0.0110 | 0.03 *** |
| Turnover | 0.814 | 0.0191 | 0.01 *** | Turnover | 0.786 | 0.0107 | 0.02 *** | |
| Nestedness | 0.060 | 0.0014 | <0.01 ns | Nestedness | 0.032 | 0.00025 | <0.01 ns | |
| Saprophytic | β-diversity | 0.775 | 0.0631 | 0.13 *** | β-diversity | 0.798 | 0.0243 | 0.11 *** |
| Turnover | 0.660 | 0.1020 | 0.17 *** | Turnover | 0.744 | 0.0318 | 0.09 *** | |
| Nestedness | 0.115 | −0.0388 | 0.06 *** | Nestedness | 0.054 | −0.00748 | 0.01 *** | |
| Pathogenic | β-diversity | 0.940 | 0.0332 | 0.06 *** | β-diversity | 0.934 | 0.0299 | 0.08 *** |
| Turnover | 0.890 | 0.0548 | 0.07 *** | Turnover | 0.901 | 0.0473 | 0.07 *** | |
| Nestedness | 0.050 | 0.0217 | 0.03 *** | Nestedness | 0.033 | −0.0174 | 0.03 *** | |
| Soil Properties | Total Fungi | Symbiotic Fungi | Saprophytic Fungi | Pathogenic Fungi |
|---|---|---|---|---|
| EC | 0.799 *** | 0.701 *** | 0.713 *** | 0.609 *** |
| WFPS | 0.787 *** | 0.742 *** | 0.718 *** | 0.757 *** |
| TP | 0.618 *** | 0.602 *** | 0.464 *** | 0.265 *** |
| pH | 0.604 *** | 0.595 *** | 0.550 *** | 0.591 *** |
| NO3−-N | 0.454 *** | 0.426 *** | 0.407 *** | 0.293 *** |
| NH4+-N | 0.377 *** | 0.390 *** | 0.402 *** | 0.513 *** |
| AP | 0.294 *** | 0.409 *** | 0.295 *** | 0.186 ** |
| SOC | 0.260 *** | 0.368 *** | 0.216 ** | 0.347 *** |
| TK | 0.233 *** | 0.308 ** | 0.170 ** | 0.261 *** |
| BD | 0.141 ** | 0.163 ** | 0.079 ns | 0.249 ** |
| C/P | 0.078 ns | 0.128 * | 0.046 ns | 0.098 * |
| N/P | 0.040 ns | 0.074 ns | 0.016 ns | 0.095 ns |
| C/N | 0.035 ns | 0.004 ns | 0.094 ns | 0.014 ns |
| Soil Fungi | Components | Fit Equation | R2 |
|---|---|---|---|
| Total | β-diversity | 0.0003EC + 0.0006C/P + 0.0008WFPS + 0.0066TN − 0.0007TC + 0.0009GEO | 0.2757 *** |
| Turnover | 0.0011C/P + 0.0011WFPS + 0.0005EC + 0.0019GEO | 0.2762 *** | |
| Nestedness | −0.0005WFPS | 0.0279 *** | |
| Symbiotic | β-diversity | 0.0009C/P + 0.0006WFPS | 0.0778 *** |
| Turnover | 0.0010C/P + 0.0006WFPS | 0.0662 *** | |
| Nestedness | −0.0004SOC + 0.0023TK−0.0001WFPS | 0.0240 * | |
| Saprophytic | β-diversity | 0.0121TN + 0.0005WFPS + 0.0025GEO | 0.1701 *** |
| Turnover | 0.0009EC + 0.0154TN + 0.0006WFPS + 0.0031GEO | 0.2470 *** | |
| Nestedness | 0.0006C/P | 0.0169 ns | |
| Pathogenic | β-diversity | 0.0048TN − 0.0005TC + 0.0008WFPS | 0.0659 *** |
| Turnover | 0.0033TN + 0.0014WFPS | 0.0569 *** | |
| Nestedness | 0.0009SOC − 0.0041TN − 0.0006WFPS | 0.0378 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Shi, Z.; Liu, S.; Zhang, M.; Cao, X.; Chen, M.; Xu, G.; Xing, H.; Li, F.; Feng, Q. Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau. J. Fungi 2022, 8, 807. https://doi.org/10.3390/jof8080807
Chen J, Shi Z, Liu S, Zhang M, Cao X, Chen M, Xu G, Xing H, Li F, Feng Q. Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau. Journal of Fungi. 2022; 8(8):807. https://doi.org/10.3390/jof8080807
Chicago/Turabian StyleChen, Jian, Zuomin Shi, Shun Liu, Miaomiao Zhang, Xiangwen Cao, Miao Chen, Gexi Xu, Hongshuang Xing, Feifan Li, and Qiuhong Feng. 2022. "Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau" Journal of Fungi 8, no. 8: 807. https://doi.org/10.3390/jof8080807
APA StyleChen, J., Shi, Z., Liu, S., Zhang, M., Cao, X., Chen, M., Xu, G., Xing, H., Li, F., & Feng, Q. (2022). Altitudinal Variation Influences Soil Fungal Community Composition and Diversity in Alpine–Gorge Region on the Eastern Qinghai–Tibetan Plateau. Journal of Fungi, 8(8), 807. https://doi.org/10.3390/jof8080807






