Morphological and Phylogenetic Analyses Reveal Four New Species of Gnomoniopsis (Gnomoniaceae, Diaporthales) from China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Morphology
2.2. DNA Extraction and Amplification
2.3. Phylogenetic Analyses
3. Results
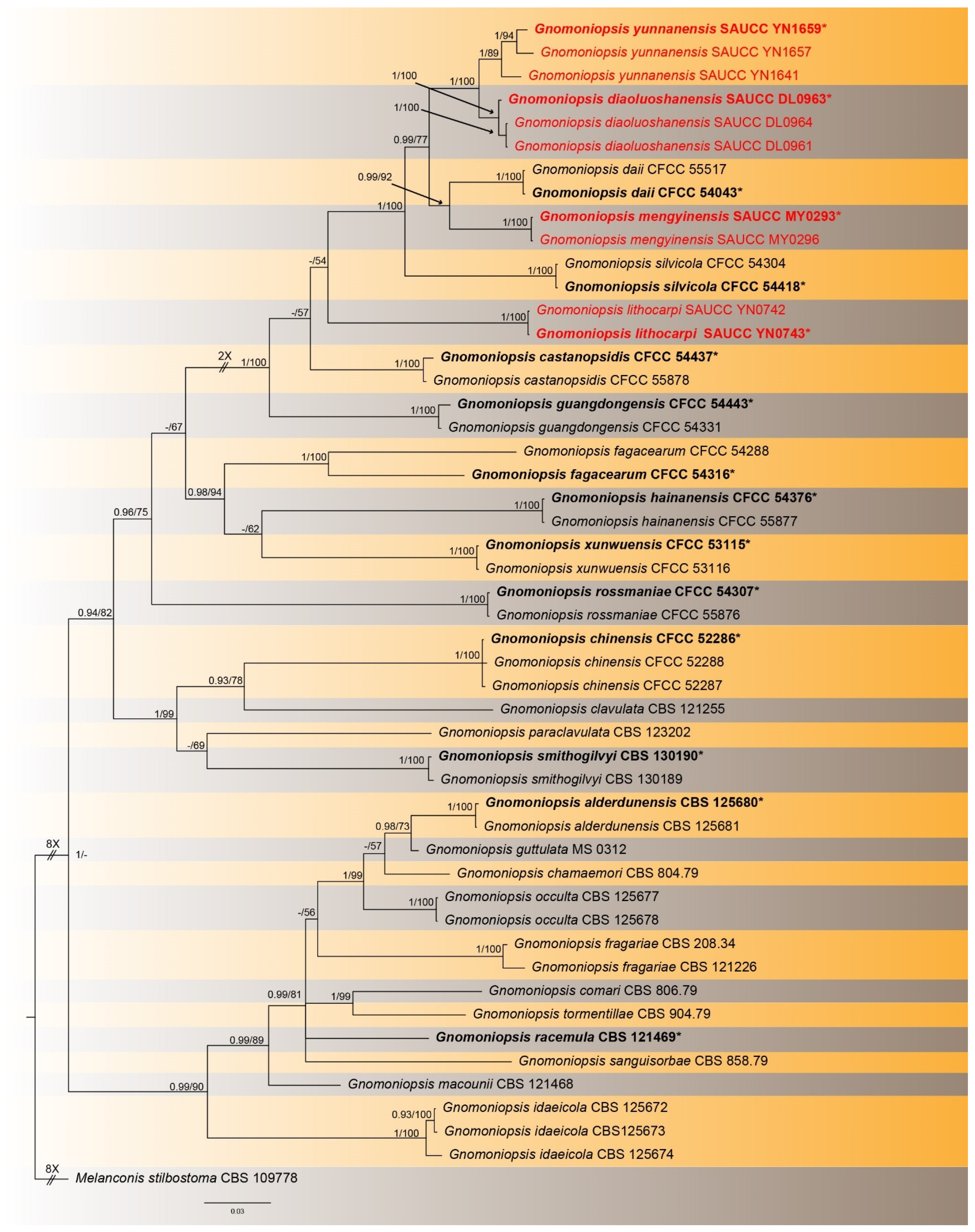
3.1. Phylogenetic Analyses
3.2. Taxonomy
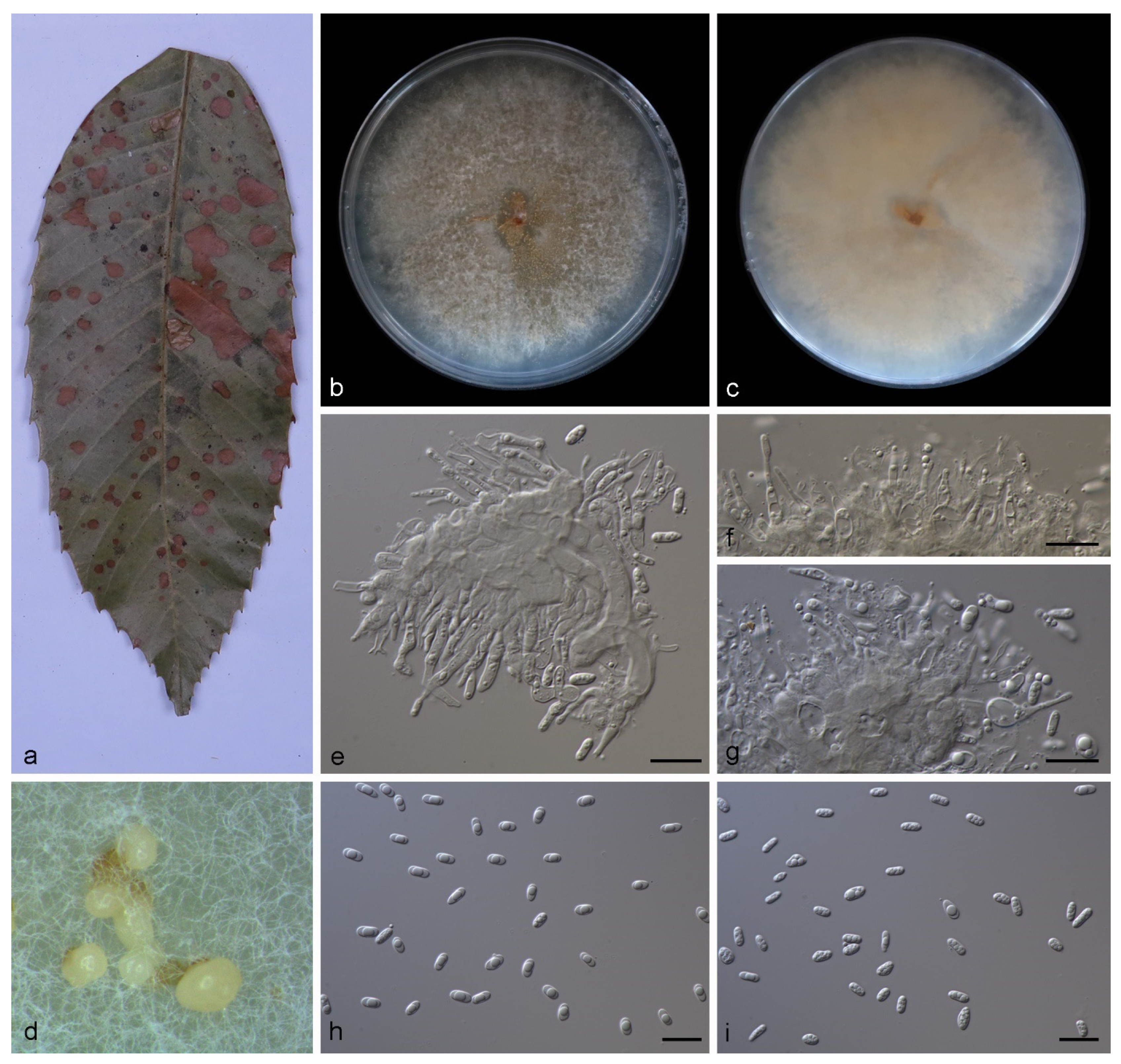
3.2.1. Gnomoniopsis diaoluoshanensis S. Wang, Z.X. Zhang, X.Y. Liu and X.G. Zhang, sp. nov.
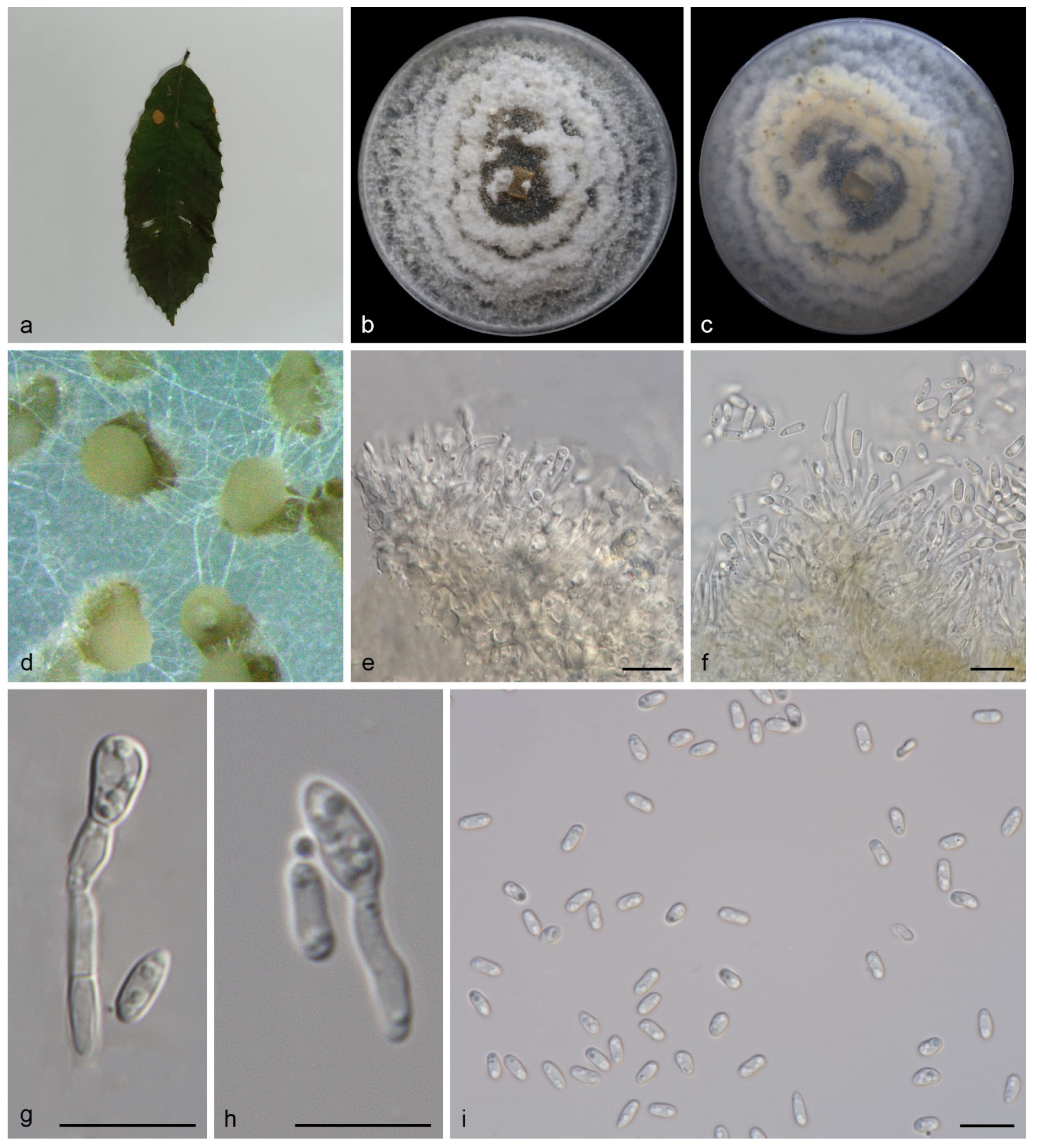
3.2.2. Gnomoniopsis lithocarpi S. Wang, Z.X. Zhang, X.Y. Liu and X.G. Zhang, sp. nov.
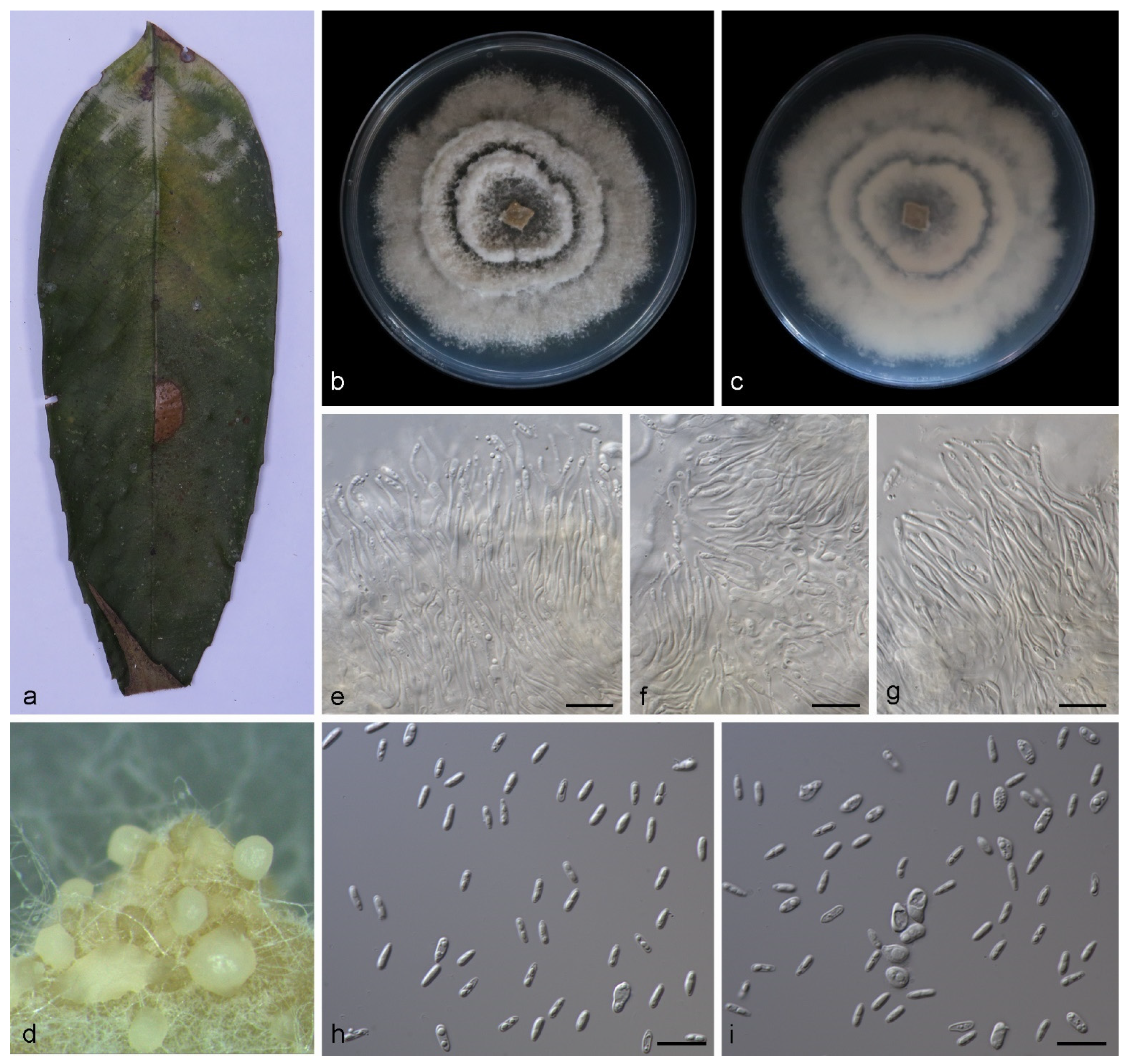
3.2.3. Gnomoniopsis mengyinensis S. Wang, Z.X. Zhang, X.Y. Liu and X.G. Zhang, sp. nov.
3.2.4. Gnomoniopsis yunnanensis S. Wang, Z.X. Zhang, X.Y. Liu and X.G. Zhang, sp. nov.
3.3. Key to the Species of Gnomoniopsis
| 1. Sexual morph known------------------------------------------------------------------------------------2 |
| 1. Sexual morph unknown-------------------------------------------------------------------------------16 |
| 2. Asci cylindrical--------------------------------------------------------------------------------------------3 |
| 2. Asci fusiform-----------------------------------------------------------------------------------------------4 |
| 3. Ascospores size 10.0–13.0 × 2.0–3.0 μm---------------------------------------------G. chamaemori |
| 3. Ascospores size 4.0–12.0 × 1.0–3.0 μm----------------------------------------------G. smithogilvyi |
| 4. Perithecia without stroma-------------------------------------------------------------------------------5 |
| 4. Perithecia with stroma-----------------------------------------------------------------------------------7 |
| 5. Perithecia immersed--------------------------------------------------------------------G. sanguisorbae |
| 5. Perithecia superficies-------------------------------------------------------------------------------------6 |
| 6. Perithecia size 110.0–150.0 × 120.0–140.0 μm-----------------------------------G. clavulata |
| 6. Perithecia size 139.0–180.0 × 156.0–241.0 μm-----------------------------G. paraclavulata |
| 7. Ascospores aseptate--------------------------------------------------------------------------------------8 |
| 7. Ascospores septate--------------------------------------------------------------------------------------10 |
| 8. Perithecia groups----------------------------------------------------------------------------G. racemula |
| 8. Perithecia solitary-----------------------------------------------------------------------------------------9 |
| 9. Ascospores size 6.0–10.0 × 1.5–3.0 μm----------------------------------------------G. tormentillae |
| 9. Ascospores size 7.0–8.0 × 1.8–2.2 μm-------------------------------------------------G. agrimoniae |
| 10. Perithecia surfaced on the host---------------------------------------------------------------------11 |
| 10. Perithecia immersed in the host--------------------------------------------------------------------12 |
| 11. Perithecia size 280.0–375.0 × 327.0–490.0 μm----------------------------------G. alderdunensis |
| 11. Perithecia size 112–330.0 × 125–500.0 μm-----------------------------------------------G. comari |
| 12. Perithecia immersed in stem------------------------------------------------------------------------13 |
| 12. Perithecia immersed in leaves----------------------------------------------------------------------14 |
| 13. Asci size 30.0–48.5 × 5.0–10.0-------------------------------------------------------------G. idaeicola |
| 13. Asci size 30.0–38.0 × 4.0–8.5--------------------------------------------------------------G. macounii |
| 14. Perithecia aggregated 2–4----------------------------------------------------------------G. guttulata |
| 14. Perithecia solitary--------------------------------------------------------------------------------------15 |
| 15. Perithecia size 150.0–475.0 × 200.0–475.0 μm----------------------------------------G. fragariae |
| 15. Perithecia size 129.0–340.0 × 147.0–428.0 μm-------------------------------------------G. occulta |
| 16. Conidiogenous cells guttulate----------------------------------------------------------------------17 |
| 16. Conidiogenous cells no guttulate------------------------------------------------------------------24 |
| 17. Conidia base circular--------------------------------------------------------G. lithocarpi sp. nov. |
| 17. Conidia base truncate---------------------------------------------------------------------------------18 |
| 18. Conidia ellipsoid or cylindrical---------------------------------------------------------------------19 |
| 18. Conidia oval or fusoid--------------------------------------------------------------------------------20 |
| 19. Conidiogenous cells size 8.0–12.0 × 1.0–2.0 μm-------------G. diaoluoshanensis sp. nov. |
| 19. Conidiogenous cells size 12.5–24.0 × 1.5–3.0 μm---------------------------G. guangdongensis |
| 20. Conidia 1-septate-------------------------------------------------------------------------G. rossmaniae |
| 20. Conidia aseptate----------------------------------------------------------------------------------------21 |
| 21. Conidia maximum length < 6.0 μm----------------------------------------------------------------22 |
| 21. Conidia maximum length > 6.0 μm----------------------------------------------------------------23 |
| 22. Conidiogenous cells 6.5–13.0 × 1.5–3.0 μm--------------------------------------G. castanopsidis |
| 22. Conidiogenous cells 7.0–15.0 × 1.5–2.5 μm--------------------------------------------G. silvicola |
| 23. Conidiogenous cells 16.0–33.5 × 2.0–5.0--------------------------------------------G. fagacearum |
| 23. Conidiogenous cells 16.5–26.0 × 2.5–4.5-------------------------------------------G. hainanensis |
| 24. Conidiogenous cells one-celled---------------------------------------------------------------------25 |
| 24. Conidiogenous cells multi-celled------------------------------------------------------------------26 |
| 25. Conidia 1-septate---------------------------------------------------------------------------G. chinensis |
| 25. Conidia aseptate-----------------------------------------------------------------------------------G. daii |
| 26. Conidiogenous cells branched-------------------------------------------------------G. xunwuensis |
| 26. Conidiogenous cells unbranched------------------------------------------------------------------27 |
| 27. Conidia maximum length < 10.0 μm--------------------------------------------------------------28 |
| 27. Conidia maximum length > 10.0 μm--------------------------------------------------------------29 |
| 28. Conidia oval to fusoid--------------------------------------------------G. mengyinensis sp. nov. |
| 28. Conidia oblong to ellipsoid---------------------------------------------G. yunnanensis sp. nov. |
| 29. Conidia subcylindrical------------------------------------------------------------------G. angolensis |
| 29. Conidia fusoid-----------------------------------------------------------------------------------G. rosae |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Voglmayr, H.; Castlebury, L.A.; Jaklitsch, W.M. Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales). Persoonia 2017, 38, 136–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.L.; Bezerra, J.D.; Tian, C.M.; Crous, P.W. Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 2018, 40, 119–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senanayake, I.C.; Jeewon, R.; Chomnunti, P.; Wanasinghe, D.N.; Norphanphoun, C.; Karunarathna, A.; Pem, D.; Perera, R.H.; Camporesi, E.; Eric, H.C.; et al. Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Divers. 2018, 93, 241–443. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Bian, D.R.; Piao, C.G.; Wang, S.K.; Li, Y. Morphology and Phylogeny of Gnomoniopsis (Gnomoniaceae, Diporthales) from Fagaceae Leaves in China. J. Fungi 2021, 7, 792. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, X.L.; Crous, P.W.; Tian, C.M. Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. MycoKeys 2019, 48, 67–96. [Google Scholar] [CrossRef]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Mejía, L.C.; White, J.F. Phylogeny and taxonomy of Ophiognomonia (Gnomoniaceae, Diaporthales), including twenty-five new species in this highly diverse genus. Fungal Divers. 2012, 57, 85–147. [Google Scholar] [CrossRef]
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Vandepitte, L.; Hobern, D.; Remsen, D.; Schalk, P.; DeWalt, R.E.; Keping, M.; et al. Catalogue of Life Checklist; (Version 2022-03-21); Catalogue of Life: Leiden, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Winter, G. Fungi Australienses. Revue Mycol. Toulouse 1886, 8, 207–213. [Google Scholar]
- Hawksworth, D.L.; Eriksson, O. Proposal to conserve 11 family names in the Ascomycotina (Fungi). Taxon 1988, 37, 190–193. [Google Scholar]
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.H.; Li, D.Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Koeltz Botanical Books: Glashütten, Germany, 2018; Volume 159. [Google Scholar] [CrossRef]
- Sogonov, M.V.; Castlebury, L.A.; Rossman, A.Y.; Mejía, L.C.; White, J.F. Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Stud. Mycol. 2008, 62, 1–77. [Google Scholar] [CrossRef]
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Phylogenetic placement and taxonomic review of the genus Cryptosporella and its synonyms Ophiovalsa and Winterella (Gnomoniaceae, Diaporthales). Mycol. Res. 2008, 112, 23–35. [Google Scholar] [CrossRef]
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, hostassociations, and a four-gene phylogeny. Stud. Mycol. 2011, 68, 211–235. [Google Scholar] [CrossRef] [PubMed]
- Mejía, L.C.; Rossman, A.Y.; Castlebury, L.A.; Yang, Z.L.; White, J.F. Occultocarpon, a new monotypic genus of Gnomoniaceae on Alnus nepalensis from China. Fungal Divers. 2012, 52, 99–105. [Google Scholar] [CrossRef]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three gene phylogeny, host associations and morphology. Mycologia 2010, 102, 1479–1496. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Struwe, L. Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae, Diaporthales). Biol. J. Linnean Soc. 2013, 111, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Barr, M.E. The Diaporthales in North America with emphasis on Gnomonia and its segregates. Mycologia 1978, 7, 43–184. [Google Scholar]
- Yang, Q.; Jiang, N.; Tian, C.M. Tree inhabiting gnomoniaceous species from China, with Cryphogonomonia gen. nov. proposed. MycoKeys 2020, 69, 71–89. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Liu, R.Y.; Liu, S.B.; Mu, T.C.; Zhang, X.G.; Xia, J.W. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 2022, 88, 171–192. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, F.J.R.M.; Lee, S.H.; Taylor, L.; Shawe-Taylor, J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W.A. greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModelTest v. 2. Program distributed by the author. In Evolutionary Biology Centre; Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the Extreme to the Campus and Beyond, Chicago, IL, USA, 16 July 2012; Association for Computing Machinery: San Diego, CA, USA, 2012; p. 8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Udayanga, D.; Miriyagalla, S.D.; Manamgoda, D.S.; Lewers, K.S.; Gardiennet, A.; Castlebury, L.A. Molecular reassessment of diaporthalean fungi associated with strawberry, including the leaf blight fungus, Paraphomopsis obscurans gen. et comb. nov. (Melanconiellaceae). IMA Fungus 2021, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Manamgoda, D.S.; Cai, L.; Bahkali, A.H.; Chukeatirote, E.; Hyde, K.D. Cochliobolus: An overview and current status of species. Fungal Divers. 2011, 51, 3–42. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; André Levesque, C.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium; Bolchacova, E.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeewon, R.; Ittoo, J.; Mahadeb, D.; Jaufeerally-Fakim, Y.; Wang, H.K.; Liu, A.R. DNA based identification and phylogenetic characterisation of endophytic and saprobic fungi from Antidesma madagascariense, a medicinal plant in Mauritius. J. Mycol. 2013, 781914, 10. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Tian, C.M. An emerging pathogen from rotted chestnut in China: Gnomoniopsis daii sp. nov. Forests 2019, 10, 1016. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Fan, X.L.; Tian, C.M. Identification and characterization of leaf-inhabiting Fungi from Castanea plantations in China. J. Fungi 2021, 7, 64. [Google Scholar] [CrossRef]
- Visentin, I.; Gentile, S.; Valentino, D.; Gonthier, P.; Tamietti, G.; Cardinale, F. Gnomoniopsis castanea sp. nov. (Gnomoniaceae, Diaporthales) as the causal agent of nut rot in sweet chestnut. J. Plant Pathol. 2012, 94, 411–419. [Google Scholar]
- Shuttleworth, L.A.; Guest, D.I. The infection process of chestnut rot, an important disease caused by Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) in Oceania and Europe. Australas. Plant Pathol. 2017, 46, 397–405. [Google Scholar] [CrossRef]
- Sakalidis, M.L.; Medina-Mora, C.M.; Kolp, M.; Fulbright, D.W. First report of Gnomoniopsis smithogilvyi causing chestnut brown rot on chestnut fruit in Michigan. Plant Dis. 2019, 103, 2134. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Alves, A.; Abdollahzadeh, J.; Phillips, A.J.L. Phylogeny, morphology and pathogenicity of Botryosphaeriaceae, Diatrypaceae and Gnomoniaceae associated with branch diseases of hazelnut in Sardinia (Italy). Eur. J. Plant Pathol. 2016, 146, 259–279. [Google Scholar] [CrossRef]

| Loci | PCR Primers | Sequence (5′—3′) | PCR Cycles | References |
|---|---|---|---|---|
| ITS | ITS5 ITS4 | GGA AGT AAA AGT CGT AAC AAG G TCC TCC GCT TAT TGA TAT GC | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | [23] |
| tef1 | EF1-728F EF-2 | CAT CGA GAA GTT CGA GAA GG GGA RGT ACC AGT SAT CAT GTT | (95 °C: 30 s, 48 °C: 30 s, 72 °C: 1 min) × 35 cycles | [24,25] |
| tub2 | Bt-2a Bt-2b | GGT AAC CAA ATC GGT GCT GCT TTC ACC CTC AGT GTA GTG ACC CTT GGC | (95 °C: 30 s, 53 °C: 30 s, 72 °C: 1 min) × 35 cycles | [26] |
| Species | Voucher | Host | Country | GenBank Accession Number | ||
|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | ||||
| Gnomoniopsis alderdunensis | CBS 125680 * | Rubus parviflorus (Rosaeace) | USA | GU320825 | GU320801 | GU320787 |
| CBS 125681 | Rubus parviflorus (Rosaeace) | USA | GU320827 | GU320802 | GU320789 | |
| G. castanopsidis | CFCC 54437 * | Castanopsis hystrix (Fagaceae) | China | MZ902909 | MZ936385 | – |
| CFCC 54438 | Castanopsis hystrix (Fagaceae) | China | MZ902910 | MZ936386 | – | |
| G. chamaemori | CBS 804.79 | Rubus chamaemorus (Rosaeace) | Finland | GU320817 | GU320809 | GU320777 |
| G. chinensis | CFCC 52286 * | Castanea mollissima (Fagaceae) | China | MG866032 | MH545370 | MH545366 |
| CFCC 52288 | Castanea mollissima (Fagaceae) | China | MG866034 | MH545372 | MH545368 | |
| CFCC 52287 | Castanea mollissima (Fagaceae) | China | MG866033 | MH545371 | MH545367 | |
| G. clavulata | CBS 121255 | Quercus falcata (Fagaceae) | USA | EU254818 | EU221934 | EU219211 |
| G. comari | CBS 806.79 | Oryza sativa (Rosaeace) | UK | EU254821 | GU320810 | EU219156 |
| G. daii | CFCC 54043 * | Castanea mollissima (Fagaceae) | China | MZ902911 | MZ936387 | MZ936403 |
| CFCC 55517 | Castanea mollissima (Fagaceae) | China | MN598671 | MN605517 | MN605519 | |
| G. diaoluoshanensis | SAUCC DL0963 * | Castanopsis chinensis (Fagaceae) | China | ON753744 | ON759769 | ON759777 |
| SAUCC DL0964 | Castanopsis chinensis (Fagaceae) | China | ON753743 | ON759768 | ON759776 | |
| SAUCC DL0961 | Castanopsis chinensis (Fagaceae) | China | ON753745 | ON759770 | ON759778 | |
| G. fagacearum | CFCC 54316 * | Lithocarpus glaber (Fagaceae) | China | MZ902916 | MZ936392 | MZ936408 |
| CFCC 54288 | Castanopsis faberi (Fagaceae) | China | MZ902913 | MZ936389 | MZ936405 | |
| G. fragariae = G. fructicola | CBS 208.34 | Fragaria sp. (Rosaeace) | USA | EU254826 | EU221968 | EU219149 |
| CBS 121226 | Fragaria vesca (Rosaeace) | USA | EU254824 | EU221961 | EU219144 | |
| G. guangdongensis | CFCC 54443 * | Castanopsis fargesii (Fagaceae) | China | MZ902918 | MZ936394 | MZ936410 |
| CFCC 54331 | Castanopsis fargesii (Fagaceae) | China | MZ902919 | MZ936395 | MZ936411 | |
| G. guttulata | MS 0312 | Agrimonia eupatoria (Rosaeace) | Bulgaria | EU254812 | – | – |
| G. hainanensis | CFCC 54376 * | Castanopsis hainanensis (Fagaceae) | China | MZ902921 | MZ936397 | MZ936413 |
| CFCC 55877 | Castanopsis hainanensis (Fagaceae) | China | MZ902922 | MZ936398 | MZ936414 | |
| G. idaeicola | CBS 125672 | Rubus sp. (Rosaeace) | USA | GU320823 | GU320797 | GU320781 |
| CBS 125673 | Rubus pedatus (Rosaeace) | USA | GU320824 | GU320798 | GU320782 | |
| CBS 125674 | Rubus sp. (Rosaeace) | France | GU320820 | GU320796 | GU320780 | |
| G. lithocarpi | SAUCC YN0743 * | Lithocarpus fohaiensis (Fagaceae) | China | ON753749 | ON759765 | ON759783 |
| SAUCC YN0742 | Lithocarpus fohaiensis (Fagaceae) | China | ON753750 | ON759764 | ON759782 | |
| G. macounii | CBS 121468 | Spiraea sp. (Rosaeace) | USA | EU254762 | EU221979 | EU219126 |
| G. mengyinensis | SAUCC MY0293 * | Castanea mollissima (Fagaceae) | China | ON753741 | ON759766 | ON759774 |
| SAUCC MY0296 | Castanea mollissima (Fagaceae) | China | ON753742 | ON759767 | ON759775 | |
| G. occulta | CBS 125677 | Potentilla sp. (Rosaeace) | USA | GU320828 | GU320812 | GU320785 |
| CBS 125678 | Potentilla sp. (Rosaeace) | USA | GU320829 | GU320800 | GU320786 | |
| G. paraclavulata | CBS 123202 | Agrostis sp. (Fagaceae) | USA | GU320830 | GU320815 | GU320775 |
| G. racemula | CBS 121469 * | Triticum aestivum (Onagraceae) | USA | EU254841 | EU221889 | EU219125 |
| G. rossmaniae | CFCC 54307 * | Castanopsis hainanensis (Fagaceae) | China | MZ902923 | MZ936399 | MZ936415 |
| CFCC 55876 | Castanopsis hainanensis (Fagaceae) | China | MZ902924 | MZ936400 | MZ936416 | |
| G. sanguisorbae | CBS 858.79 | Sanguisorba minor (Rosaeace) | Switzerland | GU320818 | GU320805 | GU320790 |
| G. silvicola | CFCC 54304 | Castanopsis hystrix (Fagaceae) | China | MZ902925 | MZ936401 | MZ936417 |
| CFCC 54418 * | Quercus serrata (Fagaceae) | China | MZ902926 | MZ936402 | MZ936418 | |
| G. smithogilvyi | CBS 130190 * | Castanea sp. (Fagaceae) | Australia | JQ910642 | JQ910645 | JQ910639 |
| CBS 130189 | Castanea sp. (Fagaceae) | Australia | JQ910644 | JQ910647 | JQ910641 | |
| G. tormentillae | CBS 904.79 | Potentilla sp. (Rosaeace) | Switzerland | EU254856 | GU320795 | EU219165 |
| G. xunwuensis | CFCC 53115 * | Castanopsis fissa (Fagaceae) | China | MK432667 | MK578141 | MK578067 |
| CFCC 53116 | Castanopsis fissa (Fagaceae) | China | MK432668 | MK578142 | MK578068 | |
| G.yunnanensis | SAUCC YN1659 * | Castanea mollissima (Fagaceae) | China | ON753746 | ON759771 | ON759779 |
| SAUCC YN1657 | Castanea mollissima (Fagaceae) | China | ON753747 | ON759772 | ON759780 | |
| SAUCC YN1641 | Castanea mollissima (Fagaceae) | China | ON753748 | ON759773 | ON759781 | |
| Melanconis stilbostoma | CBS 109778 | Betula pendula (Betulaceae) | Australia | DQ323524 | EU221886 | EU219104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Zhang, Z.; Liu, R.; Liu, S.; Liu, X.; Zhang, X. Morphological and Phylogenetic Analyses Reveal Four New Species of Gnomoniopsis (Gnomoniaceae, Diaporthales) from China. J. Fungi 2022, 8, 770. https://doi.org/10.3390/jof8080770
Wang S, Zhang Z, Liu R, Liu S, Liu X, Zhang X. Morphological and Phylogenetic Analyses Reveal Four New Species of Gnomoniopsis (Gnomoniaceae, Diaporthales) from China. Journal of Fungi. 2022; 8(8):770. https://doi.org/10.3390/jof8080770
Chicago/Turabian StyleWang, Shi, Zhaoxue Zhang, Rongyu Liu, Shubin Liu, Xiaoyong Liu, and Xiuguo Zhang. 2022. "Morphological and Phylogenetic Analyses Reveal Four New Species of Gnomoniopsis (Gnomoniaceae, Diaporthales) from China" Journal of Fungi 8, no. 8: 770. https://doi.org/10.3390/jof8080770
APA StyleWang, S., Zhang, Z., Liu, R., Liu, S., Liu, X., & Zhang, X. (2022). Morphological and Phylogenetic Analyses Reveal Four New Species of Gnomoniopsis (Gnomoniaceae, Diaporthales) from China. Journal of Fungi, 8(8), 770. https://doi.org/10.3390/jof8080770







