Digoxin Derivatives Sensitize a Saccharomyces cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p
Abstract
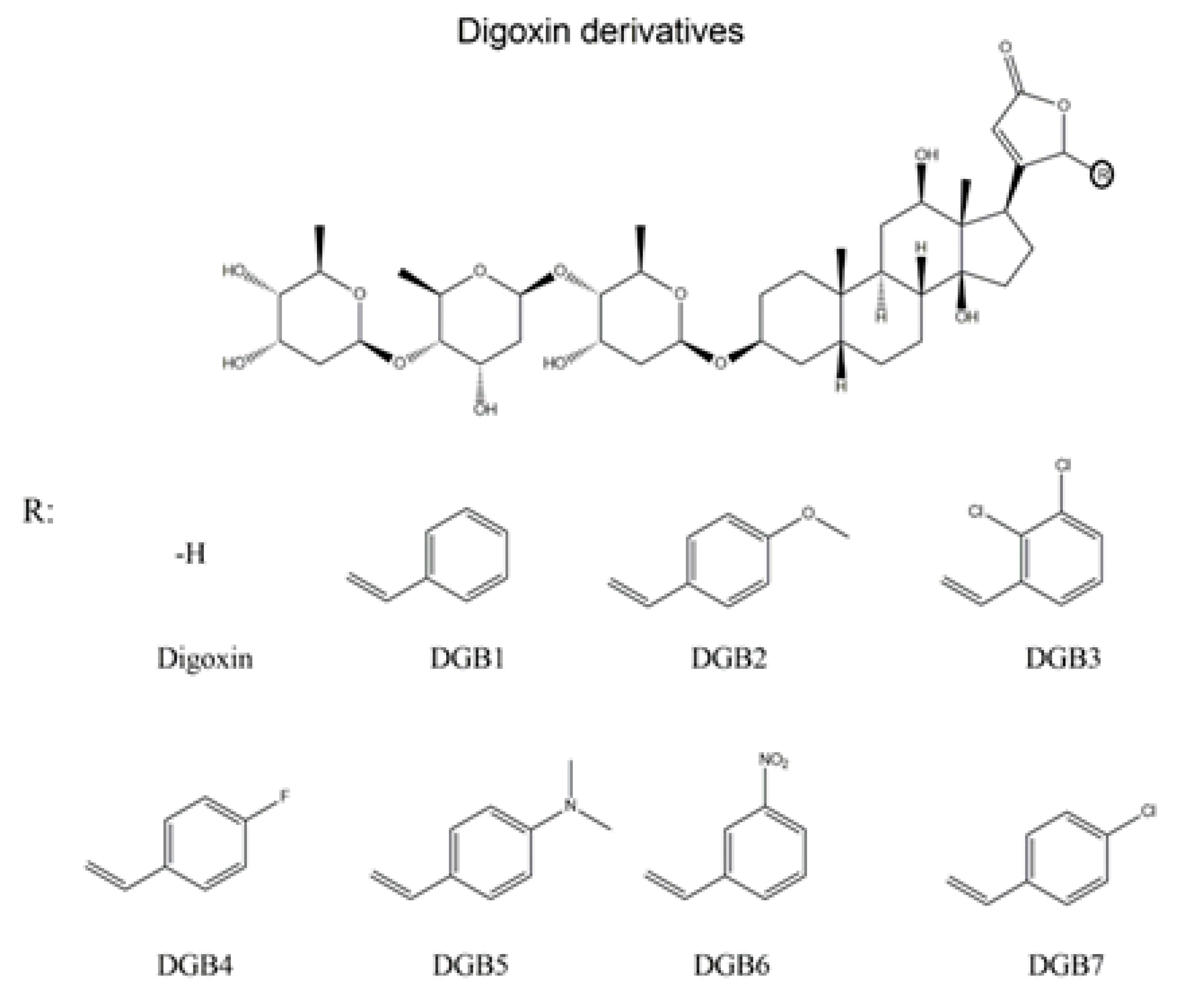
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Chemicals
2.3. Antifungal Susceptibility Test
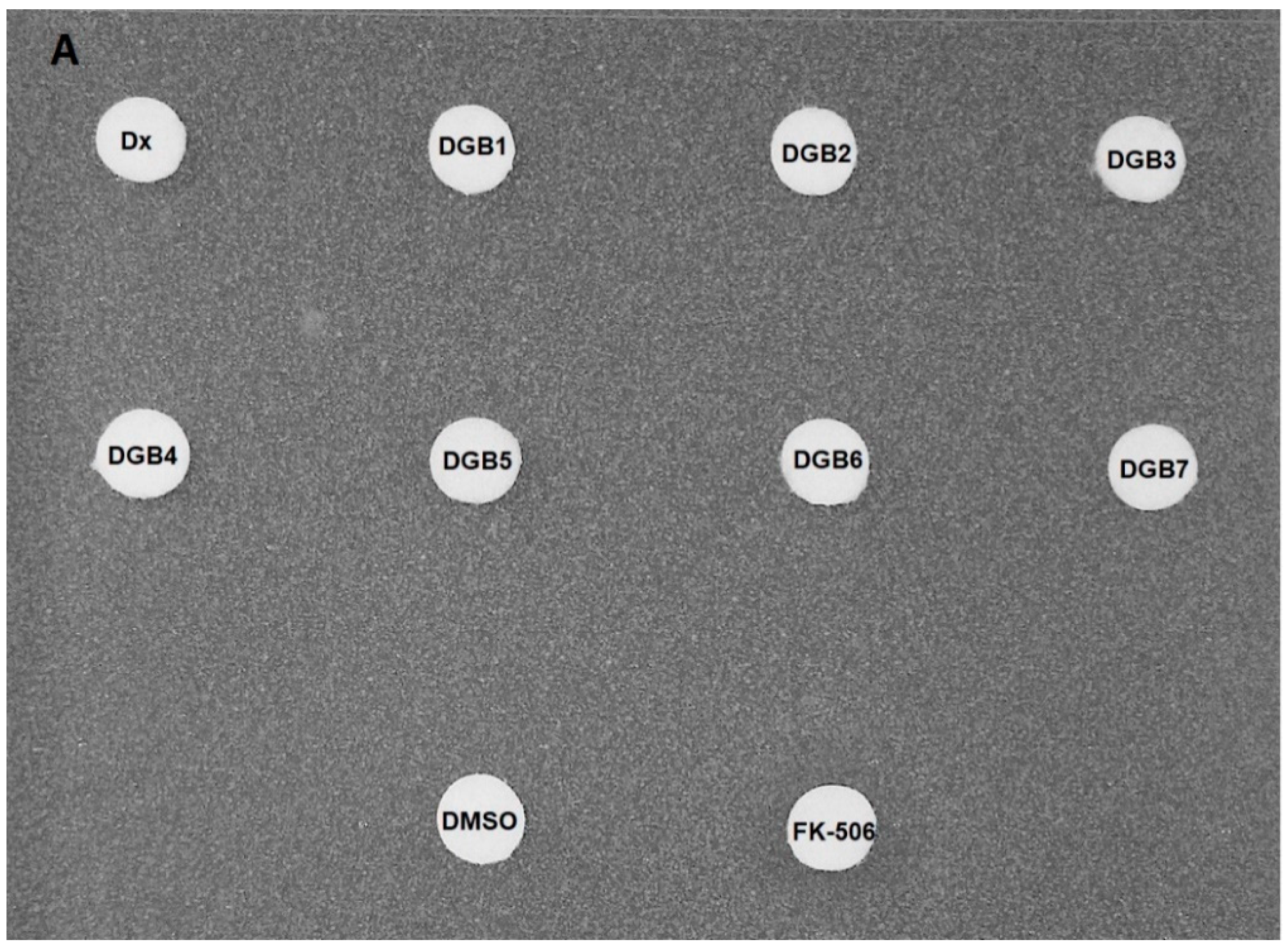
2.4. Disk Diffusion Assay
2.5. Checkerboard Assay
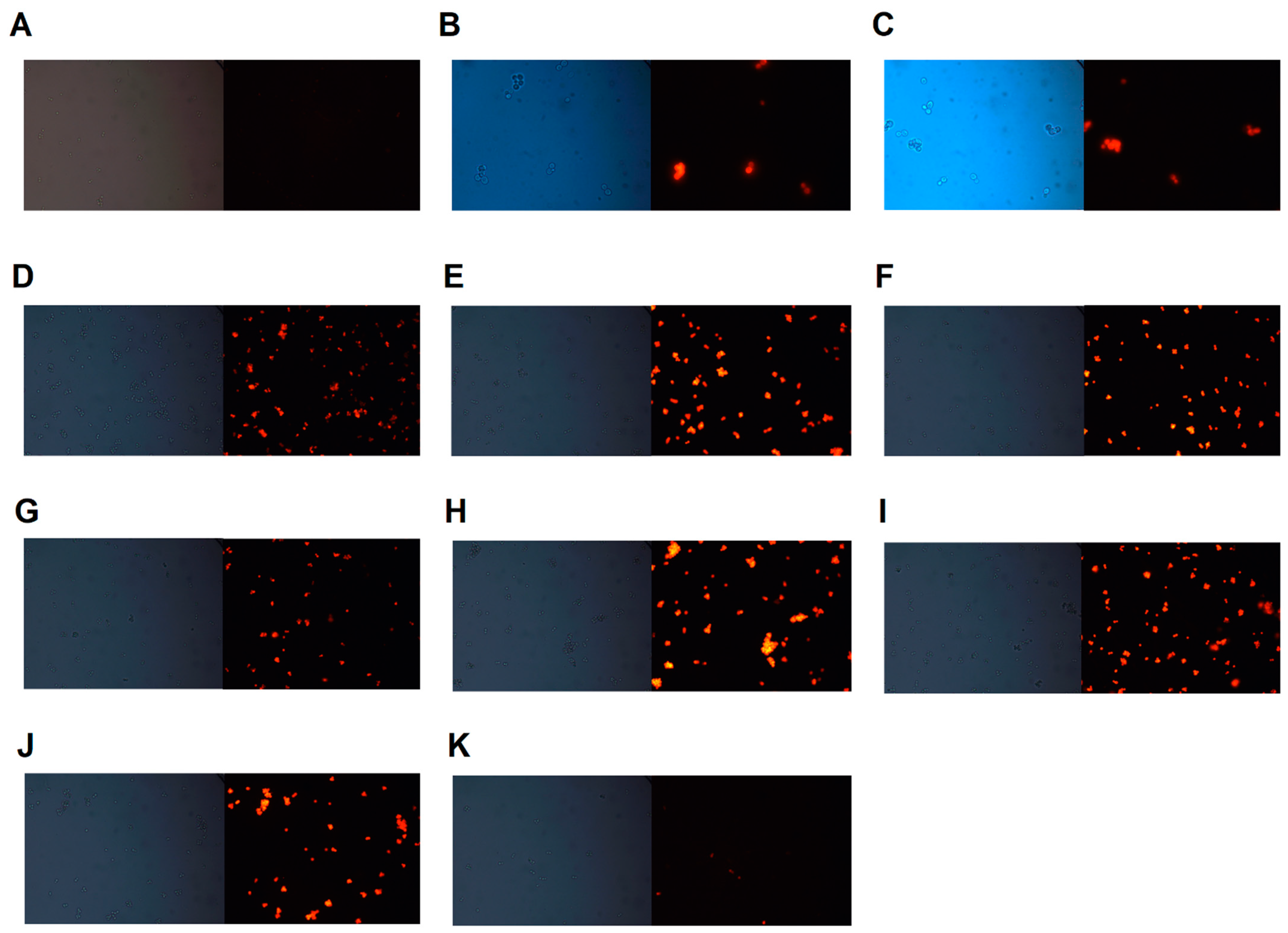
2.6. Rhodamine 6G Accumulation Assay
2.7. Preparation of Plasma Membranes
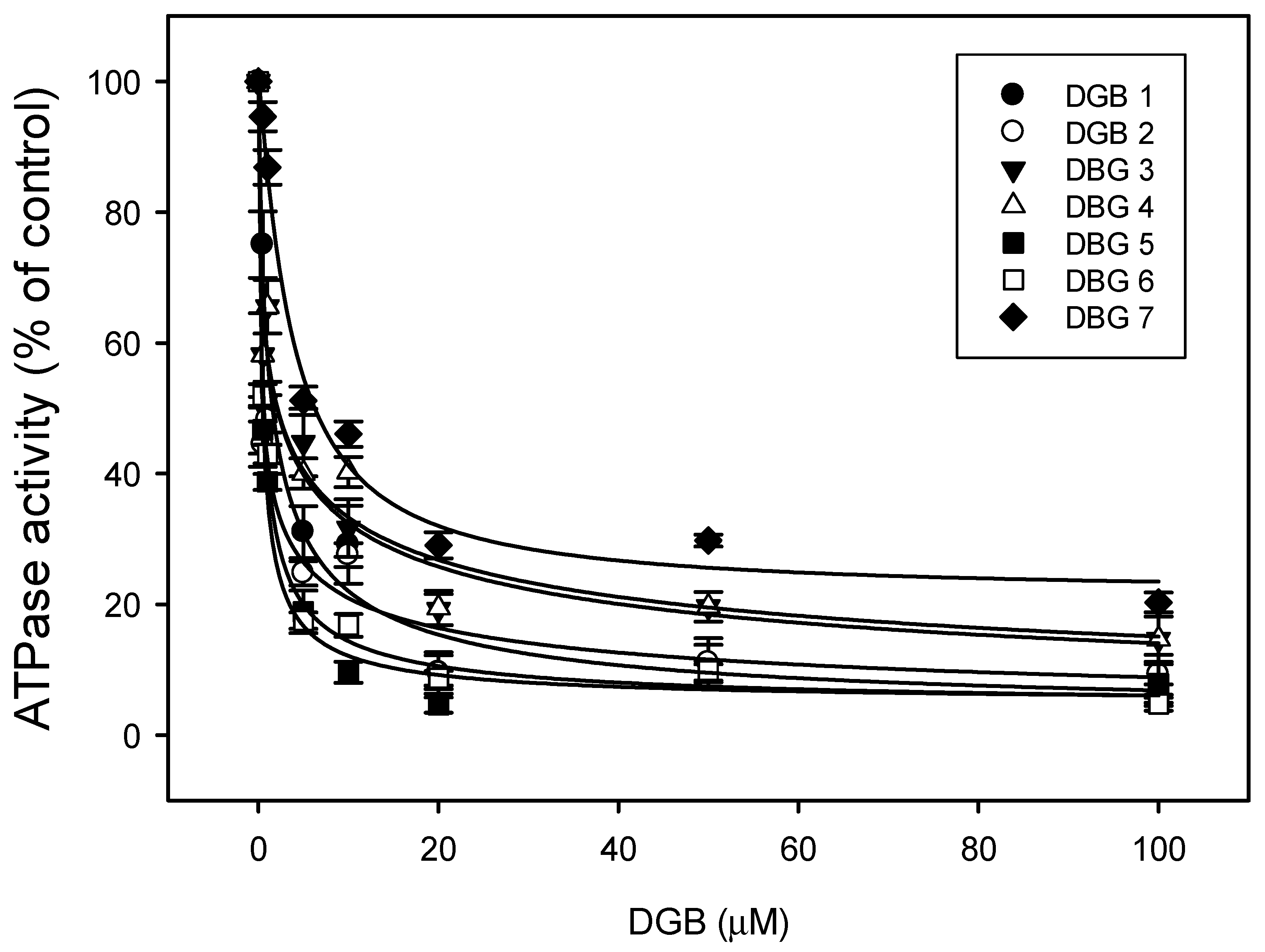
2.8. ATPase Activity
2.9. Hemolysis Assay
2.10. Statistical Analysis
3. Results
3.1. Antifungal Susceptibility Test
3.2. Disk Diffusion Assay
3.3. Checkerboard Assay
3.4. Rhodamine 6G Accumulation Assay
3.5. ATPase Activity
3.6. Hemolysis Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug resistance: An emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spampinato, C.; Leonardi, D. Candida infections, causes, targets, and resistance mechanisms: Traditional and alternative antifungal agents. Biomed. Res. Int. 2013, 2013, 204237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiei, M.; Peyton, L.; Hashemzadeh, M.; Foroumadi, A. History of the development of antifungal azoles: A review on structures, SAR, and mechanism of action. Bioorg. Chem. 2020, 104, 104240. [Google Scholar] [CrossRef]
- Kolaczkowski, M.; Kolaczkowska, A.; Luczynski, J.; Witek, S.; Goffeau, A. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 1998, 4, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Cannon, R.D.; Holmes, A.R. Learning the ABC of oral fungal drug resistance. Mol. Oral Microbiol. 2015, 30, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Redhu, A.K.; Shah, A.H.; Prasad, R. MFS transporters of Candida species and their role in clinical drug resistance. FEMS Yeast Res. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, G.; Lima, L.; Pedra, T.; Assumpção, A.; Morgado, S.; Mascarenhas, L. Bloodstream infection by Saccharomyces cerevisiae in a COVID-19 patient receiving probiotic supplementation in the ICU in Brazil. Access Microbiol. 2021, 3, 000250. [Google Scholar] [CrossRef] [PubMed]
- Ventoulis, I.; Sarmourli, T.; Amoiridou, P.; Mantzana, P.; Exindari, M.; Gioula, G.; Vyzantiadis, T. Bloodstream infection by Saccharomyces cerevisiae in two COVID-19 patients after receiving supplementation of Saccharomyces in the ICU. J. Fungi 2020, 6, 98. [Google Scholar] [CrossRef]
- Paumi, C.M.; Chuk, M.; Snider, J.; Stagljar, I.; Michaelis, S. ABC Transporters in Saccharomyces cerevisiae and Their Interactors: New Technology Advances the Biology of the ABCC (MRP) Subfamily. Microbiol. Mol. Biol. Rev. 2009, 73, 577–593. [Google Scholar] [CrossRef] [Green Version]
- Demuyser, L.; Van Dijck, P. Can Saccharomyces cerevisiae keep up as a model system in fungal azole susceptibility research? Drug Resist. Updates 2019, 42, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; De Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.N.D.; Garcia, I.J.P.; Valadares, J.M.M.; Pessoa, M.T.C.; Toledo, M.M.; Machado, M.V.; Busch, M.S.; Rocha, I.; Villar, J.A.F.P.; Atella, G.C.; et al. Evaluation of Cardiotonic Steroid Modulation of Cellular Cholesterol and Phospholipid. J. Membr. Biol. 2021, 254, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Alves, S.L.G.; Paixão, N.; Ferreira, L.G.R.; Santos, F.R.S.; Neves, L.D.R.; Oliveira, G.C.; Cortes, V.F.; Salomé, K.S.; Barison, A.; Santos, F.V.; et al. γ-Benzylidene digoxin derivatives synthesis and molecular modeling: Evaluation of anticancer and the Na,K-ATPase activity effect. Bioorg. Med. Chem. 2015, 23, 4397–4404. [Google Scholar] [CrossRef] [PubMed]
- White, T.C.; Holleman, S.; Dy, F.; Mirels, L.F.; Stevens, D.A. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 2002, 46, 1704–1713. [Google Scholar] [CrossRef] [Green Version]
- Niimi, K.; Harding, D.R.K.; Parshot, R.; King, A.; Lun, D.J.; Decottignies, A.; Niimi, M.; Lin, S.; Cannon, R.D.; Goffeau, A.; et al. Chemosensitization of Fluconazole Resistance in Saccharomyces cerevisiae and Pathogenic Fungi by a D -Octapeptide Derivative. Antimicrob. Agents Chemother. 2004, 48, 1256–1271. [Google Scholar] [CrossRef] [Green Version]
- Reis de Sá, L.F.; Toledo, F.T.; Gonçalves, A.C.; Sousa, B.A.; Dos Santos, A.A.; Brasil, P.F.; Duarte da Silva, V.A.; Tessis, A.C.; Ramos, J.A.; Carvalho, M.A.; et al. Synthetic Organotellurium Compounds Sensitize Drug-Resistant Candida albicans Clinical Isolates to Fluconazole. Antimicrob. Agents Chemother. 2017, 61, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Reis de Sa, L.F.; Toledo, F.T.; de Sousa, B.A.; Gonçalves, A.C.; Tessis, A.C.; Wendler, E.P.; Comasseto, J.V.; Dos Santos, A.A.; Ferreira-Pereira, A. Synthetic organotelluride compounds induce the reversal of Pdr5p mediated fluconazole resistance in Saccharomyces cerevisiae. BMC Microbiol. 2014, 14, 201. [Google Scholar] [CrossRef] [Green Version]
- Wernli, D.; Harbarth, S.; Levrat, N.; Pittet, D. A ‘whole of United Nations approach’ to tackle antimicrobial resistance? A mapping of the mandate and activities of international organisations. BMJ Glob. Health 2022, 7, e008181. [Google Scholar] [CrossRef]
- Gold, J.A.W.; Ahmad, F.B.; Cisewski, J.A.; Rossen, L.M.; Montero, A.J.; Benedict, K.; Jackson, B.R.; Toda, M. Increased deaths from fungal infections during the COVID-19 pandemic—National Vital Statistics System, United States, January 2020—December 2021. Clin. Infect. Dis. 2022, ciac489. [Google Scholar] [CrossRef]
- Rogers, B.; Decottignies, A.; Kolaczkowski, M.; Carvajal, E.; Balzi, E.; Goffeau, A. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 2001, 3, 207–214. [Google Scholar] [PubMed]
- Sorbo, G.D.; Andrade, A.C.; Van Nistelrooy, J.G.M.; Van Kan, J.A.L.; Balzi, E.; De Waard, M.A. Multidrug resistance in Aspergillus nidulans involves novel ATP-binding cassette transporters. Mol. Gen. Genet. 1997, 254, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Pashazadeh-Panahi, P.; Hasanzadeh, M. Digoxin as a glycosylated steroid-like therapeutic drug: Recent advances in the clinical pharmacology and bioassays of pharmaceutical compounds. Biomed. Pharmacother. 2020, 123, 109813. [Google Scholar] [CrossRef] [PubMed]
- Frohock, B.H.; Gilbertie, J.M.; Daiker, J.C.; Schnabel, L.V.; Pierce, J.G. 5-benzylidene-4-oxazolidinones are synergistic with antibiotics for the treatment of Staphylococcus aureus biofilms. Chembiochem 2020, 21, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Ledwitch, K.V.; Barnes, R.W.; Roberts, A.G. Unravelling the complex drug-drug interactions of the cardiovascular drugs, verapamil and digoxin, with P-glycoprotein. Biosci. Rep. 2016, 36, e00309. [Google Scholar] [CrossRef]
- Ferreira-Pereira, A.; Marco, S.; Decottignies, A.; Nader, J.; Goffeau, A.; Rigaud, J.L. Three-dimensional reconstruction of the Saccharomyces cerevisiae multidrug resistance protein Pdr5p. J. Biol. Chem. 2003, 278, 11995–11999. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.R.; Tessis, A.C.; Ferreira, P.F.; Rangel, L.P.; Garcia-Gomes, A.S.; Pereira, F.R.; Berlinck, R.G.S.; Muricy, G.; Ferreira-Pereira, A. Oroidin inhibits the activity of the multidrug resistance target Pdr5p from yeast plasma membranes. J. Nat. Prod. 2011, 74, 279–282. [Google Scholar] [CrossRef]
- Moraes, D.C.; Cardoso, K.M.; Domingos, L.T.S.; Pinto, M.C.F.R.; Monteiro, R.Q.; Ferreira-Pereira, A. β-lapachone enhances the antifungal activity of fluconazole against a Pdr5p-mediated resistant Saccharomyces cerevisiae strain. Braz. J. Microbiol. 2020, 51, 1051–1060. [Google Scholar] [CrossRef]
- Moraes, D.C.; Ferreira-Pereira, A. Insights on the anticandidal activity on non-antifungal drugs. J. Mycol. Med. 2019, 29, 253–259. [Google Scholar] [CrossRef]
- Oliveira, G.C.; Rocha, S.C.; Lopes, M.A.S.; Paixão, N.; Alves, S.L.G.; Pessoa, M.T.C.; Noël, F.; Quintas, L.E.M.; Barbosa, L.A.; Villar, J.A.F.P.; et al. Implications of synthetic modifications of the cardiotonic steroid lactone ring on cytotoxicity. J. Membr. Biol. 2021, 254, 487–497. [Google Scholar] [CrossRef]
- Mukhopadhyay, K.; Kohli, A.; Prasad, R. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 2002, 46, 3695–3705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanglard, D.; Ischer, F.; Monod, M.; Bille, J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: Characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 1997, 143, 405–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patocka, J.; Nepovimova, E.; Wu, W.; Kuca, K. Digoxin: Pharmacology and toxicology—A review. Environ. Toxicol. Pharmacol. 2020, 79, 103400. [Google Scholar] [CrossRef] [PubMed]



| Strain and Compound | Compound (µM) | Fluconazole (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| MIC a | MIC c | FIC | MIC a | MIC c | FIC | FICI | Outcome | |
| AD/124567 | ||||||||
| DGB1 | 200 | 50 | 0.250 | 480 | 120 | 0.250 | 0.500 | S |
| DGB2 DGB3 DGB4 DGB5 DGB6 DGB7 | 200 100 200 200 200 200 | 25 6.25 50 3.125 50 3.125 | 0.125 0.063 0.250 0.016 0.250 0.016 | 480 480 480 480 480 480 | 7.5 7.5 15 7.5 60 7.5 | 0.016 0.016 0.031 0.016 0.125 0.016 | 0.141 0.078 0.281 0.031 0.375 0.031 | S S S S S S |
| 95–142 | ||||||||
| DGB1 | 200 | 25 | 0.125 | 62.5 | 31.25 | 0.500 | 0.625 | I |
| DGB2 DGB3 DGB4 DGB5 DGB6 DGB7 | 200 200 200 200 200 200 | 200 6.25 200 200 200 200 | 1 0.031 1 1 1 1 | 62.5 62.5 62.5 62.5 62.5 62.5 | 62.5 15.63 62.5 62.5 62.5 62.5 | 1 0.250 1 1 1 1 | 2 0.281 2 2 2 2 | I S I I I I |
| Compound | Rhodamine 6G Efflux (%) | IC50 (μM) | Molecular Weight | cLogP |
|---|---|---|---|---|
| DGB1 | 6.84 | 1.25 | 869 | 3.88 |
| DGB2 | 1.31 | 0.49 | 899 | 3.81 |
| DGB3 | 0.00 | 2.13 | 938 | 5.09 |
| DGB4 | 23.35 | 2.19 | 887 | 3.98 |
| DGB5 | 8.52 | 0.41 | 912 | 3.78 |
| DGB6 | 4.66 | 0.53 | 914 | 2.96 |
| DGB7 | 2.45 | 3.72 | 903 | 4.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Moraes, D.C.; Tessis, A.C.; Rollin-Pinheiro, R.; Princival, J.L.; Villar, J.A.F.P.; Barbosa, L.A.; Barreto-Bergter, E.; Ferreira-Pereira, A. Digoxin Derivatives Sensitize a Saccharomyces cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p. J. Fungi 2022, 8, 769. https://doi.org/10.3390/jof8080769
de Moraes DC, Tessis AC, Rollin-Pinheiro R, Princival JL, Villar JAFP, Barbosa LA, Barreto-Bergter E, Ferreira-Pereira A. Digoxin Derivatives Sensitize a Saccharomyces cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p. Journal of Fungi. 2022; 8(8):769. https://doi.org/10.3390/jof8080769
Chicago/Turabian Stylede Moraes, Daniel Clemente, Ana Claudia Tessis, Rodrigo Rollin-Pinheiro, Jefferson Luiz Princival, José Augusto Ferreira Perez Villar, Leandro Augusto Barbosa, Eliana Barreto-Bergter, and Antônio Ferreira-Pereira. 2022. "Digoxin Derivatives Sensitize a Saccharomyces cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p" Journal of Fungi 8, no. 8: 769. https://doi.org/10.3390/jof8080769
APA Stylede Moraes, D. C., Tessis, A. C., Rollin-Pinheiro, R., Princival, J. L., Villar, J. A. F. P., Barbosa, L. A., Barreto-Bergter, E., & Ferreira-Pereira, A. (2022). Digoxin Derivatives Sensitize a Saccharomyces cerevisiae Mutant Strain to Fluconazole by Inhibiting Pdr5p. Journal of Fungi, 8(8), 769. https://doi.org/10.3390/jof8080769







