Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
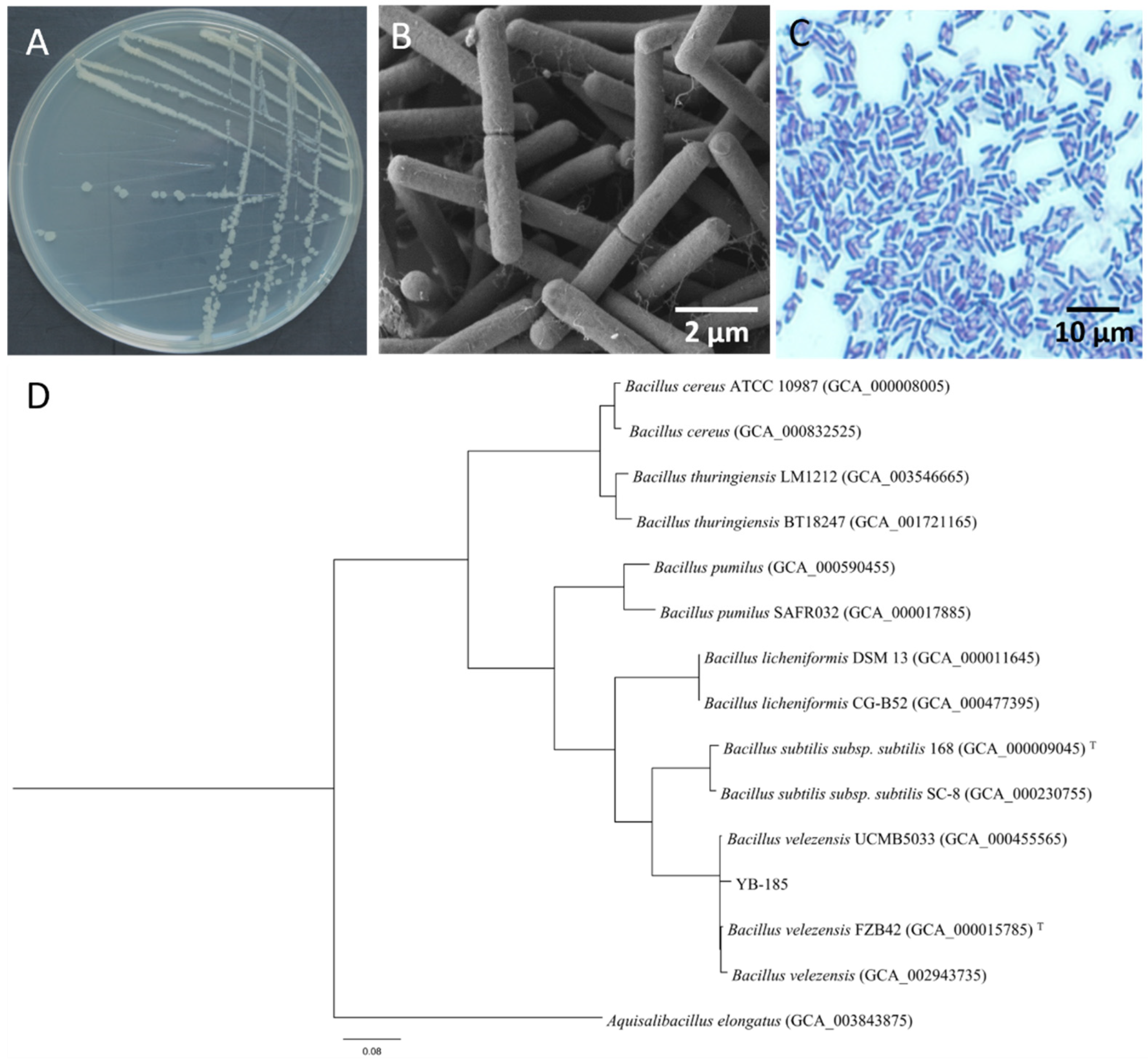
2.1. Strain YB-185 Isolation and Identification
2.2. Antagonistic Activity of Strain YB-185 against F. pseudograminearum
2.3. YB-185 Suppression of FCR in Greenhouse
2.4. YB-185 Suppression of FCR in the Field
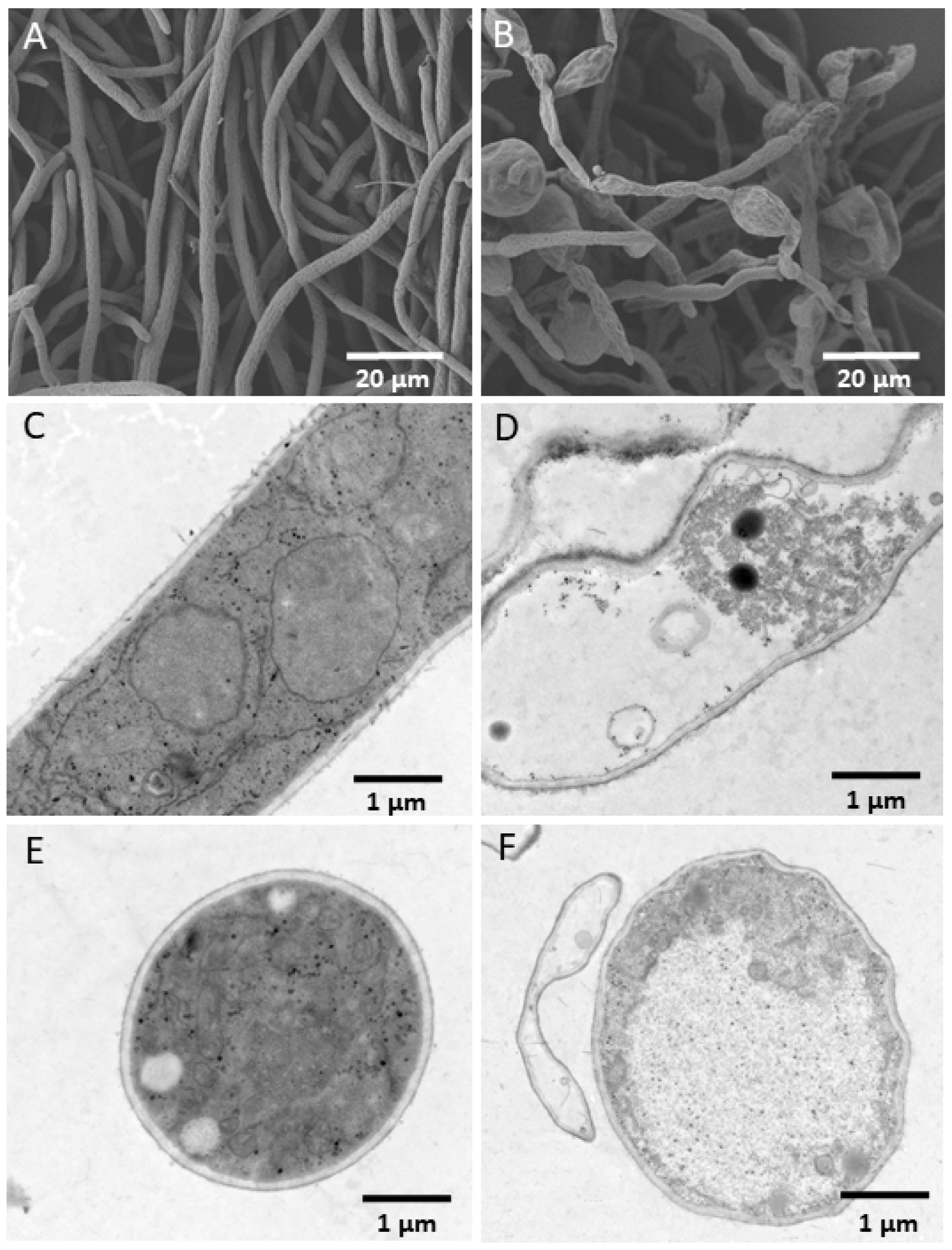
2.5. Transcriptome of Fusarium pseudograminearum Co-Cultured with YB-185
2.6. RT-PCR of Fusarium pseudograminearum Genes in Co-Cultures with YB-185
2.7. Total Antioxidant and Glutathione-S-Transferase Activity
2.8. Statistical Analysis
3. Results
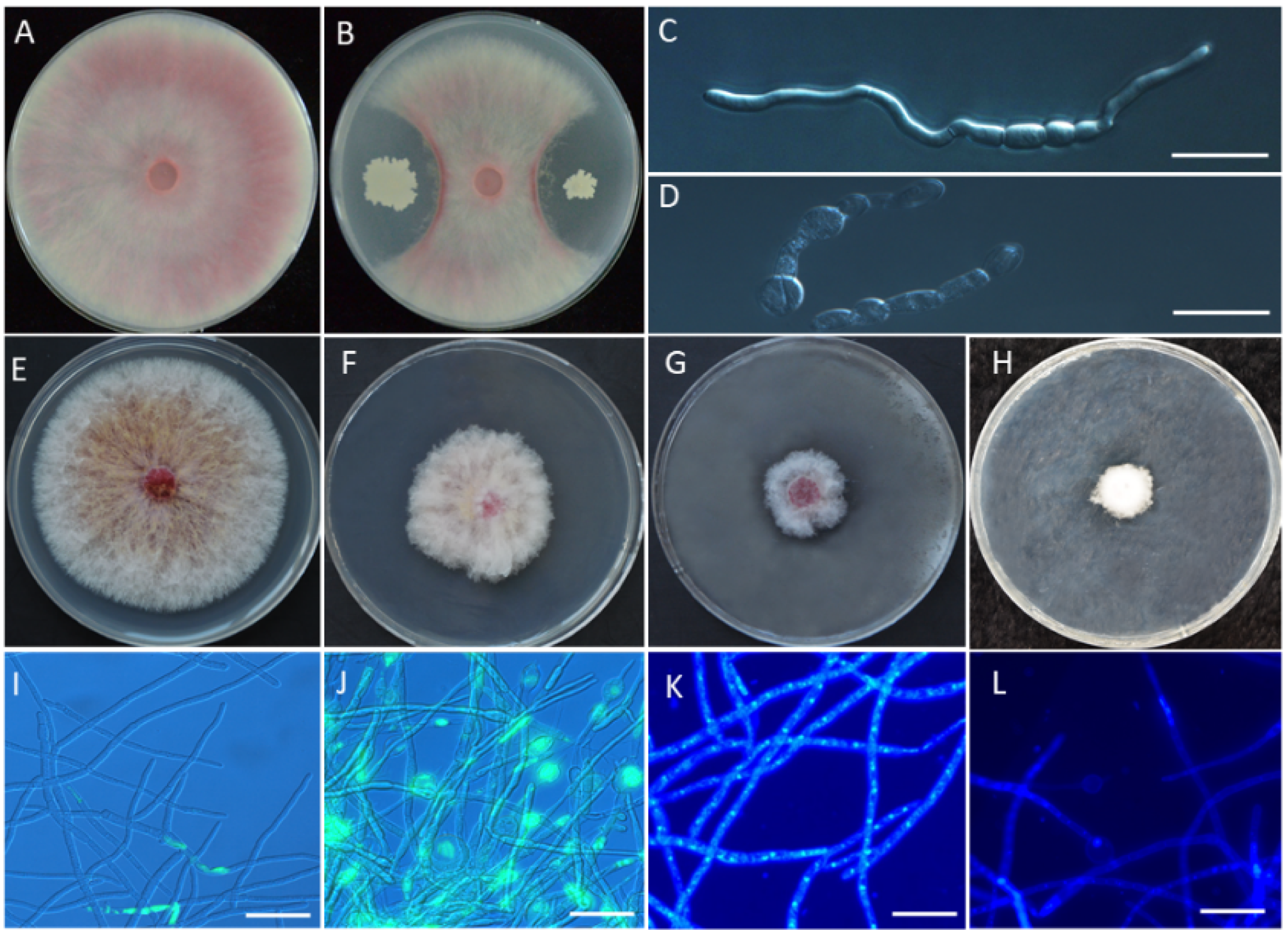
3.1. Strain YB-185 Isolation and Identification
3.2. In Vitro Antagonism of YB-185 against Fusarium pseudograminearum
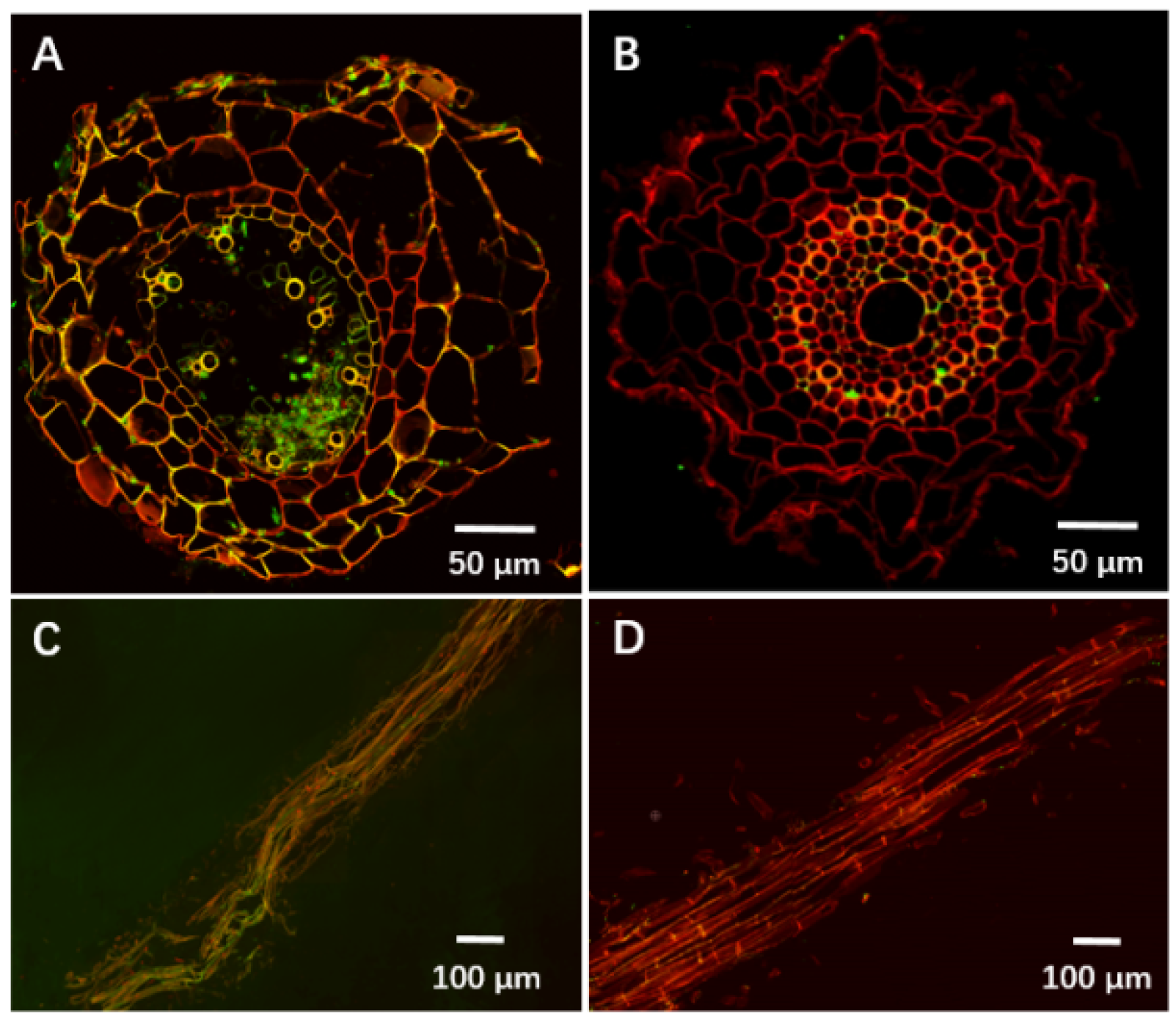
3.3. YB-185 Suppression of FCR in Greenhouse
3.4. YB-185 Suppression of FCR in the Field
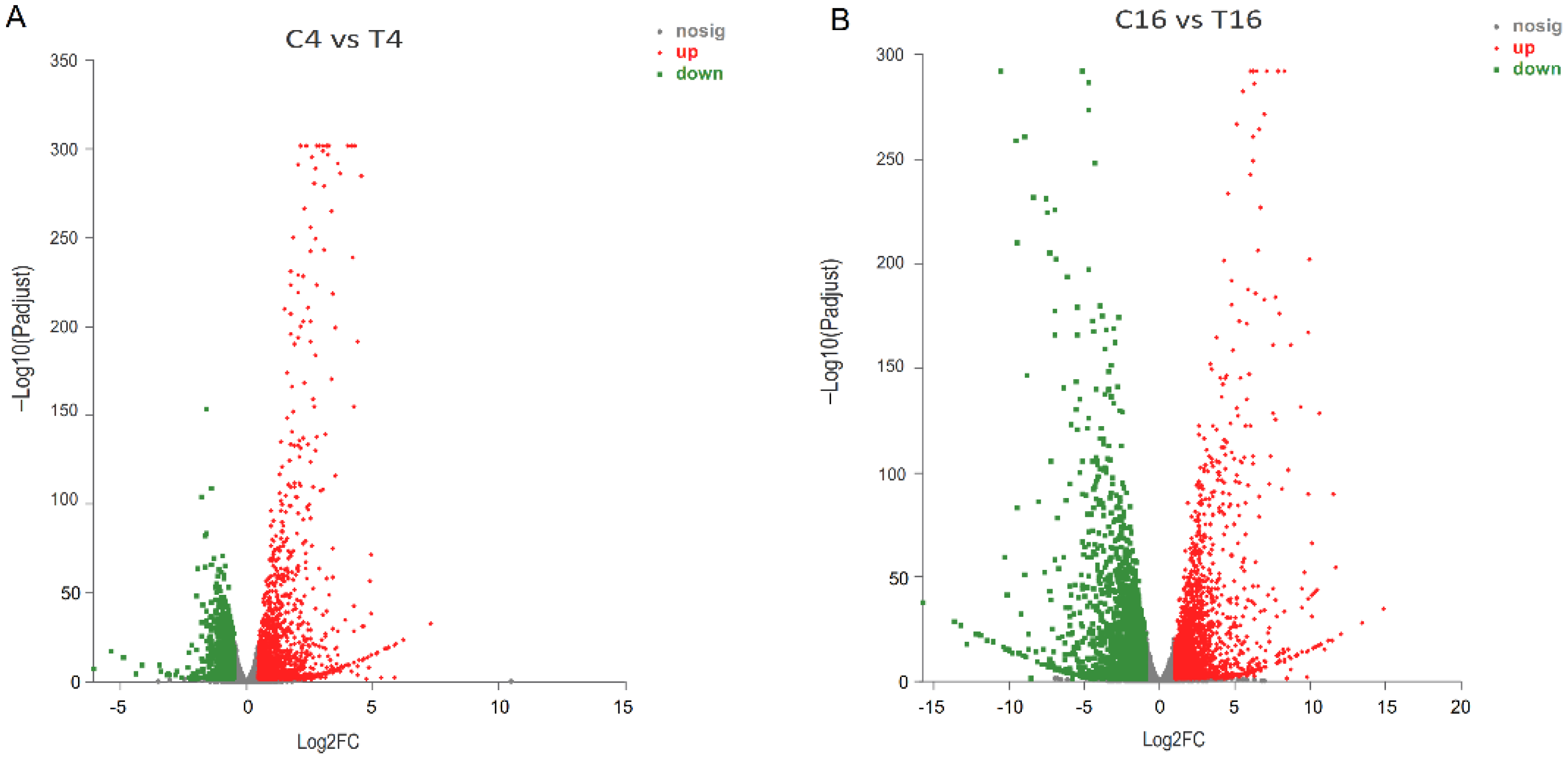
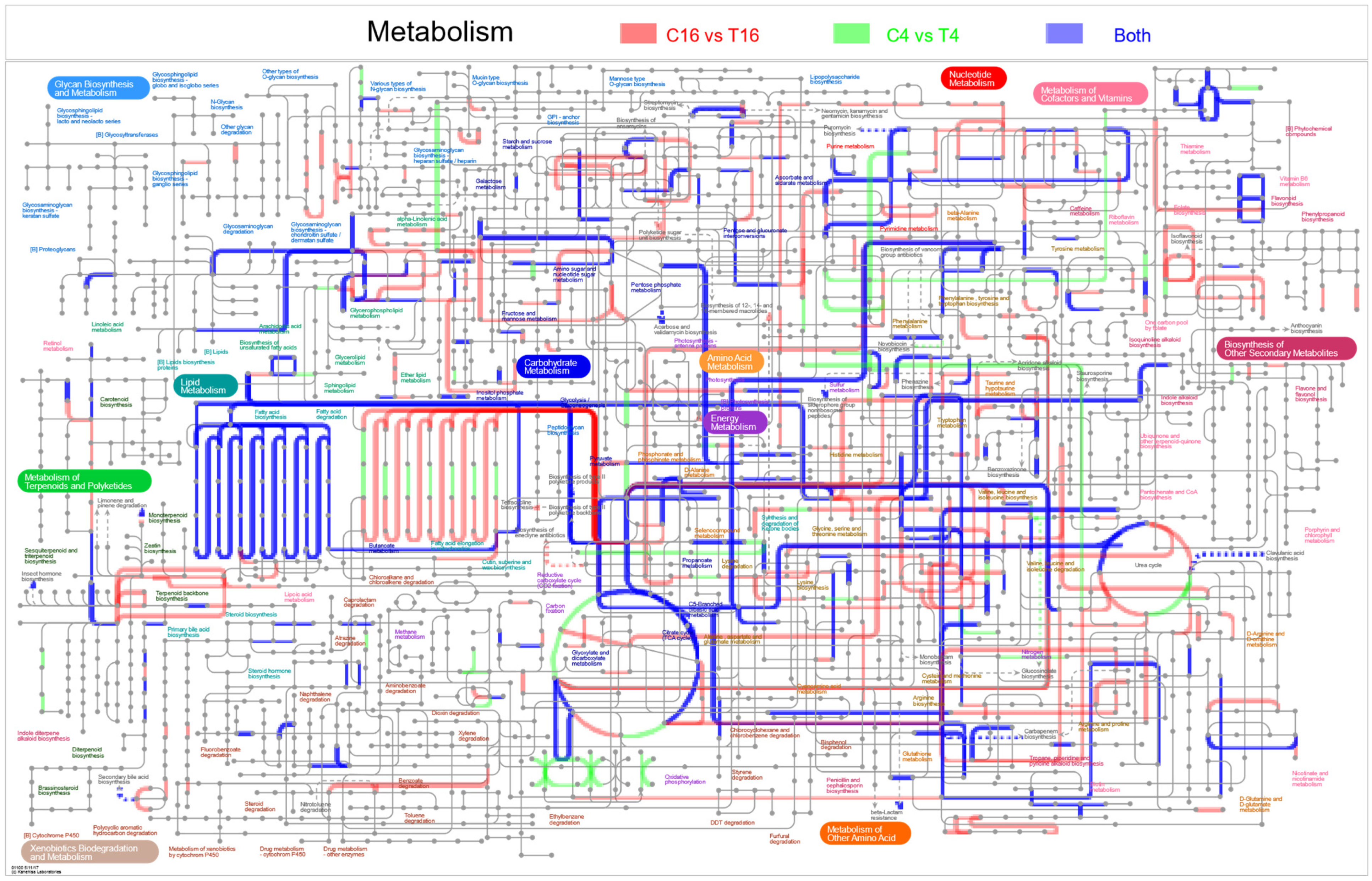
3.5. Transcriptome of F. pseudograminearum Co-Cultured with B. velezensis YB-185
3.6. DEGs of Fusarium pseudograminearum Associated with Co-Cultivation with YB-185
3.7. RT-PCR of Fusarium pseudograminearum Genes in Co-Cultures with YB-185
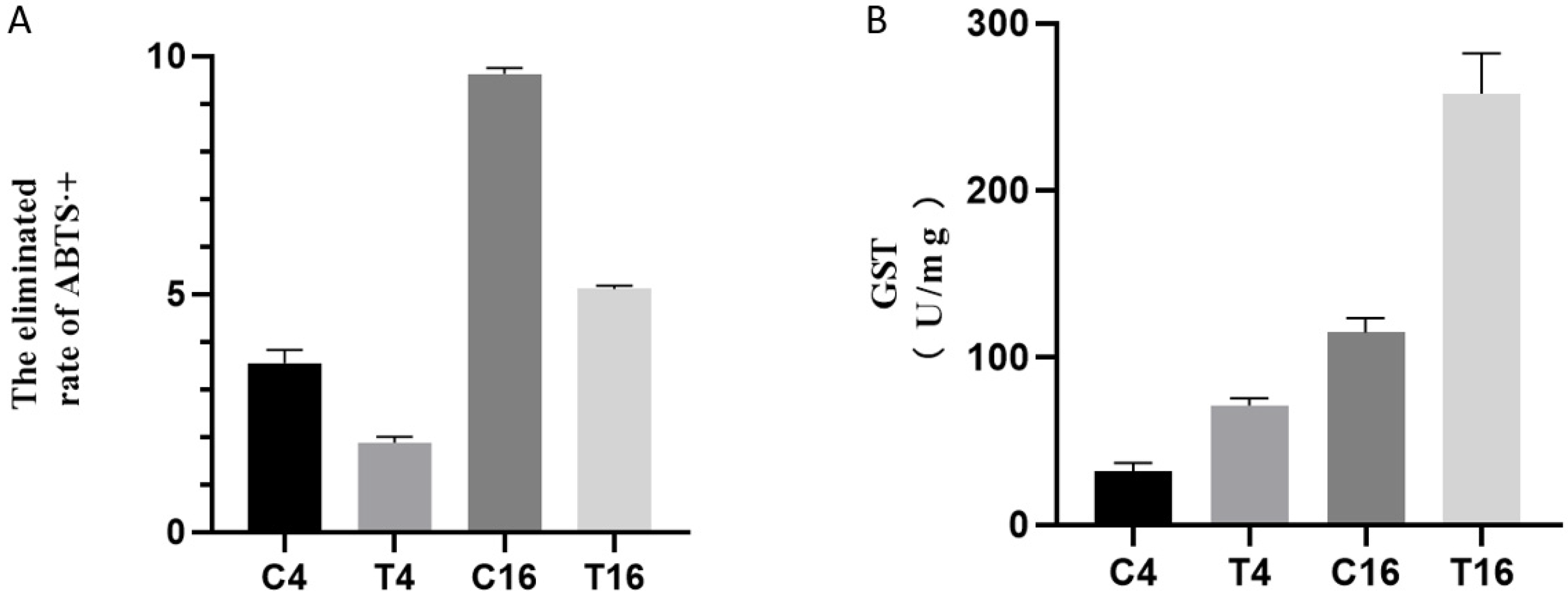
3.8. Total Antioxidant and GST Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mastrangelo, A.A.; Cattivelli, L. What makes bread and durum wheat different? Trends Plant Sci. 2021, 26, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 2010, 38, 558–570. [Google Scholar] [CrossRef]
- Mudge, A.M.; Ruth, D.; Yanhong, D.; Gardiner, D.M.; White, R.G.; Manners, J.M. A role for the mycotoxin deoxynivalenol in stem colonisation during crown rot disease of wheat caused by Fusarium graminearum and Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2006, 69, 73–85. [Google Scholar] [CrossRef]
- Dyer, A.T.; Johnston, R.H.; Hogg, A.C.; Johnston, J.A. Comparison of pathogenicity of the fusarium crown rot (FCR) complex (F. culmorum, F. pseudograminearum and F. graminearum) on hard red spring and durum wheat. Eur. J. Plant Pathol. 2009, 125, 387–395. [Google Scholar] [CrossRef]
- Li, H.L.; Yuan, H.X.; Fu, B.; Xing, X.P.; Tang, W.H. First report of Fusarium pseudograminearum causing crown rot of wheat in Henan, China. Plant Dis. 2012, 96, 1065. [Google Scholar] [CrossRef]
- Zhou, H.; He, X.; Wang, S.; Ma, Q.; Sun, B. Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China. Environ. Microbiol. 2019, 29, 2740–2754. [Google Scholar] [CrossRef]
- Blyuss, K.B.; Fatehi, F.; Tsygankova, V.A.; Biliavska, L.O.; Iutynska, G.O.; Yemets, A.I.; Blume, Y.B. RNAi-based biocontrol of wheat nematodes using natural poly-component biostimulants. Front. Plant Sci. 2019, 17, 483–494. [Google Scholar] [CrossRef]
- Obanor, F.; Neate, S.; Simpfendorfer, S.; Sabburg, R.; Chakraborty, S. Fusarium graminearum and Fusarium pseudograminearum caused the 2010 head blight epidemics in Australia. Plant Pathol. 2013, 62, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Ji, X.; Ge, Y.; Li, J.; Qi, W.; Qiao, K. Characterization of antagonistic Bacillus methylotrophicus isolated from rhizosphere and its biocontrol effects on maize stalk rot. Phytopathology 2019, 109, 571–581. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wong, P. Effect of Burkholderia (Pseudomonas) cepacia and soil type on the control of crown rot in wheat. Plant Soil 1998, 203, 103–108. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Q.; Xie, X.; Goodwin, P.H.; Deng, X.; Zhang, J.; Sun, R.H.; Wang, Q.; Xia, M.C.; Wu, C.; et al. Genomic and phenotypic insights into the potential of Bacillus subtilis YB-15 isolated from rhizosphere to biocontrol against crown rot and promote growth of wheat. Biology 2022, 11, 778. [Google Scholar] [CrossRef] [PubMed]
- Moya-Elizondo, E.A.; Jacobsen, B.J. Integrated management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of systemic acquired resistance (SAR). Biol. Control. 2016, 92, 153–163. [Google Scholar] [CrossRef]
- Ji, S.H.; Paul, N.C.; Deng, J.X.; Kim, Y.S.; Yu, S.H. Biocontrol activity of Bacillus amyloliquefaciens CNU114001 against fungal plant diseases. Mycobiology 2013, 41, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.C.; Hussey, M.A. Gram stain protocols. In ACM Microbelibrary-Laboratory Protocols; American Society for Microbiology: Washington, DC, USA, 2005; Volume 6, p. 21. Available online: http://www.microbelibrary.org (accessed on 12 January 2022).
- Marchesi, J.R.; Sato, T.; Weightman, J.A.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.J. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Bae, K.S. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 2000, 78, 123–127. [Google Scholar] [CrossRef]
- Yun, Y.; Liu, Z.; Yin, Y.; Jiang, J.; Chen, Y.; Xu, J.R.; Ma, Z. Functional analysis of the Fusarium graminearum phosphatome. New Phytol. 2015, 207, 119–134. [Google Scholar] [CrossRef]
- Akpinar, I.; Unal, M.; Sar, T. Potential antifungal effects of silver nanoparticles (AgNPs) of different sizes against phytopathogenic Fusarium oxysporum f. sp. radicis-lycopersici (FORL) strains. SN Appl. Sci. 2021, 3, 506–514. [Google Scholar] [CrossRef]
- Domachowske, J.B.; Bonville, C.A.; Mortelliti, A.J.; Colella, C.B.; Kim, U.; Rosenberg, H.F. Respiratory syncytial virus infection induces expression of the anti-apoptosis gene IEX-1L in human respiratory epithelial cells. J. Infect. Dis. 2000, 181, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Redkar, A.; Jaeger, E.; Doehlemann, G. Visualization of growth and morphology of fungal hyphae in planta using WGA-AF488 and propidium iodide co-staining. Bio-Protocl 2018, 8, 2492. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, H.X.; Xia, M.C.; Han, X.Y.; Xie, L.H.; Paul, H.G.; Quan, X.; Sun, R.H.; Wu, C.; Yang, L.R. Wheat root transcriptional responses against Gaeumannomyces graminis var. tritic. Phytopathol. Res. 2020, 2, 23. [Google Scholar] [CrossRef]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A.; Patterson, L.M. Pathogenicity of fungi associated with the wheat crown rot complex in Oregon and Washington. Plant Dis. 2005, 89, 949–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, G.J.; Smiley, R.W.; Paulitz, T.C.; Walker, C.A.; Carter, A.H.; See, D.R.; Garland-Campbell, K. Identification of quantitative trait loci (QTL) for resistance to Fusarium crown rot (Fusarium pseudograminearum) in multiple assay environments in the Pacific Northwestern US. Theor. Appl. Genet. 2012, 125, 91–107. [Google Scholar] [CrossRef] [Green Version]
- Lindgreen, S. Adapter removal: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Meth 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Chen, J.; Zhao, Y.; Cai, H.; Guo, C. Bacillus subtilis HSY21 can reduce soybean root rot and inhibit the expression of genes related to the pathogenicity of Fusarium oxysporum. Pestic. Biochem. Phys. 2021, 178, 104916. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, H.; Zhao, S.; Huang, J.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Functional redundancy of two ABC transporter proteins in mediating toxicity of Bacillus thuringiensis to cotton bollworm. PLoS Pathog. 2020, 16, e1008427. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Fu, L.; Xu, G.; Lin, X.; Qiao, J.; Xia, Y. Biocontrol of wheat crown rot using Bacillus halotolerans QTH8. Pathogens 2022, 11, 595. [Google Scholar] [CrossRef]
- Sasani, M.; Ahmadzadeh, M.; Jahansuz, M.R.; Navid, S. Bioprimee of seed with Bacillus velezensis UTB96 to control the fungal pathogen of root and crown rot (Fusarium pseudograminearum) and improving some growth indicators of wheat. Iran. J. Seed Sci. Technol. 2021, 10, 85–102. [Google Scholar]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Hassen, A.I.; Labuschagne, N. Rhizobacteria-induced systemic resilience in Sorghum bicolor L. moench against Fusarium pseudograminearum crown rot under drought stress conditions. Biol. Control 2020, 151, 104395. [Google Scholar] [CrossRef]
- Heydari, A.; Pessarakli, M. A review on biological control of fungal plant pathogens using microbial antagonists. J. Biol. Sci. 2010, 10, 273–290. [Google Scholar] [CrossRef] [Green Version]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.H.; Chen, P.Y.; Yang, Y.L.; Kan, S.C.; Hsieh, F.C.; Liu, Y.C. Clarification of the antagonistic effect of the lipopeptides produced by Bacillus amyloliquefaciens BPD1 against Pyricularia oryzae via In Situ MALDI-TOF IMS Analysis. Molecules 2016, 21, 1670. [Google Scholar] [CrossRef] [Green Version]
- Gong, A.D.; Li, H.P.; Yuan, Q.S.; Song, X.S.; Yao, W.; He, W.J.; Zhang, J.B.; Liao, Y.C. Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef] [Green Version]
- Toral, L.; Rodríguez, M.; Béjar, V.; Sampedro, I. Antifungal activity of lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea. Front. Microbiol. 2018, 9, 1315. [Google Scholar] [CrossRef]
- Wang, Q.H.; Ji, Y.P.; Qu, Y.Y.; Qi, Y.K.; Li, D.W.; Liu, Z.Y.; Wu, X.Q. The response strategies of Colletotrichum gloeosporioides s.s. due to the stress caused by biological control agent Bacillus amyloliquefaciens deciphered by transcriptome analyses. Biol. Control 2020, 150, 104372. [Google Scholar] [CrossRef]
- Tian, W.H.; Cheng, Z.R.; Wang, J.X.; Cheng, F.F.; Li, L.P.; Huo, C.X.; Li, W.X.; Han, S.Y.; Guo, X.Y.; Wang, A.Y. A transcriptome profile reveals the regulatory mechanism of Verticillium dahliae against Bacillus. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, L.; Xiang, Y.; Du, L.; Huang, X.; Liu, Y. Comparative transcriptome analysis of Sclerotinia sclerotiorum revealed its response mechanisms to the biological control agent, Bacillus amyloliquefaciens. Sci. Rep. 2020, 10, 12576. [Google Scholar] [CrossRef]
- Samaras, A.; Karaoglanidis, G.S.; Tzelepis, G. Insights into the multitrophic interactions between the biocontrol agent Bacillus subtilis MBI 600, the pathogen Botrytis cinerea and their plant host. Microbiol. Res. 2021, 248, 126752. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Ortiz-Castellanos, L. Cell wall glucans of fungi. A review. Cell Surf. 2019, 5, 100022. [Google Scholar] [CrossRef] [PubMed]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sant, D.G.; Tupe, S.G.; Ramana, C.V.; Deshpande, M.V. Fungal cell membrane-promising drug target for antifungal therapy. J. Appl. Microbiol. 2016, 121, 1498–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreri, C.; Masi, A.; Sansone, A.; Giacometti, G.; Larocca, A.V.; Menounou, G.; Scanferlato, R.; Tortorella, S.; Rota, D.; Conti, M.; et al. Fatty acids in membranes as homeostatic, metabolic and nutritional biomarkers: Recent advancements in analytics and diagnostics. Diagnostics 2016, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Ann. Rev. Plant Biol. 2015, 266, 113–137. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Perkins, A.; Nelson, K.J.; Parsonage, D.; Poole, L.B.; Karplus, P.A. Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015, 40, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Collet, J.F.; Messens, J. Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 2010, 13, 1205–1216. [Google Scholar] [CrossRef]
- Valenzuela-Cota, D.F.; Buitimea-Cantúa, G.V.; Plascencia-Jatomea, M.; Cinco-Moroyoqui, F.J.; Martínez-Higuera, A.A.; Rosas-Burgos, E.C. Inhibition of the antioxidant activity of catalase and superoxide dismutase from Fusarium verticillioides exposed to a Jacquinia macrocarpa antifungal fraction. J. Environ. Sci. Health B 2019, 54, 647–654. [Google Scholar] [CrossRef]
- Ramsay, E.E.; Dilda, P.J. Glutathione S-conjugates as prodrugs to target drug-resistant tumors. Front. Pharmacol. 2014, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.L.; Zhang, K.; Wang, G.L.; Liu, W.D. Comparative transcriptome analysis of Setosphaeria turcica revealed its responses mechanisms to the biological control agent Bacillus amyloliquefaciens. Sci. Sin. Vitae 2016, 46, 627–636. [Google Scholar]
- Chang, Z.; Jiang, N.; Zhang, Y.; Lu, H.; Yin, J.; Wahlgren, M.; Chen, Q. The TatD-like DNase of Plasmodium is a virulence factor and a potential malaria vaccine candidate. Nat. Commun. 2016, 7, 11537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, Z.; Zhu, W.; Su, H.; Zhao, Y.; Zhang, K.Q.; Yang, J. Recent advances in genes involved in secondary metabolite synthesis, hyphal development, energy metabolism and pathogenicity in Fusarium graminearum (teleomorph Gibberella zeae). Biotechnol. Adv. 2014, 32, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Gaffoor, I.; Brown, D.W.; Plattner, R.; Proctor, R.H.; Qi, W.; Trail, F. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae(anamorph Fusarium graminearum). Eukaryot. Cell 2005, 4, 1926–1933. [Google Scholar] [CrossRef] [Green Version]
- Frandsen, R.J.N.; Nielsen, N.J.; Maolanon, N.; Sørensen, J.C.; Olsson, S.; Nielsen, J.; Giese, H. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol. Microbiol. 2006, 61, 1069–1080. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Sondergaard, T.E.; Covarelli, L.; Fuertes, P.R.; Giese, H. Identification of the biosynthetic gene clusters for the lipopeptides fusaristatin a and W493 B in Fusarium graminearum and F. pseudograminearum. J. Nat. Prod. 2014, 77, 2619–2625. [Google Scholar] [CrossRef]
- Song, Z.; Cox, R.J.; Lazarus, C.M.; Simpson, T.J. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. Chembiochem 2004, 5, 1196–1203. [Google Scholar] [CrossRef]
- Kim, D.W.; Shin, Y.K.; Lee, S.W.; Wimonmuang, K.; Kang, K.B.; Lee, Y.S.; Yun, S.H. FgPKS7 is an essential player in mating-type-mediated regulatory pathway required for completing sexual cycle in Fusarium graminearum. Environ. Microbiol. 2021, 23, 1972–1990. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.A.; López, V.E.L.y. Nonribosomal peptides synthetases and their applications in industry. Sustain. Chem. Process. 2016, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Oide, S.; Berthiller, F.; Wiesenberger, G.; Adam, G.; Turgeon, B.G. Individual and combined roles of malonichrome, ferricrocin, and TAFC siderophores in Fusarium graminearum pathogenic and sexual development. Front. Microbiol. 2015, 5, 759. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.T.; Sorensen, J.L.; Giese, H.; Sondergaard, T.E.; Frandsen, R.J.N. Quick guide to polyketide synthase and nonribosomal synthetase genes in Fusarium. Int. J. Food Microbiol. 2012, 155, 128–136. [Google Scholar] [CrossRef]
- Oide, S.; Moeder, W.; Krasnoff, S.; Gibson, D.; Haas, H.; Yoshioka, K.; Turgeon, B.G. NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 2006, 18, 2836–2853. [Google Scholar] [CrossRef] [Green Version]
- Valette-Collet, O.; Cimerman, A.; Reignault, P.; Levis, C.; Boccara, M. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant-Microbe Interact. 2003, 16, 360–367. [Google Scholar] [CrossRef] [Green Version]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schäfer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Kimura, M.; Kaneko, I.; Komiyama, M.; Takatsuki, A.; Koshino, H.; Yoneyama, K.; Yamaguchi, I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. J. Biol. Chem. 1998, 273, 1654–1661. [Google Scholar] [CrossRef] [Green Version]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Gen. Genet. 1999, 261, 977–984. [Google Scholar] [CrossRef]


| Sequence ID | Gene Name | Putative Function | log2 FC(T4/C4) | log2 FC(T16/C16) |
|---|---|---|---|---|
| Cell wall synthesis | ||||
| gene11337 | FPSE_10797 | 1,3-beta-glucan synthase | −0.98 | −1.36 * |
| gene2172 | FPSE_05113 | 1,3-beta-glucanosyltransferase ARB_07487 | −1.43 * | −1.02 * |
| gene3142 | FPSE_03879 | 1,3-beta-glucanosyltransferase gel1 | −1.38 * | −2.32 * |
| gene8431 | FPSE_02936 | 1,3-beta-glucanosyltransferase gel2 | 0.03 | −1.90 * |
| gene9847 | FPSE_01706 | chitin synthase 6 | 3.60 * | 0.62 |
| gene2063 | FPSE_09193 | chitin synthase 8 | 1.06 * | −0.24 |
| Cell membrane synthesis | ||||
| gene7320 | FPSE_12291 | ergosterol biosynthesis protein ERG3 | 1.26 * | 1.56 * |
| gene5907 | FPSE_01847 | ergosterol biosynthesis protein ERG5 | 2.85 * | 2.93 * |
| gene7662 | FPSE_07169 | fatty acid synthase subunit alpha FAS2 | −2.35 * | −3.55 * |
| gene7663 | FPSE_07168 | fatty acid synthase subunit beta FAS1 | −2.18 * | −4.06 * |
| gene10219 | FPSE_11763 | fatty acid elongase (ELOA) | −1.01 * | −2.14 * |
| gene10703 | FPSE_04157 | fatty acid elongase (ELO2) | −1.33 * | −1.07 * |
| gene8365 | FPSE_03004 | mechanosensitive ion channel protein Msy1 | −1.17 * | −1.25 * |
| Antioxidants | ||||
| gene4661 | FPSE_07706 | SOD1 | −0.27 | −1.15 * |
| gene10897 | FPSE_10302 | peroxiredoxin PRX1 | −2.36 * | −2.33 * |
| gene3471 | FPSE_12380 | peroxiredoxin DOT5 | −1.28 * | −2.21 * |
| gene4615 | FPSE_11702 | peroxiredoxin | −0.26 | −1.24 * |
| gene3597 | FPSE_07430 | thioredoxin | −0.71 | −4.21 * |
| gene6188 | FPSE_05329 | thioredoxin | −0.7 | −2.34 * |
| gene2370 | FPSE_00230 | catalase | 5.85 * | 4.56 * |
| gene6030 | FPSE_04574 | glutathione S-transferase | 2.12 * | 3.90 * |
| gene6184 | FPSE_05333 | glutathione S-transferase | 3.79 * | 3.99 * |
| gene11189 | FPSE_07916 | glutathione S-transferase-like protein FUS3 | 2.99 * | 5.34 * |
| gene12001 | FPSE_09673 | Glutathione S-transferase | 3.33 * | 2.36 * |
| gene6812 | FPSE_10852 | glutathione S-transferase-like protein OpS6 | 1.01 * | 1.16 * |
| gene780 | FPSE_01282 | glutathione S-transferase-like protein ustS | 1.02 * | 1.79 * |
| gene11533 | FPSE_04311 | ABC transporter F family member 4 | 6.37 * | 6.92 * |
| gene3848 | FPSE_05712 | ABC multidrug transporter MDR2 | 1.73 * | 1.91 * |
| gene10087 | FPSE_11895 | ABC multidrug transporter mdr1 | 2.95 * | 3.54 * |
| gene8542 | FPSE_00902 | ABC multidrug transporter B | 1.41 * | 1.93 * |
| Apoptosis | ||||
| gene6144 | FPSE_05373 | TatD_DNase | 9.93 * | 11.2 * |
| gene8111 | FPSE_08319 | TatD_DNase | 2.03 * | - |
| Secondary metabolites | ||||
| gene2473 | PKS2 | highly reducing polyketide synthase azaB | −1.57 * | 3.38 * |
| gene2057 | PKS6 | highly reducing polyketide synthase 40 | 4.28 * | 9.86 * |
| gene4735 | PKS7 | reducing polyketide synthase FUB1 | 1.06 * | 2.32 * |
| gene11187 | PKS10 | polyketide synthase dehydratase | −0.07 | 6.02 * |
| gene2491 | PKS12 | non-reducing polyketide synthase PKS12 | −1.75 * | −4.57 * |
| gene7713 | NPS2 | nonribosomal peptide synthetase 2 | 0.61 | −2.45 * |
| gene5972 | NPS6 | nonribosomal peptide synthetase 6 | −1.40 * | −4.50 * |
| Virulence | ||||
| gene9294 | FPSE_04467 | cellulase/esterase CelE | −0.24 | −3.48 * |
| gene8741 | FPSE_01167 | cellulase (glycosyl hydrolase family 5) | −0.27 | −1.67 * |
| gene4558 | FPSE_06033 | cellulase (glycosyl hydrolase family 5) | −2.36 * | −1.41 * |
| gene3142 | FPSE_03879 | cellulase (glycosyl hydrolase family 5) | −1.38 * | −2.32 * |
| gene10918 | FPSE_10579 | lipase 4 | 0.27 | −1.15 * |
| gene11906 | FPSE_05068 | lipase_GDSL_2 | −2.4 * | −1.89 * |
| gene1293 | FPSE_07610 | lipase_3 | 0.11 | −2.22 * |
| gene2706 | FPSE_08639 | lipase_3 | 0.28 | −1.51 * |
| gene4905 | FPSE_08884 | lipase_3 | 0.26 | −1.21 * |
| gene9418 | FPSE_08802 | lipase_GDSL_2 | −1.02 | −4.36 * |
| gene1287 | FPSE_01774 | alpha-amylase | −0.88 | −1.25 * |
| gene6132 | FPSE_06584 | alpha-amylase | −0.45 | −6.94 * |
| gene3826 | FPSE_05690 | alpha-amylase | −1.37 * | −3.29 * |
| gene5852 | FPSE_01902 | endo-beta-1,4-glucanase D | 0.54 | −5.95 * |
| gene2343 | FPSE_00619 | endoglucanase-4 | −0.38 | −2.26 * |
| gene2125 | FPSE_05752 | endo-1,4-beta-xylanase 4 | 0.13 | −3.14 * |
| gene3590 | FPSE_07423 | endo-1,4-beta-xylanase 1 | −1.19 * | −0.85 |
| gene9426 | FPSE_08810 | laccase-1 (Multicopper oxidase) | - | −4.87 * |
| gene11048 | FPSE_07047 | laccase | −1.64 * | −1.54 * |
| gene6701 | FPSE_09929 | pectinesterase | 1.33 * | 1.12 * |
| gene11282 | FPSE_11049 | trichothecene 3-O-acetyltransferase TRI101 | 6.44 * | 1.10 * |
| gene5181 | FPSE_02231 | trichothecene 3-O-acetyltransferase TRI101 | 2.12 * | 3.10 * |
| gene3830 | FPSE_05694 | fungal trichothecene efflux pump (TRI12) | 4.52 * | 3.79 * |
| gene6437 | FPSE_10392 | fungal trichothecene efflux pump (TRI12) | 5.25 * | 5.67 * |
| gene6518 | FPSE_08720 | fungal trichothecene efflux pump (TRI12) | 2.35 * | 5.90 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhu, W.; Goodwin, P.H.; Lin, Q.; Xia, M.; Xu, W.; Sun, R.; Liang, J.; Wu, C.; Li, H.; et al. Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis. J. Fungi 2022, 8, 763. https://doi.org/10.3390/jof8080763
Zhang J, Zhu W, Goodwin PH, Lin Q, Xia M, Xu W, Sun R, Liang J, Wu C, Li H, et al. Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis. Journal of Fungi. 2022; 8(8):763. https://doi.org/10.3390/jof8080763
Chicago/Turabian StyleZhang, Jie, Wenqian Zhu, Paul H. Goodwin, Qitong Lin, Mingcong Xia, Wen Xu, Runhong Sun, Juan Liang, Chao Wu, Honglian Li, and et al. 2022. "Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis" Journal of Fungi 8, no. 8: 763. https://doi.org/10.3390/jof8080763
APA StyleZhang, J., Zhu, W., Goodwin, P. H., Lin, Q., Xia, M., Xu, W., Sun, R., Liang, J., Wu, C., Li, H., Wang, Q., & Yang, L. (2022). Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis. Journal of Fungi, 8(8), 763. https://doi.org/10.3390/jof8080763






