Abstract
Emerging and unregulated contaminants end up in soils via stabilized/composted sewage sludges, paired with possible risks associated with the development of microbial resistance to antimicrobial agents or an imbalance in the microbial communities. An enrichment experiment was performed, fortifying the sewage sludge with carbamazepine, ketoprofen and diclofenac as model compounds, with the aim to obtain strains with the capability to transform these pollutants. Culturable microorganisms were obtained at the end of the experiment. Among fungi, Cladosporium cladosporioides, Alternaria alternata and Penicillium raistrickii showed remarkable degradation rates. Population shifts in bacterial and fungal communities were also studied during the selective pressure using Illumina MiSeq. These analyses showed a predominance of Ascomycota (Dothideomycetes and Aspergillaceae) and Actinobacteria and Proteobacteria, suggesting the possibility of selecting native microorganisms to carry out bioremediation processes using tailored techniques.
1. Introduction
Many different technologies have been developed to guarantee the efficient municipal and industrial wastewater recycling of an increasing population, improving the capacity and efficiency of wastewater treatment plants (WWTPs) [1,2]. Currently, the accuracy of technology in detecting chemicals at trace levels is crucial in identifying emerging contaminants (ECs) in sewage sludge. ECs have been detected in primary and secondary sewage sludge after sewage sludge treatment by anaerobic digestion, and even at higher concentrations due to concentration processes. Included in this variety of compounds are novel and old types of contaminants, human and veterinary drugs, nanomaterials, personal care products, paints and coatings, among several others [3,4]. Amid these chemical components, pharmaceutical active compounds (PhACs) represent one of the most important groups, since they can exert changes in microbial populations. They can also promote resistance to drugs in potential pathogen microorganisms, with enormous consequences to human health [5,6]. Hormones and non-steroidal anti-inflammatory drugs (NSAIDs) represent two of the main groups of contaminants with a proven potential risk for environmental organisms and humans, some of which have recently been included in the priority list of pollutants. NSAIDs, such as diclofenac and ketoprofen, and antiepileptic drugs, such as carbamazepine, have been frequently detected in wastewater, sewage sludge and soils [7]. In activated sludge, these pollutants endure the biodegradation process because of their nature, low concentration and retention status in the matrix of the sludge. These remain untreated and in continuous accumulation [8,9]. Deep concern for the possible harmful effects from long term exposure to these organic compounds has started to grow in society, as these molecules have been detected in WWTPs, air, soil and water of clean environments. The sewage sludge from WWTs is usually stabilized in anaerobic digestion to obtain energy, and, after that, the dried residue is further composted for its application as a soil amendment [10]. Even though these compounds may be completely or partially removed in WWTPs, the recalcitrant and bioaccumulative properties of some of these pollutants enable their entrance in the food chain [11]. Thus, the development of new strategies and technologies are needed to deal with the dispersion of these compounds into the environment via sewage sludge.
Physical and chemical treatments have been considered for their high efficiency in the removal of ECs from WWTPs effluents [12]. Nonetheless, biological treatments have been widely preferred and applied for the removal of ECs. This process involves biochemical transformations in which ECs are degraded by microorganisms such as algae, bacteria, and fungi into smaller molecules [13]. Microorganisms use organic compounds as primary substrates for their cell growth, inducing enzymes for their assimilation [14] and, finally, these molecules can be mineralized into carbon dioxide and water.
At present, information on the diversity of microorganisms and enzymes with the capacity to degrade ECs is quite limited. Therefore, future studies must be carried out to identify the microorganisms acting on one of the most dominant new pollutants, exploring in further depth to what extent autochthonous microorganisms can degrade ECs in the environment. As sewage sludge is one of the main sources of ECs and a rich environment of adapted microorganisms, this residue could constitute a source of ECs-degraders. Previous studies have been performed using sewage sludge to obtain microorganisms with biodegradation capabilities. For instance, selective pressure has been applied to the study of antibiotic resistance genes (ARGs) [15]. Furthermore, other studies have shown an increment in diversity under non-lethal selective pressure of the target pollutant (e.g., Bisphenol-A) in addition to carbon source. These processes can promote the growth of microorganisms and their adaptation to the concentration of contaminants and can contribute to the establishment of cooperative relationships that improve the degradation of the target compound [16,17]. The treatment of sewage sludge by composting is widely applied to revalorize this residue. Moreover, composting is an optimal ecosystem in which microorganisms with different metabolic capabilities can be found. Previous studies have shown effectiveness in degrading triclosan and antibiotics in sewage sludge composting by native microorganisms [18,19].
In addition to this, combining high-throughput sequencing technologies and isolation strategies has led to a greater understanding of the microbiome that inhabits polluted environments, underlying organismal rarity, metabolism profile and its relevance in their own ecology [20,21,22]. Even though there is a level of specialization between the decomposers from the microbial communities, information about specialist taxa that degrade specific compounds is limited. In addition, limited information is available about the dominance of specialist taxa that perform the degradation of compounds of a specific chemical composition.
In this study, the occurrence of ECs in undigested and digested sewage sludge was determined. Additionally, the genomic profile of native fungi and bacteria in both residues were studied during a selective pressure experiment with carbamazepine, ketoprofen and diclofenac. Finally, microorganisms were isolated to discover the degradation capabilities for further native degrader consortium construction to be used in further biotechnological processes.
2. Materials and Methods
2.1. Sample Collection
Sewage sludge was collected in the WWTPs “Los Vados” (coordinates in decimal degrees: 37.19121, −3.67639) Granada (Spain) in August 2018. Two types of sewage sludge were used: raw sewage sludge before anaerobic digestion (rSS) and sewage sludge after the anaerobic digestion process, digested sludge (dSS). Samples were disposed following biosafety protocols inside amber sterile glass bottles (~2 L) and transported to the lab in containers with ice. Half of each sample was taken to immediately begin the selective pressure experiments. The other half was stored at −20 °C for DNA extraction and for lyophilization to perform ECs analytical analysis. Bulking samples (B) were provided by the biosolid plant Biomasa del Guadalquivir S.A (Granada, Spain), composed of pruning residues. These are conventionally used in the composting system carried out in the same biosolid plant for the sewage sludge composting processes [23].
2.2. Selective Pressure Experiments
2.2.1. Setting the Flask Content for Selective Pressure Experiments
A bulking agent, which contributed to the composition of the microbiota [24], was included in addition to rSS and dSS. This was carried out to approximate the conditions of the composting process in the selective pressure experiments. For this, a quantity of 9 g of rSS and dSS samples were mixed with 1 g of B (previously grinded to obtain a homogeneous particle size) in sterilized Erlenmeyer flasks. They were filled with 60 mL of a modified Kirk and Bushnell Haas (BH) media [25,26] (Tables S1 and S2) to analyze the differences between fungal and bacterial growth. The selective media Kirk and BH were used for stimulating the actual communities of fungi and bacteria, respectively; they had also been selected in previous research projects focusing on the degradation and transformation of xenobiotics [27,28]. Carbamazepine (CBZ), ketoprofen (KET) and diclofenac (DCF), all purchased from Sigma Aldrich (St. Louis, MO, USA. ≥98% purity), were added to each flask to reach a final concentration of 50 μM for each compound. For each condition, three flasks were prepared to perform the analysis in triplicate, plus an additional control flask before incubation, i.e., time = 0 (DNA isolation, see Section 2.3) (Figure S1). The prepared flasks (rSS+B in Kirk, rSS+B in BH, dSS in Kirk and dSS in BH) were incubated for 7 days at 28 °C and 120 rpm to start the selective pressure experiment.
2.2.2. Setting the Pharmaceutical Pressure of the Flask Experiment
After 1 week of incubation, a weekly transfer was performed, transferring 10 mL of each flask into 50 mL of appropriate fresh media. To create the conditions for the fungal community, a booster of the three compounds was added weekly. At the beginning and at the end of each week-old flask, an aliquot of media was taken and immediately centrifuged twice at 14,000 rpm for 5 min. The supernatant was transferred into an HPLC glass vial and stored at -20 °C for further analysis to determine the PhACs concentration (Section 2.5). This transfer was repeated each week for a total of 9 weeks with the predecessor flask. During weeks 0, 5 and 8, a sample of 2 mL of the contents of the flask was frozen at −20 °C for total genomic DNA extraction.
2.3. Analysis of Fungal and Bacterial Communities by Illumina MiSeq Sequencing
2.3.1. Sample Preparation and DNA Isolation
To analyze the microbial communities based on DNA analysis, total genomic DNA extraction of the starting materials B, rSS and dSS, as well as from the selective pressure experiments (flasks after 0, 5 and 8 weeks of selective pressure), was performed. For that, a pre-treatment was performed for solid samples (rSS, dSS and B), adding a 0.9% saline solution in a ratio 1:3 (w/v), sonicated for 10 min and centrifuged twice 800 rpm for 10 min to remove the water phase. The bottom part was subsequently centrifuged at a pulse of 1 min/14,000 rpm to obtain a concentrated pellet. For liquid samples (flask content), the total volume was centrifuged at 5000 rpm for 10 min. The biomass was recovered with a 0.9% saline solution and sonicated for 10 min and continued as described before. A pellet of ~500 mg was obtained for each sample and DNA was extracted using the FastDNA™ Spin kit for Soil (Palex Medical, SA, Sant Cugat del Vallès, Barcelona, Spain) following the manufacturer’s instructions. DNA concentration and purity were determined by NanoDrop microspectrophotometry (ND-1000, Thermo Fisher Scientific, Waltham, MA, USA).
2.3.2. Sequencing Analysis
The Earth Microbiome Project pipeline (www.earthmicrobiome.org, 18 December 2018) was used for Illumina library preparation, sequencing, and core amplicon data analyses. The V4 region from the 16S rRNA using a 515F–806R primer pair was used for bacteria [29]. The ITS2 region using the primer pairs ITS1f-ITS2 was used for fungi [30]. PCR master mix and thermocycler conditions have been previously described [31]. The pooled sample was sequenced on the Illumina MiSeq platform using the Illumina MiSeq Reagent v3 600-cycle, at the Vincent J. Coates Genomics Sequencing Laboratory (UC Berkeley, Berkeley, CA, USA).
2.3.3. Data and Bioinformatic Analysis
QIIME2 version V2018.8 was used to analyze bacterial and fungal raw sequences. DADA2 was used for denoising and clustering (quality filtering) [32]. For fungal and bacteria analyses, UNITE (v7.2) and A 16S rRNA Greengenes database (http://greengenes.lbl.gov, 10 December 2018) were used. Thresholds of 96% and 100% were used for fungal and bacterial gene sequence identity, respectively. The obtained raw bacterial and fungal sequences associated with this study were deposited in the GenBank SRA database under BioProject accession number PRJNA780876. Statistical analysis was performed using the R Core Team (https://CRAN.R-project.org/package=here, 10 December 2018) with the vegan package.
2.4. Fungal and Bacterial Strains Isolation after Selective Pressure Experiment
2.4.1. Isolation and Molecular Identification of Fungal and Bacterial Strains
After 9 weeks of selective pressure, a total of 6 flasks (3 from Kirk media and 3 from BH media) were used for the isolation of fungal and bacterial strains according to the methodology described by Waksman [33]. For each flask, 1 mL of content was mixed with 9 mL of sterilized saline solution (0.9% NaCl) and serial diluted. The dilutions were plated on Petri dishes containing the solid version of the same media (by the addition of 15 g/L of agar). In Kirk media, streptomycin and tetracycline were used as antibiotics (50 and 25 μg/mL) to avoid bacterial growth. Inoculated Petri dishes were incubated at 28 °C for fungi and 30 °C for bacteria for 15 days. All distinct morphotypes were isolated and subcultured on Malt Extract Agar (MEA) (28 °C) and Tryptic Soy Agar (TSA) (30 °C) for fungal and bacterial strains, respectively. The pure cultures were stored at room temperature and glycerol was prepared for long-term storage.
Genomic DNA of fungal and bacterial isolated strains was extracted using the PrepMan®Ultra Kit following the manufacturer’s instructions. Then, a fresh colony was lysed in 100 mL of reactant and was heated at 100 °C for 10 min. Later, it was cooled down at room temperature for 2 min, centrifuged at 12,000 rpm for another 2 min, and finally 50 mL of the supernatant was transferred to a new tube to be stored at −20 °C. DNA obtained from isolates were amplified using pairs of primers. For fungi, ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [30] from the intragenic 5.8S region, and for bacteria, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 699R (5′-RGGTTGCGCTCGTT-3′ [34] to amplify 16S rRNA according to the conditions previously described [24]. Nucleotide sequences were submitted to the online BLAST search engine (http://www.ncbi.nlm.nih.gov/BLAST, accessed on 17 July 2020) of the National Centre for Biotechnology Information (NCBI).
2.4.2. Biodegradation Experiments of the Isolated Microorganisms
The isolated fungal and bacterial strains were individually tested to remove CBZ, KET and DCF. A fungal suspension for each strain was prepared and adjusted in a Neubauer chamber to inoculate 1 × 105 spores/mL in 150 mL amber glass bottles containing 50 mL of the medium [35]. For bacterial strains, a pre-inoculum of each isolate was prepared in Tryptone Soy Broth (TSB). When maximum OD (λ = 600 nm) was reached, 2 mL of the culture was transferred into sterile Eppendorf tubes. These were then centrifuged at 14,000 rpm and washed with a sterile saline solution (0.85%), which was recovered in sterilized distilled water. Finally, 1 mL of each bacterial suspension was inoculated in 150 mL amber glass bottles containing 50 mL of BH medium. Flasks were cultured under agitation (120 rpm) at 28 °C and 30 °C for fungi and bacteria, respectively, for 48 h. After that, 100 μΜ of CBZ, KET and DCF was added to three systems: (a) active strain (biodegradation test), (b) inactivated strain by autoclaving (absorption control) (c) non-inoculated flask (abiotic control). Each determination was performed in triplicate. To analyze degradation kinetics, sacrificial flasks from each system were taken every seven days. Samples were centrifuged twice at 14,000 rpm for ten minutes and the collected supernatants were stored at −20 °C for HPLC analysis.
2.5. Chromatographic Analyses
2.5.1. Characterization of Emerging Contaminants by LC/MS-QTOF in Sewage Sludge Samples
Lyophilized samples rSS and dSS (Section 2.1) were processed with a modified QuEChERS (Quick, Easy, Cheap, Effective, Rugged and Safe) extraction method [36]. One g of each sample was transferred into a 15 mL polypropylene tube. Following that, 2.2 mL of EDTA-McIlvaine buffer was added to the mixture that was shaken in a vortex for 1 min for homogenization. Subsequently, 5 mL of acetonitrile were added and then shaken for 1 min. After that, 1.25 g of (NH4)2SO4 was added to this mixture, which was vortexed for 1 min and centrifuged at 9000 rpm for 5 min. To continue, 5.5 mL of the supernatant were transferred into polypropylene tubes with 50 mg of C18, for the clean-up step, shaken again in vortexed for 1 min and centrifuged for 5 min at 5000 rpm. Finally, 5 mL of the supernatant were transferred into vials and dried with a gentle N2 steam at 40 °C. Final residue was re-dissolved with 0.5 mL of H2O-MeOH (95:5, v/v), vortexed for 2 min, filtered through 0.20 µm nylon syringe filters and injected into the SCIEX X500R QTOF system. Chromatographic separation was achieved on an Hibar® HR Purospher® STAR RP-C18 column (100 mm × 2.1 mm i.d., 2 µm particle size, Merck) using a mobile phase consisting of 0.05% aqueous formic acid solution (solvent A) and MeOH (solvent B) at a flow rate of 0.5 mL min−1. The gradient profile was as follows: 0 min, 5% B; 2.5 min, 5% B; 4.0 min, 100% B; 4.5 min, 100% B; 5.0 min, 5% B; 6.0 min, 5% B. The temperature of the column was 30 °C and the injection volume was 5 µL. A Sequential Window Acquisition of All Theoretical fragment ionic spectra (SWATH) was constructed as a second degree of data quantification at the Institute for Water Research Foundation of Catalonia (IDEA-ICRA). Samples were analyzed in triplicate.
2.5.2. HPLC Analyses of the Removal of PhACs during Pressure Experiment and Biodegradation Experiments
An Agilent® 1050 HPLC system (Waldbronn, Germany) was used to determine the efficiency in removing CBZ, KET and DCF in selective pressure experiments (Section 2.2.2) and in degradation experiments by microbial isolates (Section 2.4.2). This system was provided with a diode array detector (DAD; 190–700 nm) and a Synergi Fusion RP C18 column (4 μm, 4.6 Å~150 mm; Phenomenex®, Madrid, Spain). A volume of 10 μL of each sample was injected at a gradient flow rate of 0.9 mL/min, using a buffer of 85% of acetonitrile and 15% of H2O-H3PO4 (0.01%) for elution. Peak areas from determined absorbance at 278 nm were interpolated into a generated standard curve using 10–100 μM from each compound.
3. Results and Discussion
3.1. Occurrence of Emerging Pollutants in Sewage Sludge Samples
The percentages found in rSS and dSS by LC/MS QTOF (raw and digested, respectively) are shown in Figure 1. After targeting 80 substances related to PhACs and potential candidates listed as priority substances [37] in raw and digested sludge, it was noticed that 34 compounds were present in one or both samples ranging from 1.0 to 1100 ng/g d.w. (Table 1). As shown in Figure 1, the profile of both sludges was very similar. The similar profile regarding the concentration of these compounds may indicate the inefficiency of anaerobic digestion under our conditions in removing pharmaceutical compounds. Indeed, there were a few compounds where the concentration of the content increased 70% more after digestion than before digestion (Table 1). These are: amlodipine, 1,2,3-benzotriazole, dexamethasone, diclofenac, fenofibrate, fluoxetine, metoprolol paroxetine and valsartan, whereas the concentration of acridone, atenolol, diltiazem, estrone, loratadine, mephedrone, valsartan acid and venlafaxine were reduced by more than 80% in the dSS. The most abundant compounds were sertraline (rSS 1100.0 ng/g d.w., dSS 864.4 ng/g d.w.), mephedrone (rSS 313.0 ng/g d.w., dSS 18.1 ng/g d.w.) and 1,2,3-benzotriazole (rSS 448.8 ng/g d.w., dSS 773.9 ng/g d.w.). Only three of the analyzed compounds were not detected (benzafibrate, citalopram and trimethoprim). The determination of PhACs will depend on different factors, such as: sampling strategy, precipitation levels, dilution of wastewater discharges, degradation within the sewer system [38], frequent industrial/agriculture activities, social trends [39,40,41,42] and population health condition [43,44]. A review of ECs content in wastewaters and raw sludge [45] showed rather different results in concentration ranges of ECs in wastewater influents, reporting higher concentrations for diclofenac (1500 ng/g d.w.), ketoprofen (102 ng/g d.w.) and carbamazepine (2593 ng/g d.w.) and were almost absent in raw sludge; only diclofenac was detected at 70 ng/kg in this matrix. Nevertheless, it is important to note that the lengthy chemical process of sludge digestion (20–30 days) does not help in the degradation of ECs, as most of the reported values stood at a similar range. Thus, after the digestion and storage of sewage sludge, some ECs will still persist [46]. More recently, the increase in the retention time of the anaerobic bioreactor or the two-phase process seems to be an efficient strategy in removing several PhACs [47].
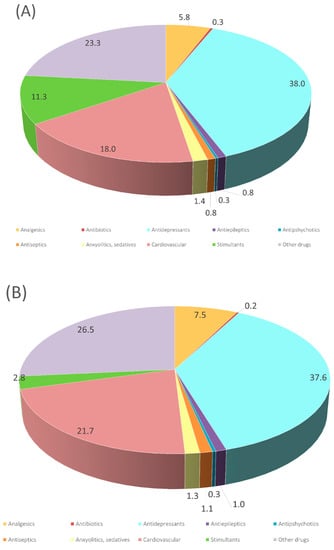
Figure 1.
Summary of the different families of chemicals identified in (A) raw sewage sludge and (B) digested sewage sludge, with the normalized number of compounds per category.

Table 1.
Pharmaceutical active compounds content expressed as ng/dw found in Sewage sludge (raw and digested sewage sludge) using LC/MS/QTOF.
3.2. Shift of Microbial Population during Selective Pressure Experiments
The analysis of the microbial communities was performed in the selective pressure experiment at weeks 0, 5 and 8 (Figure 2, Tables S3 and S4). The details of the sequence processing during the bioinformatics pipeline are shown in the SMT1 and SMT2. Regarding the fungal community, differences in the diversity between rSS and dSS were not found due to a high percentage of unidentified sequences. Representatives of the genera Rhodotorula and Penicillium were present in the dSS (more than 1%) and the genera Apiotrichum in the case of rSS (more than 1%). At the end of the experiment, Rhodotorula was the predominant genus in BH medium and dSS. Its abundance at the end of the experiment may indicate the potential of this genus to use pharmaceutical compounds as a carbon source. The percentage of Rhodotorula at the end of the experiment was higher than 60%. In the case of dSS, the abundance of Rhodotorula was higher compared to rSS. The rSS sample in the BH medium was more diverse than in the one cultivated in the Kirk medium. It contained genus such as Penicillium, Stemphylium, Cladosporium, Aspergillus, Alternaria, but also Rhodotorula (Figure 2A). The highest bacterial diversity was found in the starting material at the start of the experiment. After 5 weeks, the relative abundance showed a selection of microbial communities. Bacterial diversity in the dSS and rSS was very similar at the beginning of the experiment to that of the selective experiment, after the addition of the pharmaceutical compounds. The most abundant order was Clostridiales in rSS (more than 20% of abundance); Bacteroidales and Synergistales in dSS (more than 30% between both) (Figure 2B). Similar bacterial groups were detected in sludges after mesophilic anaerobic digestion, where the phylum Bacteroidetes, Chloroflexi, Firmicutes, and Proteobacteria were predominant [47].
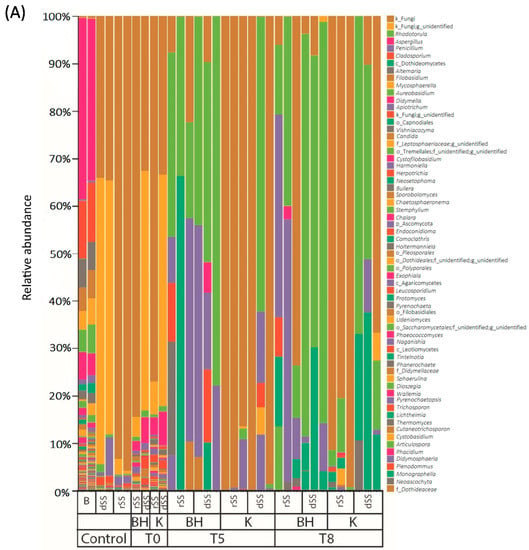
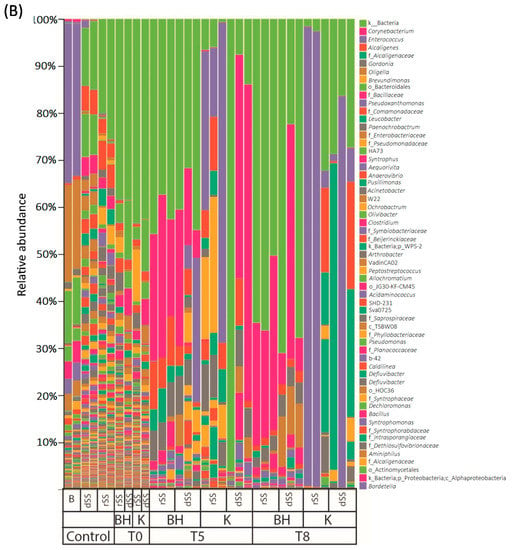
Figure 2.
Relative abundance (%) in different media over time in the selective pressure experiments: (A) Fungal diversity and (B) bacterial diversity. B: Bulking, dSS: Digested Sewage Sludge, rSS: Raw Sewage Sludge, BH: Bushnell Hass Medium, K: Kirk medium, T: Time in weeks.
BH medium favored the proliferation of Actinomycetales and Burkholderiales in rSS. The most abundant orders in Kirk medium were Lactobacillales, Caudobacteriales and Rhizobiales for the rSS. However, the bacterial structure in both media for dSS was similar. The diversity of the bacterial community decreased over time. After 8 weeks of experiments, less diversity was found in both sludges. The changes in the community were more affected by the medium used. For instance, in Kirk medium the genera found at the end of the experiment were Enterococcus, Alcaligenes or members of the Alcaligenaceae family. A different trend was observed in BH medium where no glucose was provided and Corynebacterium and Oligella were the most representative genera after 8 weeks of the experiment. This suggests that these genera could be more tolerant to the provided concentration of PhACs and, in the case of BH medium, be able to grow without any glucose addition, apart from the carbon source provided by the starting material. Therefore, both communities were different from the beginning and were selective according to the medium used.
3.3. Effect of the Sewage Sludge and the Medium for the Selection of Microorganisms
Figure 3 shows the effect of the selected medium in the microbial communities. The MDS for fungal (Figure 3A) and bacterial (Figure 3B) communities did not show changes according to the medium at the starting condition used. Therefore, the use of BH or Kirk media did not affect the microbial communities that are grouped in the same clusters in Figure 3A. Time was the main factor that influenced the communities. The ones most affected were those that were contained in the starting material. However, in the case of bacterial communities, the use of different media shows different clusters (Figure 3B). This was also influenced by time. The bacterial community in Kirk medium at 5 weeks was similar to the BH media at 8 weeks. This may indicate the absence of a carbon source (such as in BH medium), which could stimulate the bacterial community with degradation abilities faster than in the presence of an extra carbon source (Kirk medium).
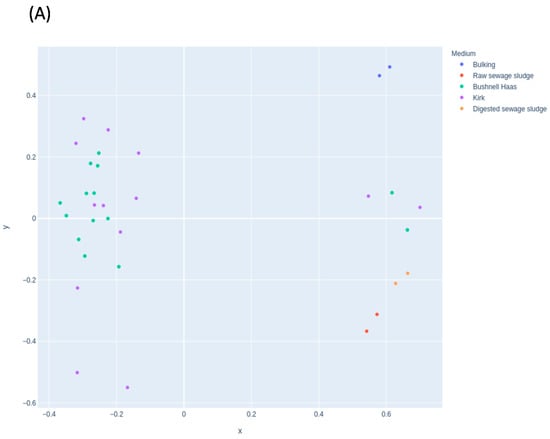
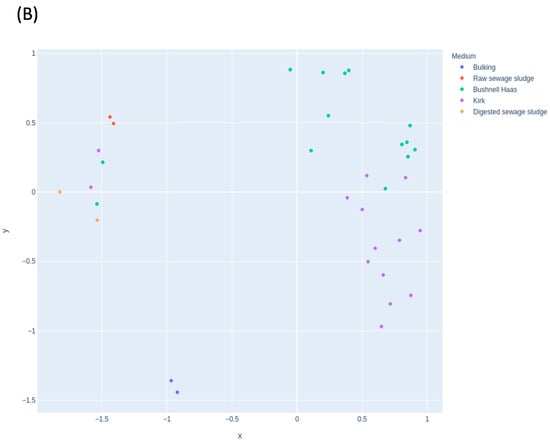
Figure 3.
Multidimensional scaling (MDS) plot showing the distribution of samples in the enrichment experiment according to the medium used for (A) fungal and (B) bacterial communities.
3.4. Pharmaceutical Active Compounds (PhACs) Removal during Selective Pressure Experiment
The removal of PhACs by the autochthonous microbiota in the selective pressure experiment is shown in Figure 4. The figures display the concentration of pharmaceutical compounds CBZ, KET and DCF detected during the eighth week of selective pressure. These results show a concentration range of 50–110 μM for each compound, indicating no accumulative effect over time, as well as the presence of microorganisms’ abilities to remove the added compounds. The concentration of PhACs increased after 5 weeks, as was expected, since every week the same concentration of PhACs was added. This suggests that microbial activity partially degraded these three compounds, since these were not accumulated after 8 weeks. In contrast, the concentration of PhACs in control treatments increased over time due to the absence of microbial activity. It also indicates a positive effect in the degradation by native microorganisms in both sludges (rSS and dSS). In a similar way, Baratpour and Moussavi [48] used a 90-day selective pressure experiment of a bacterial biofilm (a mixture of Pseudomonas spp. and Bacillus spp.) with 100 mg/L of acetaminophen (ACT) for biomass acclimation and 6 mM of H2O2 as a stimulation for accelerating the removal efficiency, completely removing ACT on a fixed bed reactor every 10 days. Another approach of a three-step enrichment procedure was made by Iranzo et al. [18], in which a compost made of WWTP sludge and rice straw was suspended in Yeast Extract Peptone Dextrose (YPD) and a complete minimal medium with and without a carbon and nitrogen source to aim for the best outcome of biodegrading. PhACs such as azithromycin, benzylpenicillin, citalopram, fluconazole, fluoxetine, ibuprofen, irbesartan, olanzapine, telmisartan, and venlafaxine were best degraded at a C/N ratio of 20. Azithromycin levels were reduced by up to 50%, citalopram was reduced by 10%, and fluoxetine was completely biodegraded over 15 days of treatment.
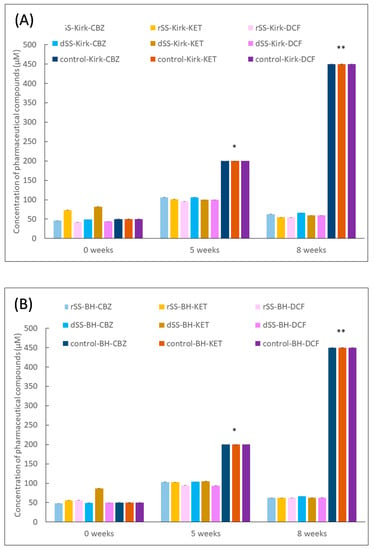
Figure 4.
Overall concentration of PhACs (μM) after eight weeks of enrichment experiment with raw (rSS) and digested sewage sludge (dSS) in (A) modified Kirk medium and (B) BH medium. Mean values of triplicate measurements were calculated, 1 bar = standard deviation (n = 3). Number of asterisks (*, **) indicated significant differences p < 0.05 calculated by three-way ANOVA (Sigmaplot v.12.0).
3.5. Isolation of Microorganisms after Selective Pressure
After 63 days of selective pressure with CBZ, KET and DCF, the isolation of fungal and bacterial strains was performed. At the end of the experiment, a total of 7 fungi and 11 bacteria were isolated and identified through the amplification of the ITS and 16S rRNA genes. Table 2 shows fungi and bacteria isolates. The obtained sequences of fungal and bacterial isolates associated with this study were deposited in the GenBank database under the accession number (Table 2). Fungal species found were: Cladosporium cladosporioides, Cladosporium limoniforme, Cladosporium halotolerans, Alternaria alternata, Aspergillus montevidensis, Penicillium raistrickii and Purpureocillium lilacinum. All of them belong to the Ascomycota phylum, the biggest taxonomic group of the fungal kingdom. This includes industrial, medical and biological model species and also the highest percentage found by non-culture techniques. These isolated fungal strains found at the end of the experiment were the most representative found by non-culture techniques.

Table 2.
Identified isolated microorganisms according to the 99% of database similarity, Gene bank ascension number.
Bacterial species identified were Bacillus simplex, Corynebacterium efficiens, Corynebacterium humireducens, Gordonia hirsute, Alcaligenes faecalis, Micrococcus yunnanensis, Enterococcus faecium, Paenalcaligenes hominis, Oligella ureolytica, Sphingobacterium jejuense and Staphylococcus hominis. These showed a mixture of different phyla such as Actinobacteria and Proteobacteria (Table 2). These co-existing bacteria had rarely been reported as xenobiotic degraders, whereas other bacteria belonging to the same phylum had remarkable results with a contingency of PhACs. One example of this capability was described by Thelusmond et al. [49], showing a partial degradation of CBZ, triclocarban and triclosan in soil after 80 days of biostimulation with native microorganisms. Illumina amplicon sequencing results showed that Rhodococcus sp., Streptomyces sp. (both Actinobacteria), Pseudomonas sp., Sphingomonas sp. and Methylobacillus sp. (the last three are Proteobacteria) were the most abundant during the process. The adaptation and tolerance of these isolates to a stable concentration of PhACs open the possibility of exploring metabolic profiles in order to perform enzyme extraction, as has been previously suggested [50,51].
3.6. Biodegradation Experiments Using the Isolated Strains
To evaluate the capacity of the isolated strains to degrade the studied compounds, and in order to select potential candidates for consortia degradation, a biodegradation experiment was performed with the isolates. The potential of degradation by the total number of isolates (18) were evaluated individually. Only 3 out of 18 fungal strains showed positive results. Cladosporium cladosporioides, Alternaria alternata and Penicillium raistrickii showed partial and total degradation activity after 21 days of incubation (Figure 5). CBZ had a minor reduction (25.2%) by C. cladosporioides, KET was mostly consumed (90%) by the same fungal isolate (Figure 5A) and partially degraded (37.8%) by Alternaria alternata; DCF was almost totally consumed by Alternaria alternata (83.1%) (Figure 5B) and Penicillium raistrickii (99.6%) (Figure 5C). The use of fungi previously isolated from polluted environments has been studied for the removal of PhACs, such as those performed by Conejo-Saucedo et al. [35], in which Talaromyces gossypii, T. verruculosus, Aspergillus terreus, A. cejpii and Syncephalastrum monosporum were able to degrade between 14.6 and 84.6% of DCF in 3 days. The main mechanisms in degrading these compounds are the intra and extracellular enzymes and/or sorption processes on fungal biomass. For instance, Trametes versicolor and Gymnopilus luteofolius have shown a good degradation of carbamazepine (90 and 95%, respectively), which is considered a very recalcitrant compound [52]. Bjerkandera adusta has shown a removal efficiency of 100% for diclofenac using versatile peroxidase [53]. The effectiveness of removal found by the strain P. raistrickii (99.6%) for diclofenac and by C. cladosporioides for ketoprofen (90%) is higher than the degradation of T. versicolor for the same compounds (95 and 48%, respectively) [54]. To our knowledge, enrichment experiments with PhACs have not been carried out using fungi. They allow us to find species that are not often studied in the degradation of PhACs which have a high potential of degradation. In some cases, these may be used as good candidates for bioreactor strategies amongst others. For instance, Phanerochaete chrysosporium isolated from polluted sites removed between 50 and 60% of diclofenac and other NSAIDS in a bioreactor after 100 days (5 mg L−1) [55]. Other studies with fungi have shown better degradation in immobilized systems than in suspensions systems [56]. This is similar to the degradation percentage caused by T. versicolor, other systems such as bio-slurry, and solid phase systems using exogenous fungi. In this case, the application of this white-rot fungus, which assisted the biodegradation experiment of CBZ and naproxen, achieved partial degradation (57 and 47%, respectively) after 24 h [57].
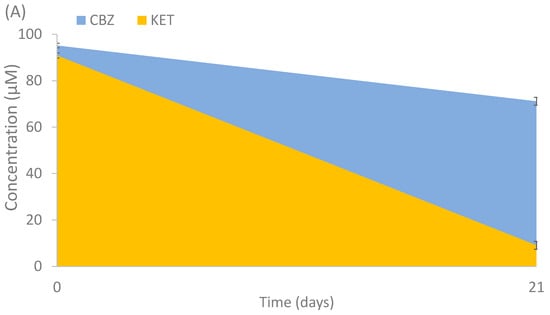
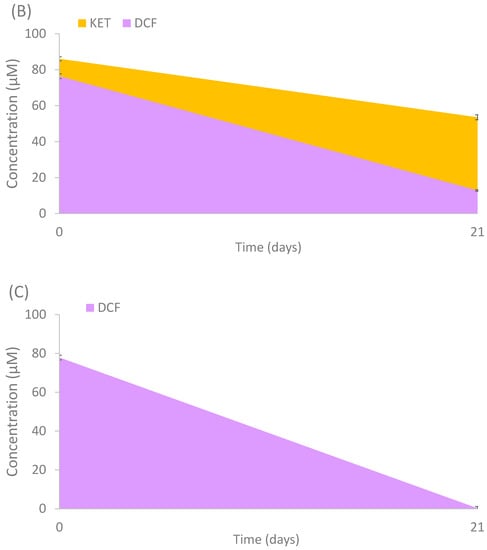
Figure 5.
Degradation of selected PhACs by (A) Cladosporium cladosporioides, (B) Alternaria alternata and (C) Penicillium raistrickii. Mean values of triplicate measurements were calculated, and error bars represent standard deviation (n = 3).
4. Conclusions
Sewage sludge represents a significant source of PhACs and adapted microbial populations. Selective pressure systems allow us to obtain and elucidate the most tolerant microbial communities and identify those microorganisms with potential abilities to degrade emerging pollutants. Despite further studies being needed, pressure selective systems may be considered a first step to implant bioaugmentation strategies in sewage sludge composting and deal with the problem of emerging pollutants. Further experiments should focus on studying the degradation potential of emerging pollutants by culturable consortia in order to use them as an integral biodegradation strategy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8070668/s1, Figure S1: Workflow of the selective pressure experiment set-up; Table S1: Composition of modified Kirk medium used in the selective pressure experiment Table S2: Composition of BH medium used in the selective pressure experiment. Table S3: Summary metric of bacterial community analyses in the bioinformatic pipeline. Table S4: Summary metric of bacterial community analyses in the bioinformatic pipeline.
Author Contributions
A.L.-V.: Investigation, Methodology, Writing—Original draft. T.R.-M.: Investigation, Methodology, Writing—Original draft. C.G.-S.: Formal analyses, Writing- Reviewing and Editing. G.A.-D.P.: Formal analysis, Writing—Reviewing and Editing. C.P.: Methodology, Writing—Reviewing and Editing. M.M.: Conceptualization, Funding acquisition, C.C.: Supervision, Writing—Reviewing and Editing, E.A.: Conceptualization, Funding acquisition, Supervision, Writing—Reviewing and Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness and European Research Founds [grant number CTM2017-84332-R] and the Junta de Andalucía [grant number B-RNM-204-UGR20]. The CONACyT Mexico for Alejandro Ledezma [grant number 377965] and Gabriela Ángeles [grant number 739637]. Additionally, the contract of Tatiana Robledo granted by Spanish Ministry of Universities (María Zambrano Program) funded by Next Generation EU (NGEU).
Acknowledgments
We gratefully thank Nicola Montemuro from the Institute of Environmental Assessment and Water Research (IDAEA-CSIC) and the group RNM-270. Additionally, we thank Shi Wang for her assistance during fungal sequencing. Graphical abstract has been made in BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bianchi, E.; Biancalani, A.; Berardi, C.; Antal, A.; Fibbi, D.; Coppi, A.; Lastrucci, L.; Bussotti, N.; Colzi, I.; Renai, L.; et al. Improving the Efficiency of Wastewater Treatment Plants: Bio-Removal of Heavy-Metals and Pharmaceuticals by Azolla filiculoides and Lemna minuta. Sci. Total Environ. 2020, 746, 141219. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Qiang, Z.; Ben, W.; Zhu, B.; Liu, J. Rapid Detection of Multiple Class Pharmaceuticals in Both Municipal Wastewater and Sludge with Ultra High Performance Liquid Chromatography Tandem Mass Spectrometry. J. Environ. Sci. 2014, 26, 1949–1959. [Google Scholar] [CrossRef] [PubMed]
- Castro, G.; Ramil, M.; Cela, R.; Rodríguez, I. Identification and Determination of Emerging Pollutants in Sewage Sludge Driven by UPLC-QTOF-MS Data Mining. Sci. Total Environ. 2021, 778, 146256. [Google Scholar] [CrossRef] [PubMed]
- Comtois-Marotte, S.; Chappuis, T.; Vo Duy, S.; Gilbert, N.; Lajeunesse, A.; Taktek, S.; Desrosiers, M.; Veilleux, É.; Sauvé, S. Analysis of Emerging Contaminants in Water and Solid Samples Using High Resolution Mass Spectrometry with a Q Exactive Orbital Ion Trap and Estrogenic Activity with YES-Assay. Chemosphere 2017, 166, 400–411. [Google Scholar] [CrossRef]
- Kümmerer, K. The Presence of Pharmaceuticals in the Environment Due to Human Use—Present Knowledge and Future Challenges. J. Environ. Manag. 2009, 90, 2354–2366. [Google Scholar] [CrossRef]
- Michalska, M.; Zorena, K.; Marks, R.; Wąż, P. The Emergency Discharge of Sewage to the Bay of Gdańsk as a Source of Bacterial Enrichment in Coastal Air. Sci. Rep. 2021, 11, 20959. [Google Scholar] [CrossRef]
- Ericson, H.; Thorsén, G.; Kumblad, L. Physiological Effects of Diclofenac, Ibuprofen and Propranolol on Baltic Sea Blue Mussels. Aquat. Toxicol. Amst. Neth. 2010, 99, 223–231. [Google Scholar] [CrossRef]
- Radjenović, J.; Petrović, M.; Barceló, D. Fate and Distribution of Pharmaceuticals in Wastewater and Sewage Sludge of the Conventional Activated Sludge (CAS) and Advanced Membrane Bioreactor (MBR) Treatment. Water Res. 2009, 43, 831–841. [Google Scholar] [CrossRef]
- Ramos, R.L.; Lebron, Y.A.R.; Moreira, V.R.; de Souza Santos, L.V.; Amaral, M.C.S. Phenolic Compounds in Surface Water: Methodology and Occurrence in Doce River, Brazil. Environ. Monit. Assess. 2021, 193, 687. [Google Scholar] [CrossRef]
- Tomasi Morgano, M.; Leibold, H.; Richter, F.; Stapf, D.; Seifert, H. Screw Pyrolysis Technology for Sewage Sludge Treatment. Waste Manag. 2018, 73, 487–495. [Google Scholar] [CrossRef]
- Rocha, A.C.; Camacho, C.; Eljarrat, E.; Peris, A.; Aminot, Y.; Readman, J.W.; Boti, V.; Nannou, C.; Marques, A.; Nunes, M.L.; et al. Bioaccumulation of Persistent and Emerging Pollutants in Wild Sea Urchin Paracentrotus lividus. Environ. Res. 2018, 161, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Khan, S.U.; Ahmed, S.; Farooqi, I.H.; Yousefi, M.; Mohammadi, A.A.; Changani, F. Recent Trends in Disposal and Treatment Technologies of Emerging-Pollutants—A Critical Review. TrAC Trends Anal. Chem. 2020, 122, 115744. [Google Scholar] [CrossRef]
- Garcia-Rodríguez, A.; Sagristà, E.; Matamoros, V.; Fontàs, C.; Hidalgo, M.; Salvadó, V. Determination of Pharmaceutical Compounds in Sewage Sludge Using a Standard Addition Method Approach. Int. J. Environ. Anal. Chem. 2014, 94, 1199–1209. [Google Scholar] [CrossRef]
- Tran, N.H.; Urase, T.; Ngo, H.H.; Hu, J.; Ong, S.L. Insight into Metabolic and Cometabolic Activities of Autotrophic and Heterotrophic Microorganisms in the Biodegradation of Emerging Trace Organic Contaminants. Bioresour. Technol. 2013, 146, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Feng, J.; Liu, J.; Fu, W.; Li, X.; Li, B. Deciphering of Microbial Community and Antibiotic Resistance Genes in Activated Sludge Reactors under High Selective Pressure of Different Antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, S.; Yang, X.-L.; Yang, Y.-L.; Xu, H.; Li, X.-N.; Song, H.-L. Enhanced Degradation of Bisphenol A and Ibuprofen by an Up-Flow Microbial Fuel Cell-Coupled Constructed Wetland and Analysis of Bacterial Community Structure. Chemosphere 2019, 217, 599–608. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, Z.; Wang, X.; Li, J.; Shen, J.; Xu, H. Remediation of Pharmaceuticals and Personal Care Products Using an Aerobic Granular Sludge Sequencing Bioreactor and Microbial Community Profiling Using Solexa Sequencing Technology Analysis. Bioresour. Technol. 2015, 179, 104–112. [Google Scholar] [CrossRef]
- Iranzo, M.; Gamón, M.; Boluda, R.; Mormeneo, S. Analysis of Pharmaceutical Biodegradation of WWTP Sludge Using Composting and Identification of Certain Microorganisms Involved in the Process. Sci. Total Environ. 2018, 640–641, 840–848. [Google Scholar] [CrossRef]
- Zheng, G.; Yu, B.; Wang, Y.; Ma, C.; Chen, T. Removal of Triclosan during Wastewater Treatment Process and Sewage Sludge Composting—A Case Study in the Middle Reaches of the Yellow River. Environ. Int. 2020, 134, 105300. [Google Scholar] [CrossRef]
- Jia, X.; Dini-Andreote, F.; Falcão Salles, J. Community Assembly Processes of the Microbial Rare Biosphere. Trends Microbiol. 2018, 26, 738–747. [Google Scholar] [CrossRef]
- Pascoal, F.; Magalhães, C.; Costa, R. The Link Between the Ecology of the Prokaryotic Rare Biosphere and Its Biotechnological Potential. Front. Microbiol. 2020, 11, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidhu, C.; Vikram, S.; Pinnaka, A.K. Unraveling the Microbial Interactions and Metabolic Potentials in Pre- and Post-Treated Sludge from a Wastewater Treatment Plant Using Metagenomic Studies. Front. Microbiol. 2017, 8, 1382. [Google Scholar] [CrossRef] [PubMed]
- Robledo-Mahón, T.; Aranda, E.; Pesciaroli, C.; Rodríguez-Calvo, A.; Silva-Castro, G.A.; González-López, J.; Calvo, C. Effect of Semi-Permeable Cover System on the Bacterial Diversity during Sewage Sludge Composting. J. Environ. Manag. 2018, 215, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Robledo-Mahón; Gómez-Silván, C.; Andersen, G.L.; Calvo, C.; Aranda, E. Assessment of Bacterial and Fungal Communities in a Full-Scale Thermophilic Sewage Sludge Composting Pile under a Semipermeable Cover. Bioresour. Technol. 2020, 298, 122550. [Google Scholar] [CrossRef]
- Kirk, T.K.; Schultz, E.; Connors, W.J.; Lorenz, L.F.; Zeikus, J.G. Influence of Culture Parameters on Lignin Metabolism ByPhanerochaete chrysosporium. Arch. Microbiol. 1978, 117, 277–285. [Google Scholar] [CrossRef]
- Bushnell, L.; Haas, H. The Utilization of Certain Hydrocarbons by Microorganisms. J. Bacteriol. 1941, 41, 653–673. [Google Scholar] [CrossRef] [Green Version]
- Abarian, M.; Hassanshahian, M.; Esbah, A. Degradation of Phenol at High Concentrations Using Immobilization of Pseudomonas putida P53 into Sawdust Entrapped in Sodium-Alginate Beads. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2019, 79, 1387–1396. [Google Scholar] [CrossRef]
- Qin, X.; Luo, H.; Zhang, X.; Yao, B.; Ma, F.; Su, X. Dye-Decolorizing Peroxidases in Irpex lacteus Combining the Catalytic Properties of Heme Peroxidases and Laccase Play Important Roles in Ligninolytic System. Biotechnol. Biofuels 2018, 11, 302. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Olicón-Hernández, D.R.; Gómez-Silván, C.; Pozo, C.; Andersen, G.L.; González-Lopez, J.; Aranda, E. Penicillium oxalicum XD-3.1 Removes Pharmaceutical Compounds from Hospital Wastewater and Outcompetes Native Bacterial and Fungal Communities in Fluidised Batch Bioreactors. Int. Biodeterior. Biodegrad. 2021, 158, 105179. [Google Scholar] [CrossRef]
- Waksman, S.A. A Method for Counting the Number of Fungi in the Soil. J. Bacteriol. 1922, 7, 339–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, K.S.; Kim, J.-M.; Kim, J.-W.; Jung, B.Y.; Kim, W.; Kim, I.J.; Kook, Y.-H. Identification of Bacillus anthracis by RpoB Sequence Analysis and Multiplex PCR. J. Clin. Microbiol. 2003, 41, 2908–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conejo-Saucedo, U.; Ledezma-Villanueva, A.; Ángeles de Paz, G.; Herrero-Cervera, M.; Calvo, C.; Aranda, E. Evaluation of the Potential of Sewage Sludge Mycobiome to Degrade High Diclofenac and Bisphenol-A Concentrations. Toxics 2021, 9, 115. [Google Scholar] [CrossRef]
- Anastassiades, M.; Lehotay, S.J.; Stajnbaher, D.; Schenck, F.J. Fast and Easy Multiresidue Method Employing Acetonitrile Extraction/Partitioning and “Dispersive Solid-Phase Extraction” for the Determination of Pesticide Residues in Produce. J. AOAC Int. 2003, 86, 412–431. [Google Scholar] [CrossRef] [Green Version]
- Official Journal of the European Union. Directive 2013/39/EU Directive 2013/39/EU of the European Parliament and of the Council of 12 August 2013 Amending Directives 2000/60/EC and 2008/105/EC as Regards Priority Substances in the Field of Water Policy; Official Journal of the European Union: Brussels, Belgium, 2013. [Google Scholar]
- Ort, C.; Lawrence, M.G.; Rieckermann, J.; Joss, A. Sampling for Pharmaceuticals and Personal Care Products (PPCPs) and Illicit Drugs in Wastewater Systems: Are Your Conclusions Valid? A Critical Review. Environ. Sci. Technol. 2010, 44, 6024–6035. [Google Scholar] [CrossRef]
- Ort, C.; Lawrence, M.G.; Reungoat, J.; Mueller, J.F. Sampling for PPCPs in Wastewater Systems: Comparison of Different Sampling Modes and Optimization Strategies. Environ. Sci. Technol. 2010, 44, 6289–6296. [Google Scholar] [CrossRef]
- Dennhardt, A.A.; Murphy, J.G. Prevention and Treatment of College Student Drug Use: A Review of the Literature. Addict. Behav. 2013, 38, 2607–2618. [Google Scholar] [CrossRef]
- Gerrity, D.; Trenholm, R.A.; Snyder, S.A. Temporal Variability of Pharmaceuticals and Illicit Drugs in Wastewater and the Effects of a Major Sporting Event. Water Res. 2011, 45, 5399–5411. [Google Scholar] [CrossRef]
- Lai, F.Y.; Bruno, R.; Hall, W.; Gartner, C.; Ort, C.; Kirkbride, P.; Prichard, J.; Thai, P.K.; Carter, S.; Mueller, J.F. Profiles of Illicit Drug Use during Annual Key Holiday and Control Periods in Australia: Wastewater Analysis in an Urban, a Semi-Rural and a Vacation Area. Addict. Abingdon Engl. 2013, 108, 556–565. [Google Scholar] [CrossRef] [Green Version]
- Lai, F.Y.; Thai, P.K.; O’Brien, J.; Gartner, C.; Bruno, R.; Kele, B.; Ort, C.; Prichard, J.; Kirkbride, P.; Hall, W.; et al. Using Quantitative Wastewater Analysis to Measure Daily Usage of Conventional and Emerging Illicit Drugs at an Annual Music Festival. Drug Alcohol Rev. 2013, 32, 594–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasprzyk-Hordern, B.; Baker, D.R. Enantiomeric Profiling of Chiral Drugs in Wastewater and Receiving Waters. Environ. Sci. Technol. 2012, 46, 1681–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasprzyk-Hordern, B.; Baker, D.R. Estimation of Community-Wide Drugs Use via Stereoselective Profiling of Sewage. Sci. Total Environ. 2012, 423, 142–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, B.; Barden, R.; Kasprzyk-Hordern, B. A Review on Emerging Contaminants in Wastewaters and the Environment: Current Knowledge, Understudied Areas and Recommendations for Future Monitoring. Water Res. 2015, 72, 3–27. [Google Scholar] [CrossRef]
- Cortés, J.M.; Larsson, E.; Jönsson, J.Å. Study of the Uptake of Non-Steroid Anti-Inflammatory Drugs in Wheat and Soybean after Application of Sewage Sludge as a Fertilizer. Sci. Total Environ. 2013, 449, 385–389. [Google Scholar] [CrossRef]
- Gallardo-Altamirano, M.J.; Maza-Márquez, P.; Montemurro, N.; Pérez, S.; Rodelas, B.; Osorio, F.; Pozo, C. Insights into the Removal of Pharmaceutically Active Compounds from Sewage Sludge by Two-Stage Mesophilic Anaerobic Digestion. Sci. Total Environ. 2021, 789, 147869. [Google Scholar] [CrossRef]
- Baratpour, P.; Moussavi, G. The Accelerated Biodegradation and Mineralization of Acetaminophen in the H2O2-Stimulated Upflow Fixed-Bed Bioreactor (UFBR). Chemosphere 2018, 210, 1115–1123. [Google Scholar] [CrossRef]
- Thelusmond, J.-R.; Strathmann, T.J.; Cupples, A.M. Carbamazepine, Triclocarban and Triclosan Biodegradation and the Phylotypes and Functional Genes Associated with Xenobiotic Degradation in Four Agricultural Soils. Sci. Total Environ. 2019, 657, 1138–1149. [Google Scholar] [CrossRef]
- Karn, S.K.; Kumar, A. Sludge: Next Paradigm for Enzyme Extraction and Energy Generation. Prep. Biochem. Biotechnol. 2019, 49, 105–116. [Google Scholar] [CrossRef]
- Robledo-Mahón, T.; Calvo, C.; Aranda, E. Enzymatic Potential of Bacteria and Fungi Isolates from the Sewage Sludge Composting Process. Appl. Sci. 2020, 10, 7763. [Google Scholar] [CrossRef]
- Naghdi, M.; Taheran, M.; Brar, S.K.; Kermanshahi-pour, A.; Verma, M.; Surampalli, R.Y. Removal of Pharmaceutical Compounds in Water and Wastewater Using Fungal Oxidoreductase Enzymes. Environ. Pollut. 2018, 234, 190–213. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Lam, S.S.; Iqbal, H.M.N. Biocatalytic Remediation of Pharmaceutically Active Micropollutants for Environmental Sustainability. Environ. Pollut. 2022, 293, 118582. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.V.; Cabana, H. Towards High Potential Magnetic Biocatalysts for On-Demand Elimination of Pharmaceuticals. Bioresour. Technol. 2016, 200, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, M.; El-sheekh, M.; Ma, Y.; Pugazhendhi, A.; Natarajan, D.; Kandasamy, G.; Raja, R.; Saravana Kumar, R.M.; Kumarasamy, S.; Sathiyan, G.; et al. Current Status of Microbes Involved in the Degradation of Pharmaceutical and Personal Care Products (PPCPs) Pollutants in the Aquatic Ecosystem. Environ. Pollut. 2022, 300, 118922. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; de Toledo, R.A.; Wang, S.; Shim, H. Removal of Carbamazepine and Naproxen by Immobilized Phanerochaete chrysosporium under Non-Sterile Condition. New Biotechnol. 2015, 32, 282–289. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, C.E.; Marco-Urrea, E.; Caminal, G. Degradation of Naproxen and Carbamazepine in Spiked Sludge by Slurry and Solid-Phase Trametes versicolor Systems. Bioresour. Technol. 2010, 101, 2259–2266. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).