UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Strains and Growth Condition
2.3. Conidial Germination Assay
2.4. Vectors Construction and Transformation
2.5. Inoculation Assay
2.6. Subcellular Localization Analysis
2.7. Immunoblot Assay
2.8. Chromatin Immunoprecipitation (ChIP) and Sequencing
2.9. qRT-PCR (Quantitative Real-Time PCR) and RNA Sequencing
2.10. Stresses Treatments
2.11. Data Availability
3. Results
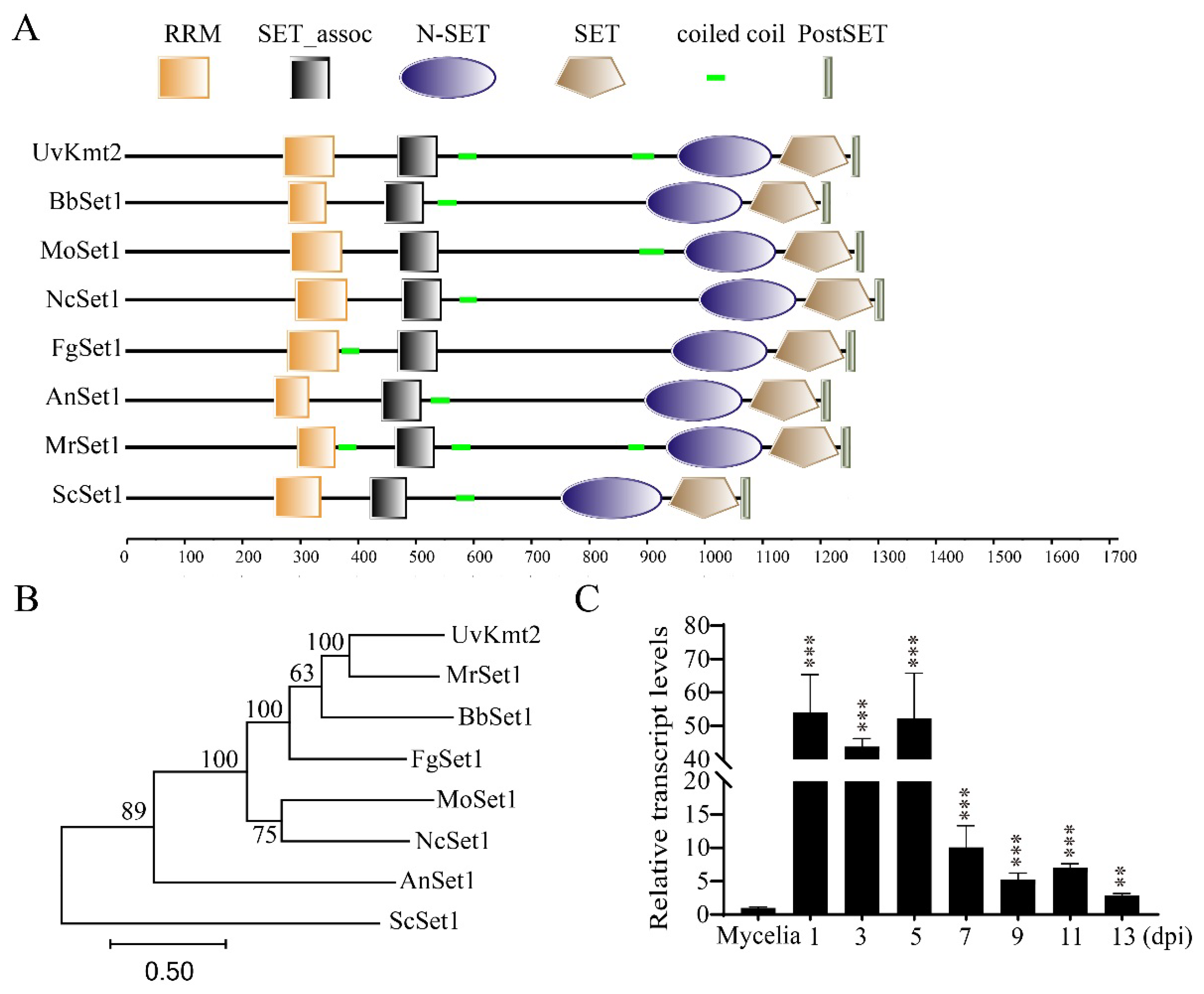
3.1. Identification of UvKMT2 in U. virens
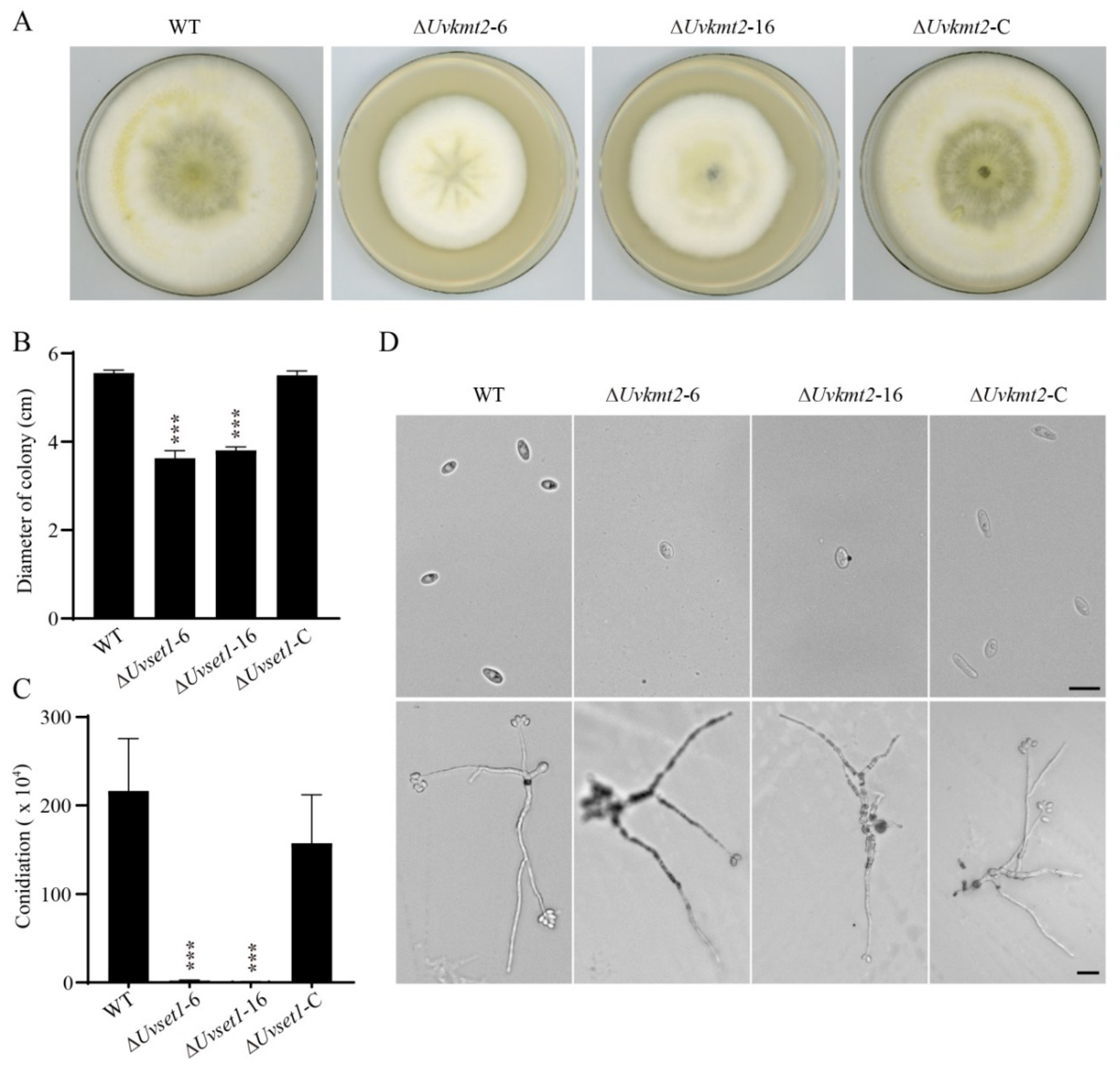
3.2. Disruption and Complementation of UvKMT2
3.3. UvKMT2 Facilitates Growth, Conidiation, and Secondary Spore Formation
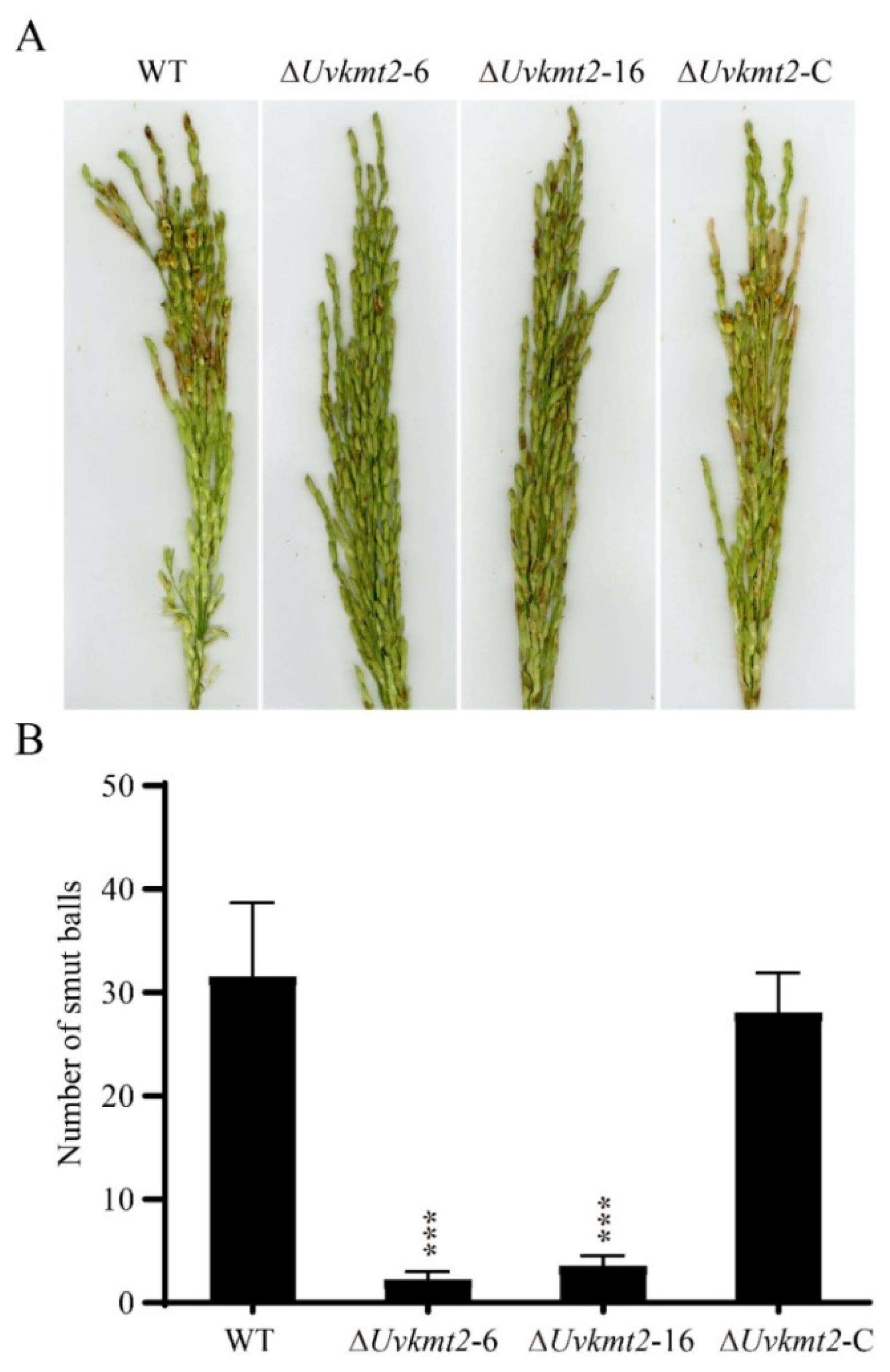
3.4. UvKMT2 Is Required for Virulence in U. virens
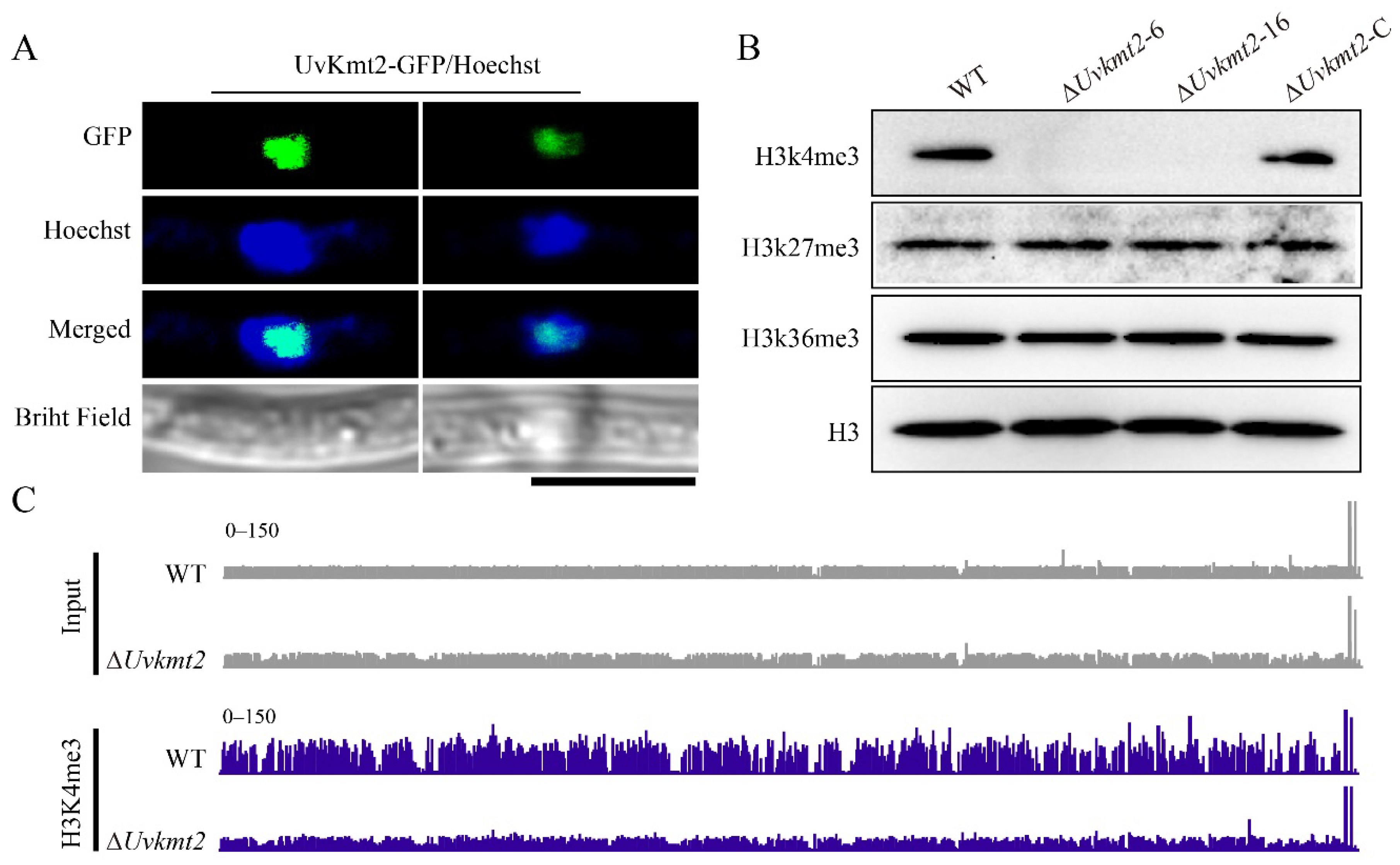
3.5. UvKmt2 Is Essential for the Establishment of Histone Modification H3K4me3
3.6. UvKmt2-Mediated H3K4me3 Plays a Critical Role in Transcriptional Activation
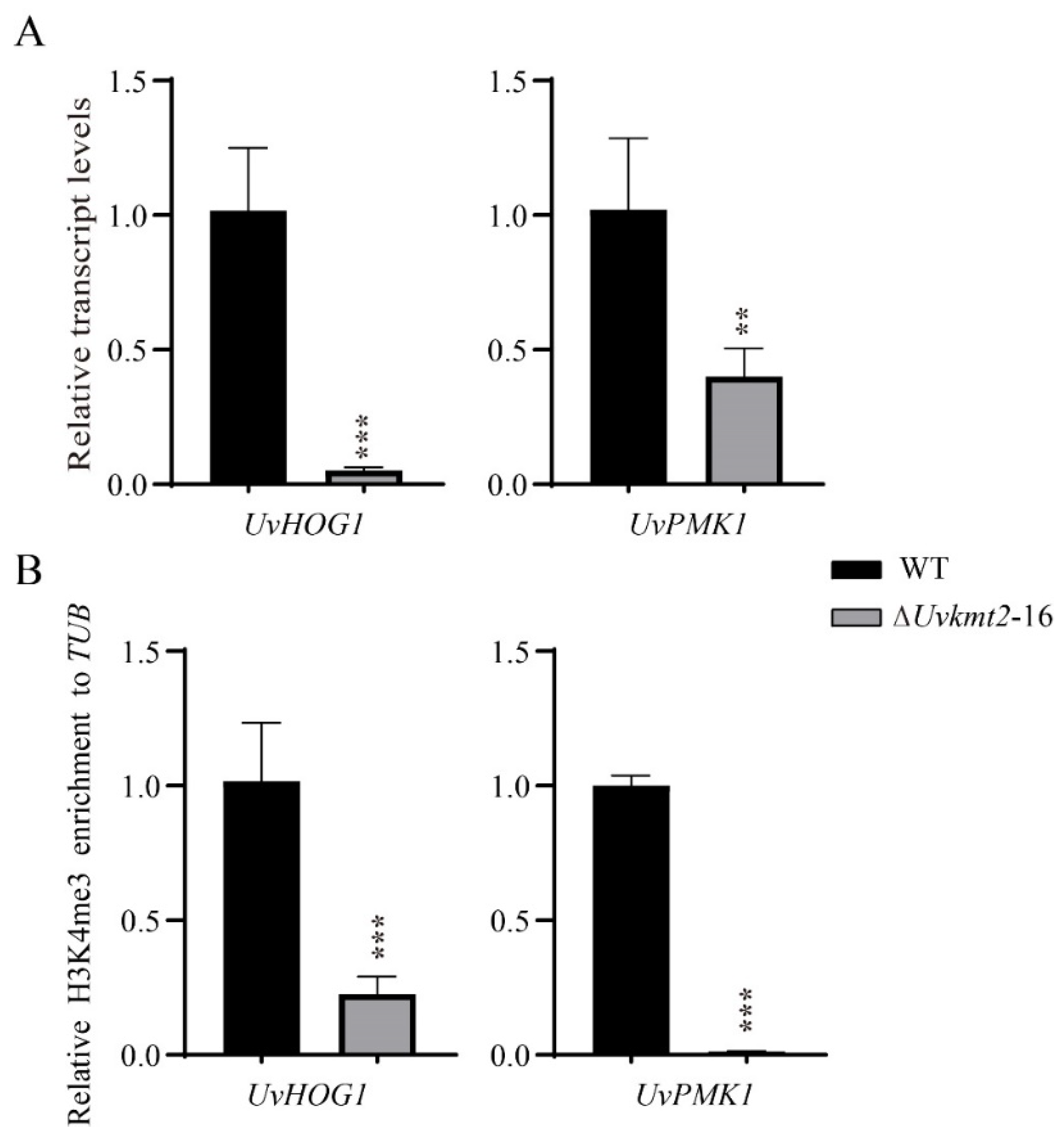
3.7. UvKmt2-Mediated H3K4me3 Modification Regulates Transcription of Conidiation Related and Pathogenic Genes
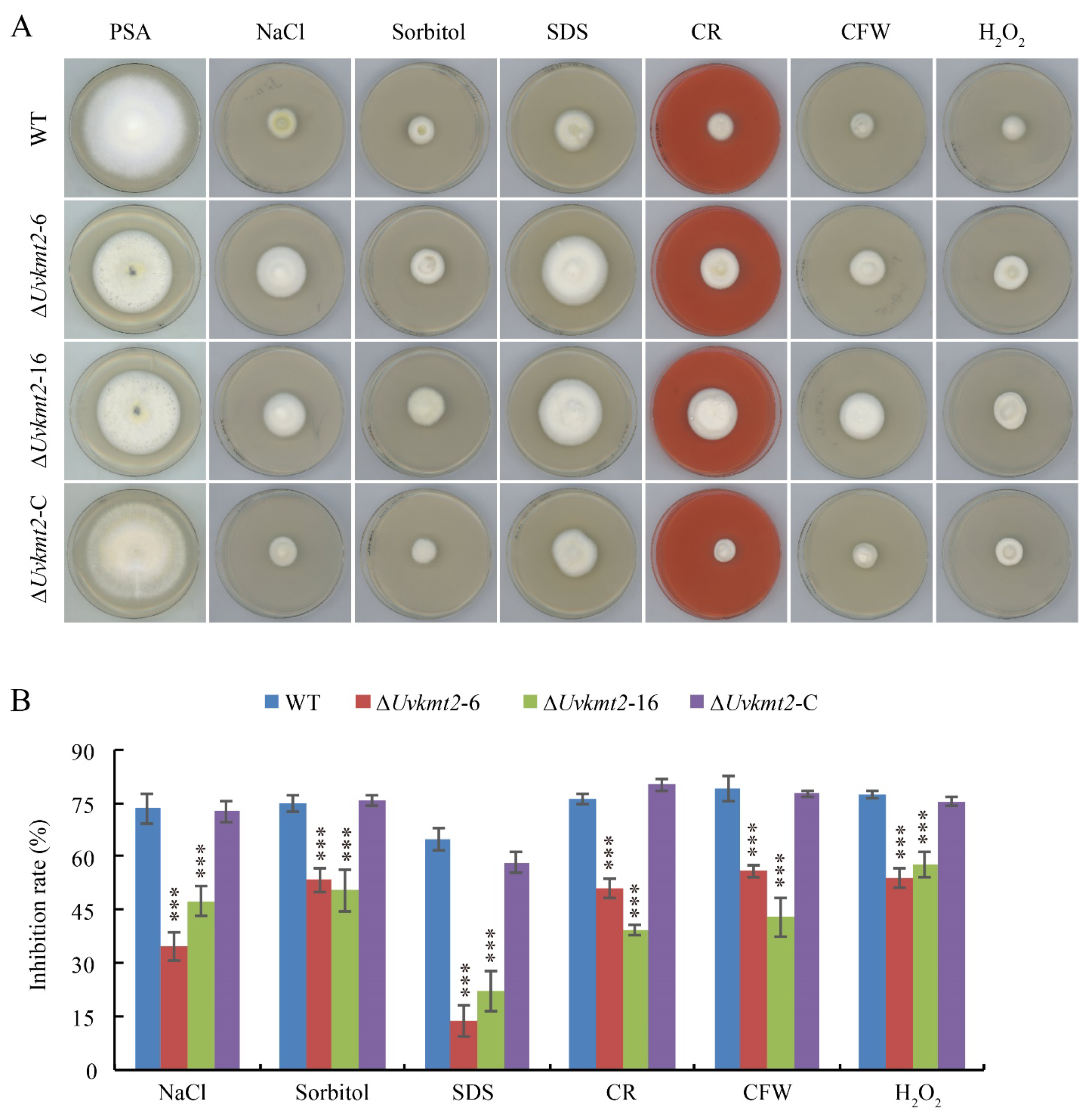
3.8. UvKMT2 Is Involved in Various Stresses Adaption
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ringrose, L.; Paro, R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu. Rev. Genet. 2004, 38, 413–443. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Eissenberg, J.C.; Shilatifard, A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010, 339, 240–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvin, C.D.; Kladde, M.P. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J. Biol. Chem. 2004, 279, 33057–33062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, J.E.; Canze, M.; Bryk, M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 2006, 173, 557–567. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, N.; Jordan, R.; Henderson Pozzi, M.; Bryk, M. Histone 3 lysine 4 monomethylation supports activation of transcription in S. cerevisiae during nutrient stress. Curr. Genet. 2022, 68, 181–194. [Google Scholar] [CrossRef]
- Roguev, A.; Schaft, D.; Shevchenko, A.; Pijnappel, W.W.M.P.; Wilm, M.; Aasland, R.; Stewart1, A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine. EMBO J. 2001, 20, 7137–7148. [Google Scholar] [CrossRef]
- Lai, Y.L.; Cao, X.; Chen, J.J.; Wang, L.; Wei, G.; Wang, S.B. Coordinated regulation of infection-related morphogenesis by the KMT2-Cre1-Hyd4 regulatory pathway to facilitate fungal infection. Microbiology 2020, 6, eaaz1659. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, M.K.; Mohd, M.S.; Ruiz, C.J.; Moore, C.L. Regulation of alternative polyadenylation in the yeast Saccharomyces cerevisiae by histone H3K4 and H3K36 methyltransferases. Nucleic Acids Res. 2020, 48, 5407–5425. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [Green Version]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berr, A.; Xu, L.; Gao, J.; Cognat, V.; Steinmetz, A.; Dong, A.; Shen, W.H. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009, 151, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Tamada, Y.; Yun, J.-Y.; Woo, S.C.; Amasino, R.M. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone h3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 2009, 21, 3257–3269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleckwehl, T.; Crispatzu, G.; Schaaf, K.; Respuela, P.; Bartusel, M.; Benson, L.; Clark, S.J.; Dorighi, K.M.; Barral, A.; Laugsch, M.; et al. Enhancer-associated H3K4 methylation safeguards in vitro germline competence. Nat. Commun. 2021, 12, 5771. [Google Scholar] [CrossRef]
- Jiang, H. The complex activities of the SET1/MLL complex core subunits in development and disease. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194560. [Google Scholar] [CrossRef]
- Raduwan, H.; Isola, A.L.; Belden, W.J. Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression. J. Biol. Chem. 2013, 288, 8380–8390. [Google Scholar] [CrossRef] [Green Version]
- Govindaraghavan, M.; Anglin, S.L.; Osmani, A.H.; Osmani, S.A. The Set1/COMPASS histone H3 methyltransferase helps regulate mitosis with the CDK1 and NIMA mitotic kinases in Aspergillus nidulans. Genetics 2014, 197, 1225–1236. [Google Scholar] [CrossRef] [Green Version]
- Pham, K.T.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar]
- Janevska, S.; Guldener, U.; Sulyok, M.; Tudzynski, B.; Studt, L. Set1 and Kdm5 are antagonists for H3K4 methylation and regulators of the major conidiation-specific transcription factor gene ABA1 in Fusarium fujikuroi. Environ. Microbiol. 2018, 20, 3343–3362. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef]
- Gu, Q.; Tahir, H.A.; Zhang, H.; Huang, H.; Ji, T.; Sun, X.; Wu, L.; Wu, H.; Gao, X. Involvement of FvSet1 in fumonisin B1 biosynthesis, vegetative growth, fungal virulence, and environmental stress responses in Fusarium verticillioides. Toxins 2017, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, M.; Xie, R.; Zhang, F.; Wang, S.; Pan, X.; Wang, S.; Zhuang, Z. The methyltransferase aflset1 is involved in fungal morphogenesis, AFB1 biosynthesis, and virulence of Aspergillus flavus. Front. Microbiol. 2020, 11, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, K.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. SET1/KMT2-governed histone H3K4 methylation coordinates the lifecycle in vivo and in vitro of the fungal insect pathogen Beauveria bassiana. Environ. Microbiol. 2021, 23, 5541–5554. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Fan, J.; Fang, A.; Li, Y.; Tariqjaveed, M.; Li, D.; Hu, D.; Wang, W.M. Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu. Rev. Phytopathol. 2020, 58, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zheng, L.; Huang, J.B.; Zhou, L.G.; Liu, C.S.; Liu, H. Ustiloxin A is produced early in experimental Ustilaginoidea virens infection and affects transcription in rice. Curr. Microbiol. 2020, 77, 2766–2774. [Google Scholar] [CrossRef]
- Li, Y.; Wang, M.; Liu, Z.; Zhang, K.; Cui, F.; Sun, W. Towards understanding the biosynthetic pathway for ustilaginoidin mycotoxins in Ustilaginoidea virens. Environ. Microbiol. 2019, 21, 2629–2643. [Google Scholar] [CrossRef]
- Umemura, M.; Kuriiwa, K.; Tamano, K.; Kawarabayasi, Y. Ustiloxin biosynthetic machinery is not compatible between Aspergillus flavus and Ustilaginoidea virens. Fungal Genet. Biol. 2020, 143, 103434. [Google Scholar] [CrossRef]
- Fan, J.; Liu, J.; Gong, Z.Y.; Xu, P.Z.; Hu, X.H.; Wu, J.L.; Li, G.B.; Yang, J.; Wang, Y.Q.; Zhou, Y.F.; et al. The false smut pathogen Ustilaginoidea virens requires rice stamens for false smut ball formation. Environ. Microbiol. 2020, 22, 646–659. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Yu, J.; Cao, H.; Song, T.; Pan, X.; Qi, Z.; Du, Y.; Zhang, R.; Huang, S.; Liu, W.; et al. SUN-family protein UVSUN1 regulates the development and virulence of Ustilaginoidea virens. Front. Microbiol. 2021, 12, 739453. [Google Scholar] [CrossRef]
- Meng, S.; Liu, Z.; Shi, H.; Wu, Z.; Qiu, J.; Wen, H.; Lin, F.; Tao, Z.; Luo, C.; Kou, Y. UvKmt6-mediated H3K27 trimethylation is required for development, pathogenicity, and stress response in Ustilaginoidea virens. Virulence 2021, 12, 2972–2988. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, K.; Fang, A.; Han, Y.; Yang, J.; Xue, M.; Bao, J.; Hu, D.; Zhou, B.; Sun, X.; et al. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 2014, 5, 3849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, A.; Han, Y.; Zhang, N.; Zhang, M.; Liu, L.; Li, S.; Lu, F.; Sun, W. Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol. Plant Microbe Interact. 2016, 29, 405–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Yang, J.; Fang, A.; Wang, J.; Li, D.; Li, Y.; Wang, S.; Cui, F.; Yu, J.; Liu, Y.; et al. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region. Mol. Plant Pathol. 2020, 21, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Tang, J.; Pei, Z.; Liu, H.; Huang, J.; Luo, C.; Tom, H.; Zheng, L. The ‘pears and lemons’ protein UvPal1 regulates development and virulence of Ustilaginoidea virens. Environ. Microbiol. 2020, 22, 5414–5432. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Xiong, M.; Jagernath, J.S.; Wang, C.; Qiu, J.; Shi, H.; Kou, Y. UvAtg8-mediated autophagy regulates fungal growth, stress responses, conidiation, and pathogenesis in Ustilaginoidea virens. Rice 2020, 13, 56. [Google Scholar] [CrossRef]
- Tang, J.; Bai, J.; Chen, X.; Zheng, L.; Liu, H.; Huang, J. Two protein kinases UvPmk1 and UvCDC2 with significant functions in conidiation, stress response and pathogenicity of rice false smut fungus Ustilaginoidea virens. Curr. Genet. 2020, 66, 409–420. [Google Scholar] [CrossRef]
- Guo, W.; Gao, Y.; Yu, Z.; Xiao, Y.; Zhang, Z.; Zhang, H. The adenylate cyclase UvAc1 and phosphodiesterase UvPdeH control the intracellular cAMP level, development, and pathogenicity of the rice false smut fungus Ustilaginoidea virens. Fungal Genet. Biol. 2019, 129, 65–73. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, Y.; Han, Y.; Xu, J.R.; Wang, C. UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci. Rep. 2016, 6, 24824. [Google Scholar] [CrossRef]
- Yan, Y.; Tang, J.; Yuan, Q.; Liu, H.; Huang, J.; Hsiang, T.; Bao, C.; Zheng, L. Ornithine decarboxylase of the fungal pathogen Colletotrichum higginsianum plays an important role in regulating global metabolic pathways and virulence. Environ. Microbiol. 2021, 24, 1093–1116. [Google Scholar] [CrossRef]
- Shi, H.; Meng, S.; Qiu, J.; Wang, C.; Shu, Y.; Luo, C.; Kou, Y. MoWhi2 regulates appressorium formation and pathogenicity via the MoTor signalling pathway in Magnaporthe oryzae. Mol. Plant Pathol. 2021, 22, 969–983. [Google Scholar] [CrossRef]
- Tao, Z.; Shen, L.; Gu, X.; Wang, Y.; Yu, H.; He, Y. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 2017, 551, 124–128. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Xu, Y.; Chen, J.; Luo, Y.; Lv, Y.; Su, J.; Kershaw, M.J.; Li, W.; Wang, J.; Yin, J.; et al. MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 2018, 14, 1543–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie. Nature Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Song, X.; Zhang, L.; Yu, Y.; Liang, Z.; Lei, Y.; Ruan, J.; Tan, B.; Liu, J.; Li, C. The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide polycomb silencing in Arabidopsis. Nucleic Acids Res. 2021, 49, 98–113. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, 160–165. [Google Scholar] [CrossRef]
- Lu, J.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog. 2014, 10, e1004432. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–555. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Zhang, H.; Liu, Y.; Liu, Y.X.; Huang, L. EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genom. 2021, 48, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, D.; Gao, X.; Morgan, M.A.; Herz, H.M.; Smith, E.R.; Shilatifard, A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell Biol. 2013, 33, 4745–4754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, L.M.; He, P.C.; Chun, Y.; Suh, H.; Kim, T.; Buratowski, S. Determinants of Histone H3K4 methylation patterns. Mol. Cell 2017, 68, 773–785.e6. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, K.; Yin, L.; Yu, Y.; Qi, J.; Shen, W.H.; Zhu, J.; Zhang, Y.; Dong, A. H3K4me2 functions as a repressive epigenetic mark in plants. Epigenetics Chromatin 2019, 12, 40. [Google Scholar] [CrossRef]
- Huh, A.; Dubey, A.; Kim, S.; Jeon, J.; Lee, Y.H. MoJMJ1, encoding a histone demethylase containing JmjC domain, is required for pathogenic development of the rice blast fungus, Magnaporthe oryzae. Plant Pathol. J. 2017, 33, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Wang, Q.; Dou, X.; Wang, W.; Zhao, Q.; Lv, R.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 2012, 13, 677–689. [Google Scholar] [CrossRef]
- Wang, R.J.; Peng, J.; Li, Q.X.; Peng, Y.L. Phosphorylation-mediated regulatory networks in mycelia of Pyricularia oryzae revealed by phosphoproteomic analyses. Mol. Cell. Proteom. 2017, 16, 1669–1682. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhong, H.M.; Yakubu, S.A.; Yang, C.D.; Wang, X.X.; Fan, Y.P.; Zhang, M.R.; Wang, Z.H.; Zhou, J.; Miao, P.F.; et al. Trehalose phosphate synthase complex-mediated regulation of trehalose 6-phosphate homeostasis is critical for development and pathogenesis in Magnaporthe oryzae. mSystems 2021, 6, e00462-21. [Google Scholar] [CrossRef]
- Yu, J.; Li, T.Y.; Tian, L.Y.; Tang, C.; Klosterman, S.J.; Tian, C.M.; Wang, Y.L. Two Verticillium dahliae MAPKKKs, VdSsk2 and VdSte11, have distinct roles in pathogenicity, microsclerotial formation, and stress adaptation. mSphere 2019, 4, e00426-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Q.L.; Meng, X.; Wang, C.; Dai, W.; Luo, Z.; Yin, Z.; Ju, Z.; Fu, X.; Yang, J.; Ye, Q.; et al. Histone H3K4me3 modification is a transgenerational epigenetic signal for lipid metabolism in Caenorhabditis elegans. Nat. Commun. 2022, 13, 768. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, F.; Xie, S.; Liu, Y.; Qu, J.; Huang, J.; Yin, W.; Luo, C. Development of rice conidiation media for Ustilaginoidea virens. PLoS ONE 2019, 14, e0217667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Hai, D.; Tang, J.; Liu, H.; Huang, J.; Luo, C.; Hsiang, T.; Zheng, L. UvCom1 is an important regulator required for development and infection in the rice false smut fungus Ustilaginoidea virens. Phytopathology 2020, 110, 483–493. [Google Scholar] [CrossRef]
- Yu, J.; Yu, M.; Song, T.; Cao, H.; Pan, X.; Yong, M.; Qi, Z.; Du, Y.; Zhang, R.; Yin, X.; et al. A homeobox transcription factor UvHOX2 regulates chlamydospore formation, conidiogenesis, and pathogenicity in Ustilaginoidea virens. Front. Microbiol. 2019, 10, 1071. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, S.; Shi, H.; Lin, C.; Wu, Z.; Lin, F.; Tao, Z.; Kou, Y. UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens. J. Fungi 2022, 8, 553. https://doi.org/10.3390/jof8060553
Meng S, Shi H, Lin C, Wu Z, Lin F, Tao Z, Kou Y. UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens. Journal of Fungi. 2022; 8(6):553. https://doi.org/10.3390/jof8060553
Chicago/Turabian StyleMeng, Shuai, Huanbin Shi, Chuyu Lin, Zhongling Wu, Fucheng Lin, Zeng Tao, and Yanjun Kou. 2022. "UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens" Journal of Fungi 8, no. 6: 553. https://doi.org/10.3390/jof8060553
APA StyleMeng, S., Shi, H., Lin, C., Wu, Z., Lin, F., Tao, Z., & Kou, Y. (2022). UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens. Journal of Fungi, 8(6), 553. https://doi.org/10.3390/jof8060553






