Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments
Abstract
1. Introduction
2. Culturable and Molecular Diversity of Marine Fungi
2.1. Current Consensus of Culturable Diversity
2.2. Mycoplankton Diversity
2.2.1. Microscopic Forms and Culturable Diversity
2.2.2. Molecular Diversity and Dynamics of Mycoplankton
2.3. Environmental Drivers of Mycoplankton Diversity
3. Abundance of Mycoplankton
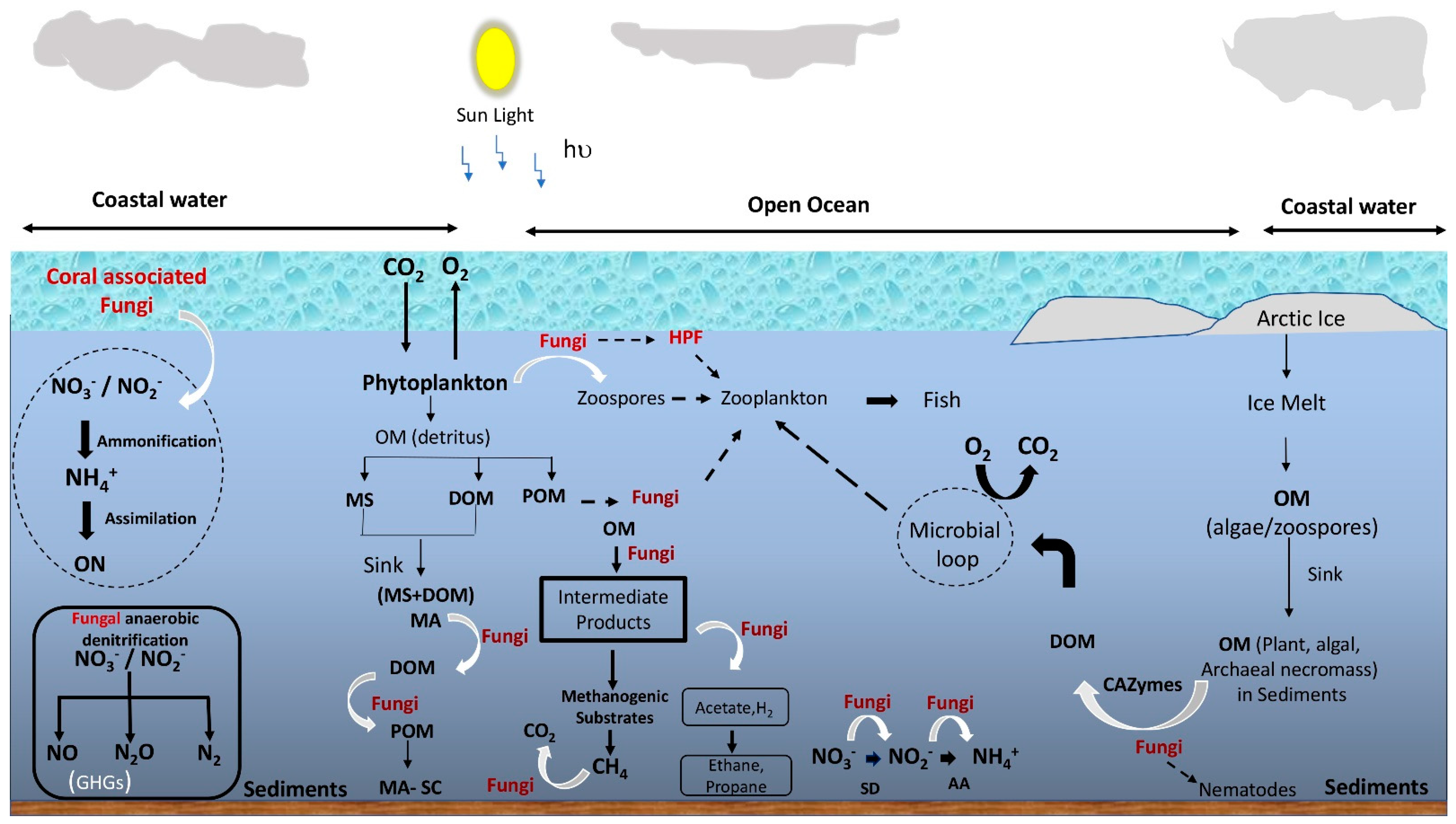
4. Ecological Roles of Mycoplankton
4.1. Biogeochemical Cycling
4.1.1. Role in Organic Matter Decomposition and Aggregation
4.1.2. Role in Nutrient Metabolism
4.2. Fungal Contribution to the Marine Food Web and Biotic Interactions
5. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vishniac, H.S. On the ecology of the lower marine Fungi. Biol. Bull. 1956, 111, 410–414. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the Marine Environment: Open Questions and Unsolved Problems. mBio 2019, 10, e01189-18. [Google Scholar] [CrossRef] [PubMed]
- Kohlmeyer, J.; Kohlmeyer, E. 1—Introduction. In Marine Mycology; Academic Press: Cambridge, MA, USA, 1979; pp. 1–6. [Google Scholar]
- Raghukumar, S.; Raghukumar, C. Marine fungi: A critique. Aquat. Microbiol. Newsl. 1999, 38, 26–27. [Google Scholar]
- Heckman, D.S.; Geiser, D.M.; Eidell, B.R.; Stauffer, R.L.; Kardos, N.L.; Hedges, S.B. Molecular Evidence for the Early Colonization of Land by Fungi and Plants. Science 2001, 293, 1129. [Google Scholar] [CrossRef] [PubMed]
- Mahé, S.; Rédou, V.; Calvez, T.L.; Vandenkoornhuyse, P.; Burgaud, G. Fungi in Deep-Sea Environments and Metagenomics. In The Ecological Genomics of Fungi; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 325–354. [Google Scholar]
- Pang, K.-L.; Overy, D.P.; Jones, E.B.G.; Calado, M.d.L.; Burgaud, G.; Walker, A.K.; Johnson, J.A.; Kerr, R.G.; Cha, H.-J.; Bills, G.F. ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biol. Rev. 2016, 30, 163–175. [Google Scholar] [CrossRef]
- Rédou, V.; Vallet, M.; Meslet-Cladière, L.; Kumar, A.; Pang, K.-L.; Pouchus, Y.-F.; Barbier, G.; Grovel, O.; Bertrand, S.; Prado, S.; et al. Marine Fungi. In The Marine Microbiome; Stal, L.J., Cretoiu, M.S., Eds.; Springer: Cham, Switzerland, 2016; pp. 99–153. [Google Scholar]
- Walker, A.K.; Vélez, P.; González, M.C. Marine Fungi. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2017. [Google Scholar]
- Damare, S.; Raghukumar, C.; Raghukumar, S. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res. Part I: Oceanogr. Res. Pap. 2006, 53, 14–27. [Google Scholar] [CrossRef]
- Singh, P.; Raghukumar, C.; Verma, P.; Shouche, Y. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers. 2010, 40, 89–102. [Google Scholar] [CrossRef]
- Zhang, T.; Fei Wang, N.; Qin Zhang, Y.; Yu Liu, H.; Yan Yu, L. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Sci. Rep. 2015, 5, 14524. [Google Scholar] [CrossRef]
- Rédou, V.; Navarri, M.; Meslet-Cladière, L.; Barbier, G.; Burgaud, G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Beaudoin, D.; Gast, R.; Biddle, J.F.; Teske, A. Marine subsurface eukaryotes: The fungal majority. Environ. Microbiol. 2011, 13, 172–183. [Google Scholar] [CrossRef]
- Zhang, L.; Kang, M.; Huang, Y.; Yang, L. Fungal communities from the calcareous deep-sea sediments in the Southwest India Ridge revealed by Illumina sequencing technology. World J. Microbiol. Biotechnol. 2016, 32, 78. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, M.; Bian, X.; Guo, J.; Cai, L. A High-Level Fungal Diversity in the Intertidal Sediment of Chinese Seas Presents the Spatial Variation of Community Composition. Front. Microbiol. 2016, 7, 2098. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Liu, X.; Jiao, N.; Wang, G. Diversity of parasitic fungi associated with phytoplankton in Hawaiian waters. Mar. Biol. Res. 2016, 12, 294–303. [Google Scholar] [CrossRef]
- Wang, Y.; Sen, B.; He, Y.; Xie, N.; Wang, G. Spatiotemporal Distribution and Assemblages of Planktonic Fungi in the Coastal Waters of the Bohai Sea. Front. Microbiol. 2018, 9, 584. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128. [Google Scholar] [CrossRef]
- Wang, Y.; Sen, K.; He, Y.; Xie, Y.; Wang, G. Impact of environmental gradients on the abundance and diversity of planktonic fungi across coastal habitats of contrasting trophic status. Sci. Total Environ. 2019, 683, 822–833. [Google Scholar] [CrossRef]
- Duan, Y.; Xie, N.; Song, Z.; Ward, C.S.; Yung, C.-M.; Hunt, D.E.; Johnson, Z.I.; Wang, G. A High-resolution Time-series Reveals Seasonal Patterns of Planktonic Fungi at a Temperate Coastal Ocean Site (Beaufort, North Carolina, USA). Appl. Environ. Microbiol. 2018, 84, e00967-18. [Google Scholar] [CrossRef]
- Morales, S.; Biswas, A.; Herndl, G.; Baltar, F. Global Structuring of Phylogenetic and Functional Diversity of Pelagic Fungi by Depth and Temperature. Front. Mar. Sci. 2019, 6, 131. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.; Burgaud, G.; Yu, H.; Cai, L. Fungal Community Composition and Potential Depth-Related Driving Factors Impacting Distribution Pattern and Trophic Modes from Epi- to Abyssopelagic Zones of the Western Pacific Ocean. Microb. Ecol. 2019, 78, 820–831. [Google Scholar] [CrossRef]
- Hassett, B.T.; Borrego, E.J.; Vonnahme, T.R.; Rämä, T.; Kolomiets, M.V.; Gradinger, R. Arctic marine fungi: Biomass, functional genes, and putative ecological roles. ISME J. 2019, 13, 1484–1496. [Google Scholar] [CrossRef]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Adholeya, A.; Barrow, C.J.; Deshmukh, S.K. Marine fungi: An untapped bioresource for future cosmeceuticals. Phytochem. Lett. 2018, 23, 15–20. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.-J.; Li, J.-J.; Liang, Z.-Z.; Zhao, C.-Q. Novel Natural Products from Extremophilic Fungi. Mar. Drugs 2018, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Mar. Drugs 2016, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Varrella, S.; Barone, G.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Corinaldesi, C. Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi. J. Fungi 2021, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Zandi, K. Potential Antiviral Agents from Marine Fungi: An Overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, S. The Pelagic Ecosystem. In Fungi in Coastal and Oceanic Marine Ecosystems: Marine Fungi; Raghukumar, S., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 185–217. [Google Scholar]
- Pang, K.-L.; Jones, E.B.G. Phylogenetic Diversity of Fungi in the Sea including the Opisthosporidia. In Biology of Microfungi; Li, D.-W., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 267–283. [Google Scholar]
- Kis-Papo, T. Marine fungal communities. In The Fungal Community: Its Organization and Role in the Ecosystem; Dighton, J., White, J.F., Oudemans, P., Eds.; Taylor & Francis: Boca Raton, NJ, USA, 2005; pp. 61–92. [Google Scholar]
- Jones, E.B.G.; Sakayaroj, J.; Suetrong, S.; Somrithipol, S.; Pang, K.L. Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers. 2009, 35, 1–187. [Google Scholar]
- Jones, E.B.G.; Suetrong, S.; Sakayaroj, J.; Bahkali, A.H.; Abdel-Wahab, M.A.; Boekhout, T.; Pang, K.-L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015, 73, 1–72. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Pang, K.-L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433. [Google Scholar] [CrossRef]
- Jones, M.D.M.; Forn, I.; Gadelha, C.; Egan, M.J.; Bass, D.; Massana, R.; Richards, T.A. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 2011, 474, 200. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Pantoja, S.; Quiñones, R.A.; González, R.R. First record of filamentous fungi in the coastal upwelling ecosystem off central Chile. Gayana (Concepción) 2010, 74, 66–73. [Google Scholar] [CrossRef]
- Lepere, C.; Ostrowski, M.; Hartmann, M.; Zubkov, M.V.; Scanlan, D.J. In situ associations between marine photosynthetic picoeukaryotes and potential parasites—A role for fungi? Environ. Microbiol. Rep. 2016, 8, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Leonard, G.; Mahé, F.; Del Campo, J.; Romac, S.; Jones, M.D.M.; Maguire, F.; Dunthorn, M.; De Vargas, C.; Massana, R.; et al. Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc. R. Soc. B: Biol. Sci. 2015, 282, 20152243. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Jara, A.M.; Pantoja, S. Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt current system off central Chile. Environ. Microbiol. 2016, 18, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Hassett, B.T.; Gradinger, R. Chytrids dominate arctic marine fungal communities. Environ. Microbiol. 2016, 18, 2001–2009. [Google Scholar] [CrossRef]
- Fell, J.W. Yeasts in marine environments. In Marine Fungi and Fungal-Like Organisms; Jones, E.B.G., Pang, K.L., Eds.; Walter de Gruyter GmbH & Co. KG: Berlin, Germany; Boston, MA, USA, 2012; pp. 91–102. [Google Scholar]
- Kutty, S.N.; Philip, R. Marine yeasts-a review. Yeast 2008, 25, 465–483. [Google Scholar] [CrossRef]
- Fotedar, R.; Chatting, M.; Kolecka, A.; Zeyara, A.; Al Malki, A.; Kaul, R.; Bukhari, S.J.; Moaiti, M.A.; Febbo, E.J.; Boekhout, T.; et al. Communities of culturable yeasts and yeast-like fungi in oligotrophic hypersaline coastal waters of the Arabian Gulf surrounding Qatar. Antonie Van Leeuwenhoek 2022, 115, 609–633. [Google Scholar] [CrossRef]
- Pham, T.T.; Dinh, K.V.; Nguyen, V.D. Biodiversity and Enzyme Activity of Marine Fungi with 28 New Records from the Tropical Coastal Ecosystems in Vietnam. Mycobiology 2021, 49, 559–581. [Google Scholar] [CrossRef]
- Roth, F.J.; Orpurt, P.A.; Ahearn, D.G. Occurrence and distribution of fungi in a subtropical marine environment. Can. J. Bot. 1964, 42, 375–383. [Google Scholar] [CrossRef]
- Vera, J.; Gutiérrez, M.H.; Palfner, G.; Pantoja, S. Diversity of culturable filamentous Ascomycetes in the eastern South Pacific Ocean off Chile. World J. Microbiol. Biotechnol. 2017, 33, 1–13. [Google Scholar] [CrossRef]
- Li, L.; Singh, P.; Liu, Y.; Pan, S.; Wang, G. Diversity and biochemical features of culturable fungi from the coastal waters of Southern China. AMB Express 2014, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.N.; Vitoreli, G.A.; de Menezes, G.C.A.; Mendes, C.R.B.; Secchi, E.R.; Rosa, C.A.; Rosa, L.H. Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 2017, 21, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Burgaud, G.; Woehlke, S.; Rédou, V.; Orsi, W.; Beaudoin, D.; Barbier, G.; Biddle, J.F.; Edgcomb, V.P. Deciphering the presence and activity of fungal communities in marine sediments using a model estuarine system. Aquat. Microb. Ecol. 2013, 70, 45–62. [Google Scholar] [CrossRef]
- Fotedar, R.; Kolecka, A.; Boekhout, T.; Fell Jack, W.; Al-Malki, A.; Zeyara, A.; Al Marri, M. Fungal diversity of the hypersaline Inland Sea in Qatar. Bot. Mar. 2018, 61, 595. [Google Scholar] [CrossRef]
- Bovio, E.; Gnavi, G.; Prigione, V.; Spina, F.; Denaro, R.; Yakimov, M.; Calogero, R.; Crisafi, F.; Varese, G.C. The culturable mycobiota of a Mediterranean marine site after an oil spill: Isolation, identification and potential application in bioremediation. Sci. Total Environ. 2017, 576, 310–318. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.; Pan, H.; Burgaud, G.; Liang, S.; Guo, J.; Luo, T.; Li, Z.; Zhang, S.; Cai, L. Highlighting patterns of fungal diversity and composition shaped by ocean currents using the East China Sea as a model. Mol. Ecol. 2018, 27, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Vargas, C.D.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Bescot, N.L.; Probert, I. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, R.; Hu, C.; Sun, P.; Jiao, N.; Warren, A. Microbial Eukaryote Diversity and Activity in the Water Column of the South China Sea Based on DNA and RNA High Throughput Sequencing. Front. Microbiol. 2017, 8, 1121. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Garcés, D.V.; Pantoja, S.; González, R.R.; Quiñones, R.A. Environmental fungal diversity in the upwelling ecosystem off central Chile and potential contribution to enzymatic hydrolysis of macromolecules in coastal ecotones. Fungal Ecol. 2017, 29, 90–95. [Google Scholar] [CrossRef]
- Gao, Z.; Johnson, Z.I.; Wang, G. Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. ISME J. 2010, 4, 111–120. [Google Scholar] [CrossRef]
- Sun, J.Y.; Song, Y.; Ma, Z.P.; Zhang, H.J.; Yang, Z.D.; Cai, Z.H.; Zhou, J. Fungal community dynamics during a marine dinoflagellate (Noctiluca scintillans) bloom. Mar. Environ. Res. 2017, 131, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Comeau, A.M.; Vincent, W.F.; Bernier, L.; Lovejoy, C. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 2016, 6, 30120. [Google Scholar] [CrossRef]
- Jeffries, T.C.; Curlevski, N.J.; Brown, M.V.; Harrison, D.P.; Doblin, M.A.; Petrou, K.; Ralph, P.J.; Seymour, J.R. Partitioning of fungal assemblages across different marine habitats. Environ. Microbiol. Rep. 2016, 8, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-P.; Liu, Z.-Z.; Wang, Y.-L.; Bi, W.-H.; Liu, L.; Wang, H.-Y.; Zheng, Y.; Zhang, L.-L.; Hu, S.-G.; Xu, S.-. Set al. Fungal community analysis in seawater of the Mariana Trench as estimated by Illumina HiSeq. RSC Adv. 2019, 9, 6956–6964. [Google Scholar] [CrossRef] [PubMed]
- Picard, K.T. Coastal marine habitats harbor novel early diverging fungal diversity. Fungal Ecol. 2017, 25, 1–13. [Google Scholar] [CrossRef]
- Wang, X.; Singh, P.; Gao, Z.; Zhang, X.; Johnson, Z.I.; Wang, G.Y. Distribution and diversity of planktonic fungi in the West Pacific Warm Pool. PLoS ONE 2014, 9, e101523. [Google Scholar] [CrossRef]
- Hassett, B.T.; Vonnahme, T.R.; Peng, X.; Jones, E.B.G.; Heuzé, C. Global diversity and geography of planktonic marine fungi. Bot. Mar. 2020, 63, 121–139. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Pang, K.-L. Tropical aquatic fungi. Biodivers. Conserv. 2012, 21, 2403–2423. [Google Scholar] [CrossRef]
- Jones, E.B.G. Ultrastructure and taxonomy of the aquatic ascomycetous order Halosphaeriales. Can. J. Bot. 1995, 73, 790–801. [Google Scholar] [CrossRef]
- Suetrong, S.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; Sakayaroj, J.; Phongpaichit, S.; Tanaka, K.; Hirayama, K.; Jones, E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009, 64, 155–173. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbruck, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- Taylor, D.L.; Hollingsworth, T.N.; McFarland, J.W.; Lennon, N.J.; Nusbaum, C.; Ruess, R.W. A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecol. Monogr. 2014, 84, 3–20. [Google Scholar] [CrossRef]
- Blackwell, M. The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Tisthammer, K.H.; Cobian, G.M.; Amend, A.S. Global biogeography of marine fungi is shaped by the environment. Fungal Ecol. 2016, 19, 39–46. [Google Scholar] [CrossRef]
- Xu, W.; Luo, Z.-H.; Guo, S.; Pang, K.-L. Fungal community analysis in the deep-sea sediments of the Pacific Ocean assessed by comparison of ITS, 18S and 28S ribosomal DNA regions. Deep Sea Res. Part I Oceanogr. Res. Pap. 2016, 109, 51–60. [Google Scholar] [CrossRef]
- Fuhrman, J.A.; Steele, J.A.; Hewson, I.; Schwalbach, M.S.; Brown, M.V.; Green, J.L.; Brown, J.H. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 7774–7778. [Google Scholar] [CrossRef]
- Rojas-Jimenez, K.; Wurzbacher, C.; Jürgens, K.; Labrenz, M.; Grossart, H.-P. A Salinity Threshold Separating Fungal Communities in the Baltic Sea. Front. Microbiol. 2019, 10, 680. [Google Scholar] [CrossRef] [PubMed]
- Kubanek, J.; Jensen, P.R.; Keifer, P.A.; Sullards, M.C.; Collins, D.O.; Fenical, W. Seaweed resistance to microbial attack: A targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 6916–6921. [Google Scholar] [CrossRef]
- Velmurugan, S.; Prasannakumar, C.; Manokaran, S.; Ajith Kumar, T.; Samkamaleson, A.; Palavesam, A. DNA barcodes for marine fungal identification and discovery. Fungal Ecol. 2013, 6, 408–418. [Google Scholar] [CrossRef]
- Krause, E.; Wichels, A.; Giménez, L.; Gerdts, G. Marine fungi may benefit from ocean acidification. Aquat. Microb. Ecol. 2013, 69, 59–67. [Google Scholar] [CrossRef]
- Xu, W.; Pang, K.L.; Luo, Z.H. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb. Ecol. 2014, 68, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Le Calvez, T.; Burgaud, G.; Mahe, S.; Barbier, G.; Vandenkoornhuyse, P. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 2009, 75, 6415–6421. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.H.; Pantoja, S.; Tejos, E.; Quiñones, R.A. The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar. Biol. 2011, 158, 205–219. [Google Scholar] [CrossRef]
- Sherr, E.B.; Sherr, B.F.; Fessenden, L. Heterotrophic protists in the Central Arctic Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 1997, 44, 1665–1682. [Google Scholar] [CrossRef]
- Mary, I.; Cummings, D.; Biegala, I.; Burkill, P.; Archer, S.; Zubkov, M. Seasonal dynamics of bacterioplankton structure at a coastal station in the western English Channel. Aquat. Microb. Ecol. 2006, 42, 119–126. [Google Scholar] [CrossRef]
- Vargas-Gastélum, L.; Riquelme, M. The Mycobiota of the Deep Sea: What Omics Can Offer. Life 2020, 10, 292. [Google Scholar] [CrossRef]
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef]
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594. [Google Scholar] [CrossRef]
- Cathrine, S.J.; Raghukumar, C. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycol. Res. 2009, 113, 100–109. [Google Scholar] [CrossRef]
- Damare, S.; Raghukumar, C. Fungi and macroaggregation in deep-sea sediments. Microb. Ecol. 2008, 56, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.E.; Quiñones, R.A. Carbon utilization profile of the filamentous fungal species Fusarium fujikuroi, Penicillium decumbens, and Sarocladium strictum isolated from marine coastal environments. Mycologia 2016, 108, 1069–1081. [Google Scholar] [PubMed]
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal-bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2017, 11, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Gulis, V.; Kuehn, K.; Suberkropp, K. The role of fungi in carbon and nitrogen cycles in freshwater ecosystems. In Fungi in Biogeochemical Cycles; Gadd, G.M., Ed.; Cambridge University Press: New York, NY, USA, 2006; pp. 404–435. [Google Scholar]
- Gessner, M.O.; Gulis, V.; Kuehn, K.; Chauvet, E.; Suberkropp, K. Fungal Decomposers of Plant Litter in Aquatic Ecosystems. In Environmental and Microbial Relationships, 2nd ed.; Kubicek, C.P., Druzhinina, I.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 301–324. [Google Scholar]
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, S. The Role of Fungi in Marine Detrital Processes. In Marine Microbiology: Facets and Opportunities; Ramaiah, N., Ed.; National Institute of Oceanography: Goa, India, 2004; pp. 91–101. [Google Scholar]
- Jones, E.B.G. Fifty years of marine mycology. Fungal Divers. 2011, 50, 73–112. [Google Scholar] [CrossRef]
- Pointing, S.B.; Hyde, K.D. Lignocellulose-degrading marine fungi. Biofouling 2000, 15, 221–229. [Google Scholar] [CrossRef]
- Kamei, I.; Daikoku, C.; Tsutsumi, Y.; Kondo, R. Saline-dependent regulation of manganese peroxidase genes in the hypersaline-tolerant white rot fungus Phlebia sp. strain MG-60. Appl. Environ. Microbiol. 2008, 74, 2709–2716. [Google Scholar] [CrossRef][Green Version]
- Bonugli-Santos, R.C.; Durrant, L.R.; da Silva, M.; Sette, L.D. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzym. Microb. Technol. 2010, 46, 32–37. [Google Scholar] [CrossRef]
- Arfi, Y.; Chevret, D.; Henrissat, B.; Berrin, J.-G.; Levasseur, A.; Record, E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat. Commun. 2013, 4, 1810. [Google Scholar] [CrossRef]
- Wang, Y.; Barth, D.; Tamminen, A.; Wiebe, M.G. Growth of marine fungi on polymeric substrates. BMC Biotechnol. 2016, 16, 3. [Google Scholar] [CrossRef]
- Mainardi, P.H.; Feitosa, V.A.; Brenelli de Paiva, L.B.; Bonugli-Santos, R.C.; Squina, F.M.; Pessoa, A., Jr.; Sette, L.D. Laccase production in bioreactor scale under saline condition by the marine-derived basidiomycete Peniophora sp. CBMAI 1063. Fungal Biol. 2018, 122, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Bucher, V.V.C.; Hyde, K.; Pointing, S.; Reddy, C.A.; Reddy, S. Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 2003, 15, 1–14. [Google Scholar]
- Leightley, L.E.; Eaton, R.A. Nia vibrissa—A marine white rot fungus. Trans. Br. Mycol. Soc. 1979, 73, 35–40. [Google Scholar] [CrossRef]
- Leightely, L.E. Wood decay activities of marine fungi. Bot. Mar. 1980, 23, 387–395. [Google Scholar]
- Jones, E.B.G.; Choeyklin, R. Chapter 10 Ecology of marine and freshwater basidiomycetes. In British Mycological Society Symposia Series; Boddy, L., Frankland, J.C., van West, P., Eds.; Academic Press: Cambridge, MA, USA, 2008; Volume 28, pp. 301–324. [Google Scholar]
- Mouzouras, R. Soft rot decay of wood by marine microfungi. J. Inst. Wood Sci. 1989, 11, 193–201. [Google Scholar]
- Hyde, K.D.; Jones, E.B.G.; Leaño, E.; Pointing, S.B.; Poonyth, A.D.; Vrijmoed, L.L.P. Role of fungi in marine ecosystems. Biodivers. Conserv. 1998, 7, 1147–1161. [Google Scholar] [CrossRef]
- Rämä, T.; Davey, M.L.; Nordén, J.; Halvorsen, R.; Blaalid, R.; Mathiassen, G.H.; Alsos, I.G.; Kauserud, H. Fungi Sailing the Arctic Ocean: Speciose Communities in North Atlantic Driftwood as Revealed by High-Throughput Amplicon Sequencing. Microb. Ecol. 2016, 72, 295–304. [Google Scholar] [CrossRef]
- Cunliffe, M.; Hollingsworth, A.; Bain, C.; Sharma, V.; Taylor, J.D. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol. 2017, 30, 135–138. [Google Scholar] [CrossRef]
- Bochdansky, A.B.; Clouse, M.A.; Herndl, G.J. Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 2017, 11, 362–373. [Google Scholar] [CrossRef]
- Zeghal, E.; Vaksmaa, A.; Vielfaure, H.; Boekhout, T.; Niemann, H. The Potential Role of Marine Fungi in Plastic Degradation–A Review. Front. Mar. Sci. 2021, 8, 738877. [Google Scholar] [CrossRef]
- Jones, E.G.; Le Campion-Alsumard, T. The biodeterioration of polyurethane by marine fungi. Int. Biodeterior. Bull. 1970, 6, 119–124. [Google Scholar]
- Alshehrei, F. Biodegradation of Low Density Polyethylene by Fungi Isolated from Red Sea Water. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1703–1709. [Google Scholar] [CrossRef]
- Orsi, W.D.; Edgcomb, V.P.; Christman, G.D.; Biddle, J.F. Gene expression in the deep biosphere. Nature 2013, 499, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Orsi, W.D.; Richards, T.A.; Francis, W.R. Predicted microbial secretomes and their target substrates in marine sediment. Nat. Microbiol. 2018, 3, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Sen, K.; Bai, M.; Sen, B.; Wang, G. Disentangling the structure and function of mycoplankton communities in the context of marine environmental heterogeneity. Sci. Total Environ. 2021, 766, 142635. [Google Scholar] [CrossRef]
- Baltar, F.; Zhao, Z.; Herndl, G.J. Potential and expression of carbohydrate utilization by marine fungi in the global ocean. Microbiome 2021, 9, 106. [Google Scholar] [CrossRef]
- Lai, X.; Cao, L.; Tan, H.; Fang, S.; Huang, Y.; Zhou, S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007, 1, 756–762. [Google Scholar] [CrossRef]
- Ruff, S.E.; Biddle, J.F.; Teske, A.P.; Knittel, K.; Boetius, A.; Ramette, A. Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. USA 2015, 112, 4015–4020. [Google Scholar] [CrossRef]
- Wolf, H.J.; Hanson, R.S. Isolation and Characterization of Methane-utilizing Yeasts. Microbiology 1979, 114, 187–194. [Google Scholar] [CrossRef]
- Rédou, V.; Ciobanu, M.C.; Pachiadaki, M.G.; Edgcomb, V.; Alain, K.; Barbier, G.; Burgaud, G. In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol. Ecol. 2014, 90, 908–921. [Google Scholar] [CrossRef]
- Drake, H.; Ivarsson, M.; Bengtson, S.; Heim, C.; Siljeström, S.; Whitehouse, M.J.; Broman, C.; Belivanova, V.; Åström, M.E. Anaerobic consortia of fungi and sulfate reducing bacteria in deep granite fractures. Nat. Commun. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Mouton, M.; Postma, F.; Wilsenach, J.; Botha, A. Diversity and characterization of culturable fungi from marine sediment collected from St. Helena Bay, South Africa. Microb. Ecol. 2012, 64, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Shoun, H.; Kim, D.-H.; Uchiyama, H.; Sugiyama, J. Denitrification by fungi. FEMS Microbiol. Lett. 1992, 94, 277–281. [Google Scholar] [CrossRef]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Wurzbacher, C.; James, T.Y.; Kagami, M. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol. 2016, 19, 28–38. [Google Scholar] [CrossRef]
- Kagami, M.; de Bruin, A.; Ibelings, B.W.; Van Donk, E. Parasitic chytrids: Their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 2007, 578, 113–129. [Google Scholar] [CrossRef]
- Tillmann, U.; Hesse, K.-J.; Tillmann, A. Large-scale parasitic infection of diatoms in the Northfrisian Wadden Sea. J. Sea Res. 1999, 42, 255–261. [Google Scholar] [CrossRef]
- Gleason, F.H.; Lilje, O.; Marano, A.V.; Sime-Ngando, T.; Sullivan, B.K.; Kirchmair, M.; Neuhauser, S. Ecological functions of zoosporic hyperparasites. Front. Microbiol. 2014, 5, 244. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, X.; Zheng, P.; Zou, S.; Li, G.; Gong, J. Distinct seasonality of chytrid-dominated benthic fungal communities in the neritic oceans (Bohai Sea and North Yellow Sea). Fungal Ecol. 2017, 30, 55–66. [Google Scholar] [CrossRef]
- Priest, T.; Fuchs, B.; Amann, R.; Reich, M. Diversity and biomass dynamics of unicellular marine fungi during a spring phytoplankton bloom. Environ. Microbiol. 2021, 23, 448–463. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Cunliffe, M. Coastal bacterioplankton community response to diatom-derived polysaccharide microgels. Environ. Microbiol. Rep. 2017, 9, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Agha, R.; Saebelfeld, M.; Manthey, C.; Rohrlack, T.; Wolinska, J. Chytrid parasitism facilitates trophic transfer between bloom-forming cyanobacteria and zooplankton (Daphnia). Sci. Rep. 2016, 6, 35039. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Wichels, A.; Panzer, K.; Krause, E.; Gimenez, L.; Gerdts, G. Impacts of a reduction in seawater pH mimicking ocean acidification on the structure and diversity of mycoplankton communities. Aquat. Microb. Ecol. 2017, 79, 221–223. [Google Scholar] [CrossRef]
- Frenken, T.; Alacid, E.; Berger, S.A.; Bourne, E.C.; Gerphagnon, M.; Grossart, H.-P.; Gsell, A.S.; Ibelings, B.W.; Kagami, M.; Küpper, F.C.; et al. Integrating chytrid fungal parasites into plankton ecology: Research gaps and needs. Environ. Microbiol. 2017, 19, 3802–3822. [Google Scholar] [CrossRef]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.M.; Wang, X.G.; Cheng, X.L.; Guo, J.J.; Bian, X.M.; Cai, L. Fungal communities in sediments of subtropical Chinese seas as estimated by DNA metabarcoding. Sci. Rep. 2016, 6, 26528. [Google Scholar] [CrossRef]
- Rojas-Jimenez, K.; Wurzbacher, C.; Bourne, E.C.; Chiuchiolo, A.; Priscu, J.C.; Grossart, H.-P. Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice-covered lakes of the McMurdo Dry Valleys, Antarctica. Sci. Rep. 2017, 7, 15348. [Google Scholar] [CrossRef]
- Kumar, V.; Sarma, V.V.; Thambugala, K.M.; Huang, J.-J.; Li, X.-Y.; Hao, G.-F. Ecology and Evolution of Marine Fungi With Their Adaptation to Climate Change. Front. Microbiol. 2021, 12, 719000. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Ramakrishna, S.; Vikineswary, S.; Das, D.; Bahkali, A.H.; Guo, S.-Y.; Pang, K.-L. How Do Fungi Survive in the Sea and Respond to Climate Change? J. Fungi 2022, 8, 291. [Google Scholar] [CrossRef]
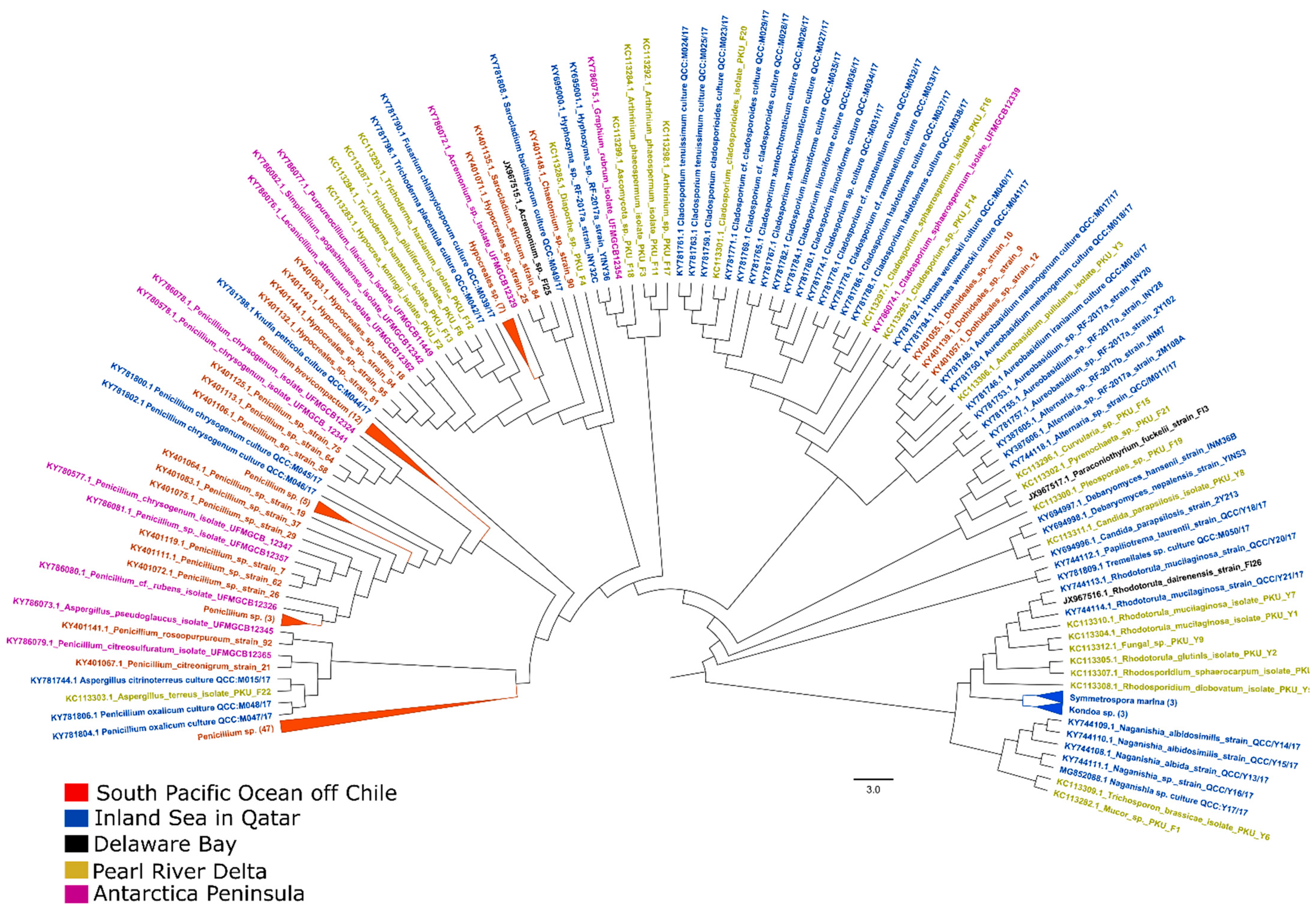
| Terrestrial Fungi | Method | Sampling Region | References |
|---|---|---|---|
| Aspergillus, Penicillium, Cladosporium, Fusarium, Sagenomella, Exophiala, Tilletiopsis, | Culture-based | Central India basin | [10,11] |
| Fusarium, Aspergillus, Phoma, Cladosporium, Mortierella, Sebacina, Alternaria | 454 pyrosequencing | Kongsfjorden (Svalbard, High Arctic) | [12] |
| Fusarium, Acremonium, Penicillium, Aspergillus Cladosporium Rhodotorulla, Paecilomyces, Exophiala, Meyerozyma | Culture-based | Canterbury Basin sediments, New Zealand | [13] |
| Malassezia | RNA- based clone library | Peru | [14] |
| Thelephoraceae, Trichophaea | Illumina MiSeq sequencing | Southwest India Ridge | [15] |
| Mycorrhizal fungi (Ambispora, Claroideoglomus, Diversispora, Glomus, Funneliformis) | Illumina HiSeq sequencing | East China Sea | [16] |
| Malassezia, Nectria, Acremonium, Leptosphaeria, Candida and Clavispora | ITS-clone library | Hawaiian waters | [17] |
| Mortierellales | Illumina HiSeq sequencing | Bohai Sea water column | [18] |
| Method | Target Region | Primers | Number of OTUs | Phyla | Sampling Region | Reference |
|---|---|---|---|---|---|---|
| 454 Pyrosequencing | 18S (V4) | TAReuk454FWD1 and TAReukREV3 | 71 | Chytridiomycota and Dikarya* | European near-shore sites | [40] |
| 454 Pyrosequencing | 18S (V4) | TAReuk454FWD1 and TAReukREV3 | 23,263 seqs. | Chytridiomycota, Dikarya, and Cryptomycota | Arctic and temperate biomes | [60] |
| 454 Pyrosequencing | ITS | ITS1F and ITS4 | - | Coastal water: Chytrids (36%) Open ocean: Rhizophydiales (30%) | Tasman Sea, and East Australian Current | [61] |
| 454 Pyrosequencing | ITS1 | ITS1F and ITS2 | 3468 | Dikarya, Chytridiomycota, Mucroromycotina, and Cryptomycota | Dongchong Bay, China | [59] |
| Illumina HiSeq | ITS1 | ITS1F and ITS2 | 1483 | Dikarya, Chytridiomycota, Mucoromycota, and Cryptomycota | Bohai Sea | [18] |
| Illumina Hiseq | ITS | 528F and 706R | 91 | Dikarya, Glomeromycota, Chytridiomycota, and Cryptomycota | Mariana Trench | [62] |
| Illumina Hiseq | ITS2 | ITS3 and ITS4 | 8701 | Dikarya, Chytridiomycota, Glomeromycota, and Rozellomycota | East China Sea water column and sediments | [54] |
| Illumina Hiseq | ITS2: | ITS3 and ITS4 | 4028 | Dikarya, Chytridiomycota, and Mucoromycota | Western Pacific Ocean (Epi-Abyssopelagic zone) | [23] |
| Illumina MiSeq | ITS | ITS1F and ITS4 | 582 | Dikarya and Chytridiomycota | Plymouth, UK | [19] |
| Illumina Miseq | ITS | ITS1F and ITS4 | 2796 | Dikarya and Chytridiomycota, Glomeromycota, and Neocallimastigomycota | Piver’s Island Coastal Observatory (PICO), USA | [21] |
| Ion-Torrent | LSU | LR0R and EDF360R | 2305 | Ascomycota, Basidiomycota, and Chytridiomycota | Piver’s Island | [63] |
| Strongly Correlated Factors | Region | Ecological Implication | Reference |
|---|---|---|---|
| Chlorophyll a, temperature, phytoplankton biomass | Hawaiian coast | Spatial variations | [58] |
| Phytoplankton, nutrients (nitrate, phosphate, nitrite), and location | West Pacific Warm Pool | Organic matter decomposition | [64] |
| Chlorophyll a, organic matter, and warm conditions | Upwelling ecosystem off the coast of Central Chile | Organic matter decomposition | [38] |
| High nitrogen availability, reduced salinity, temperature, phytoplankton, organic matter | Coastal station off Plymouth | Temporal variations, niche differentiation, and host–parasite interactions | [19] |
| Salinity, temperature, oxygen, and nutrients | Tasman Sea, East Tasman Sea, and East Australian Current | Biogeochemical cycling and spatial variations | [61] |
| Depth, dissolved oxygen, and nitrate | Across the globe | Local environmental conditions govern assemblages | [73] |
| Temperature, salinity, nitrate, nitrite, ammonium, and phosphate | Coastal region Dongchong Bay | Fungi regulate phytoplankton bloom | [59] |
| Temperature, depth, salinity, riverine input, location | Upwelling ecosystem off the coast of Central Chile | Organic matter decomposition | [57] |
| Dissolved nitrogen, particulate phosphorous silicate, pH, salinity, chlorophyll a | Coastal water column | Spatial variations | [18] |
| Dissolved oxygen and depth | East China Sea water and sediments | Ocean currents govern assemblages | [54] |
| Temperature, pH, insolation, dissolved inorganic carbon | Waters of Piver’s Island Coastal Observatory (PICO) | Temporal variations | [21] |
| Depth, temperature, and dissolved oxygen | Epi- to abyssopelagic zones of the Western Pacific Ocean | Distinct zonation of assemblages in the water column | [23] |
| Salinity | Baltic Sea | Salinity threshold separates assemblages | [76] |
| Estimation Method for Fungi | Sampling Region | Fungal Abundance | Bacterial Abundance | Reference |
|---|---|---|---|---|
| Biomass carbon | Coastal Chile | 0.03–6 μg C L−1 | - | [38] |
| Biomass carbon | Coastal Chile | 0.01–40 μg C L−1 | 10–44 μg C L−1 | [82] |
| Fatty Acid (18:2ω6) | Coastal Chile | 0.1–3 μg L−1 | 10–44 μg C L−1 | [82] |
| Ergosterol | Arctic waters | 1.02 μg C L−1 | 5 to >25 μg C L−1 | [24,83] |
| qPCR (DNA concentration) | West Pacific Warm Pool | Basidiomycota (max. 10 ng μL−1, open-ocean station) Ascomycota (max. 14 pg μL−1, coastal station) | ~10 ng μL−1 | [64] |
| qPCR (18S rRNA gene copy number) | Coastal Plymouth, Western English Channel | 5.1 × 105 to 9.9 × 107 copies L−1 | 0.2 × 106–1.6 × 106 cells mL−1 | [19,84] |
| qPCR (18S rRNA gene copy number) | Coastal region, Bohai Sea | 4.28 × 106 to 1.13 × 107 copies L−1 | ~ 2 × 106 cells L−1 | [18] |
| qPCR (18S rRNA gene copy number) | PICO | 1.0 × 107 to 7.54 × 108 copies L−1 | - | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sen, K.; Sen, B.; Wang, G. Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments. J. Fungi 2022, 8, 491. https://doi.org/10.3390/jof8050491
Sen K, Sen B, Wang G. Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments. Journal of Fungi. 2022; 8(5):491. https://doi.org/10.3390/jof8050491
Chicago/Turabian StyleSen, Kalyani, Biswarup Sen, and Guangyi Wang. 2022. "Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments" Journal of Fungi 8, no. 5: 491. https://doi.org/10.3390/jof8050491
APA StyleSen, K., Sen, B., & Wang, G. (2022). Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments. Journal of Fungi, 8(5), 491. https://doi.org/10.3390/jof8050491








