Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity
Abstract
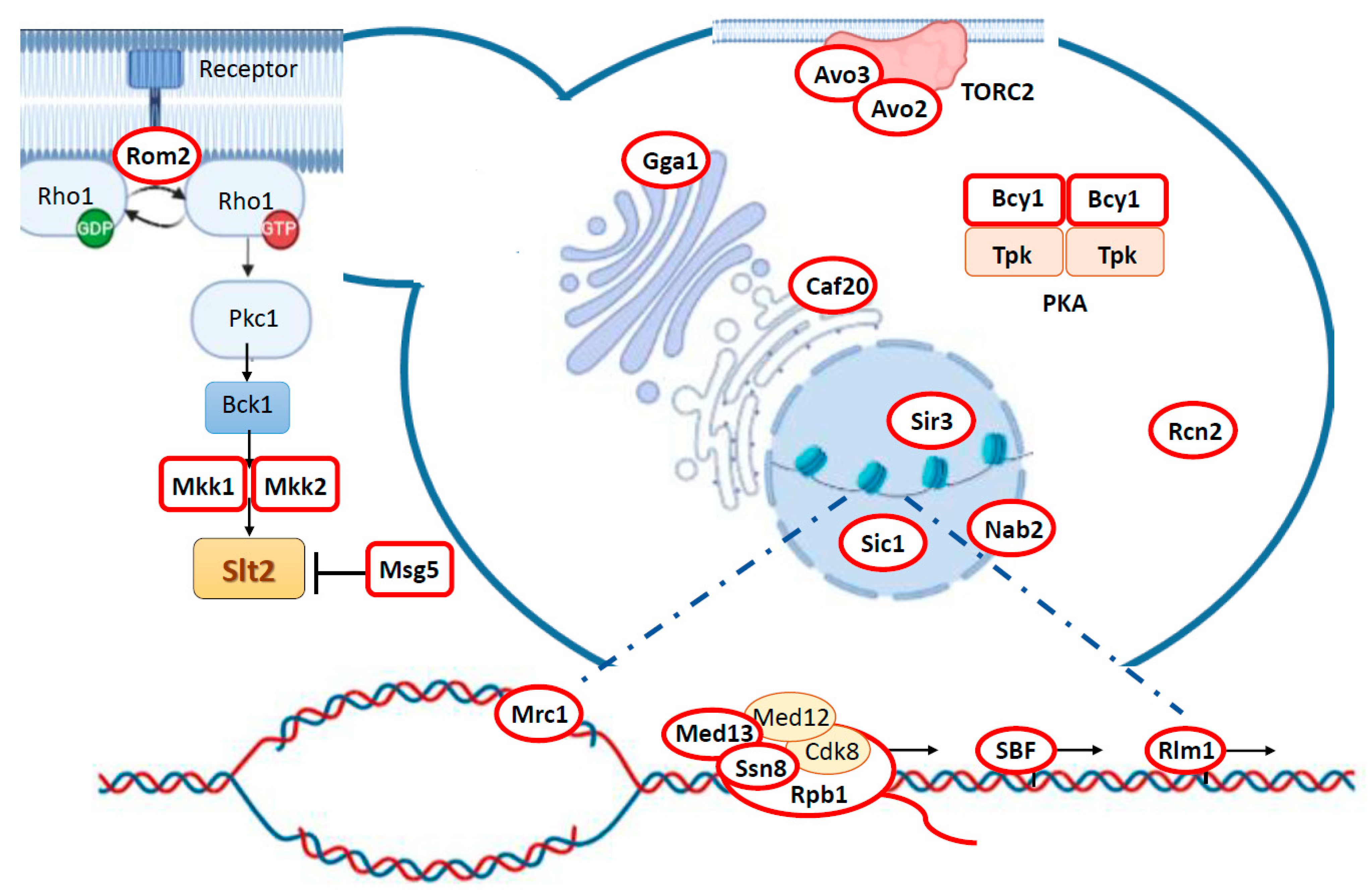
1. Cell Wall Integrity Pathway: An Introductory View
2. Substrate Fishing: Methods Used for Searching Slt2 Targets
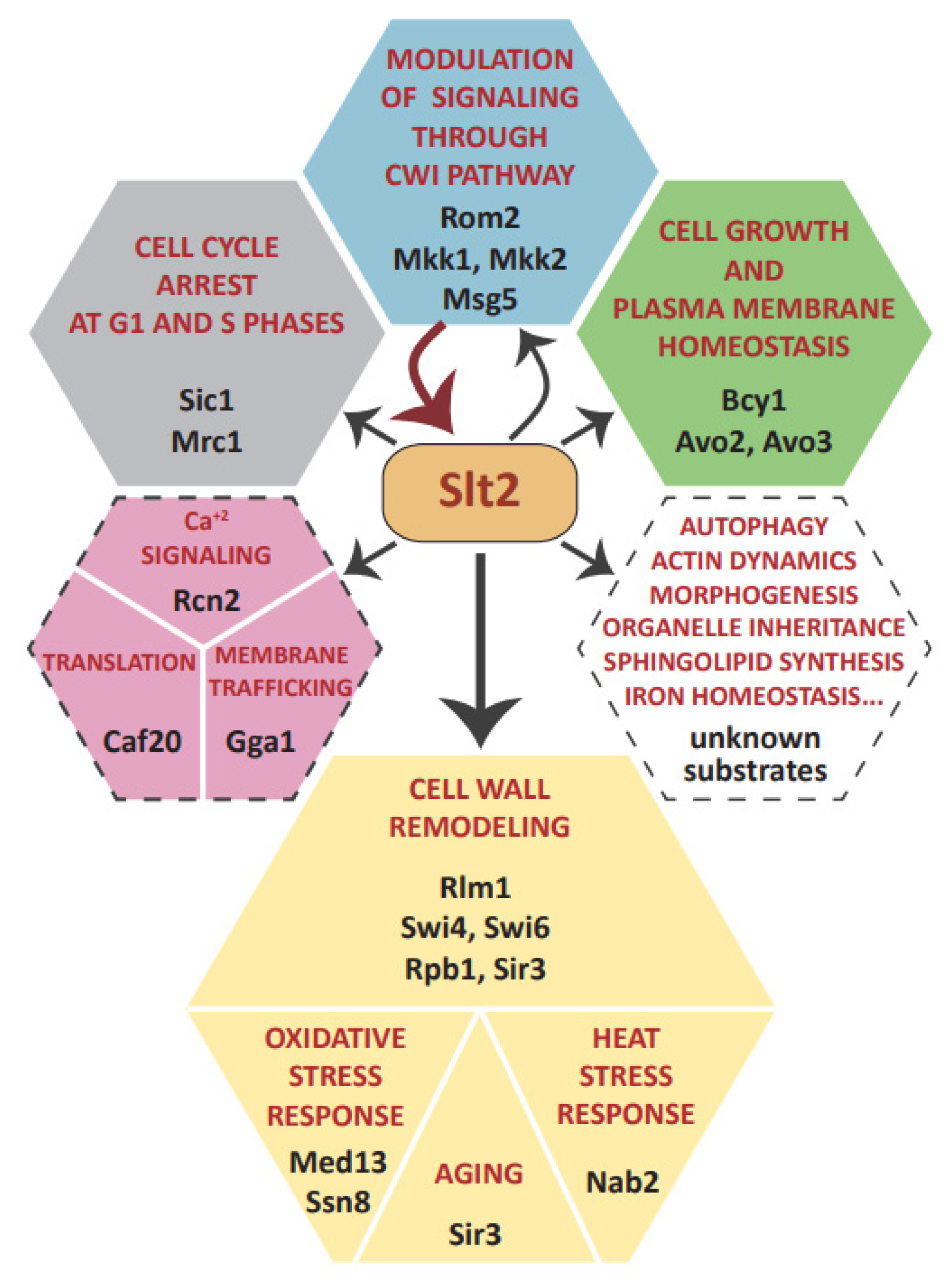
3. Targeting Different Yeast Processes: Genuine Slt2 Substrates
3.1. Feedback Regulation of the CWI Pathway: Rom2, Mkk1/2, and Msg5
3.2. Slt2 Impinges on Central Yeast Signaling Pathways via Bcy1 and Avo2/3
3.3. Cell Wall Stress-Related Gene Transcription: Rlm1 and SBF Complex
3.4. Regulation of RNA Polymerase Holoenzyme Complex: Cyclin C, Med13, and Rbp1
3.5. Epigenetic Control of Gene Expression and Yeast Life Span Extension: Sir3 Phosphorylation
3.6. Regulation of mRNA Nuclear Export: Nab2
3.7. Control of Cell Cycle: Sic1 and Mrc1
3.8. Orphan Slt2 Substrates: Caf20, Rcn2, and Gga1
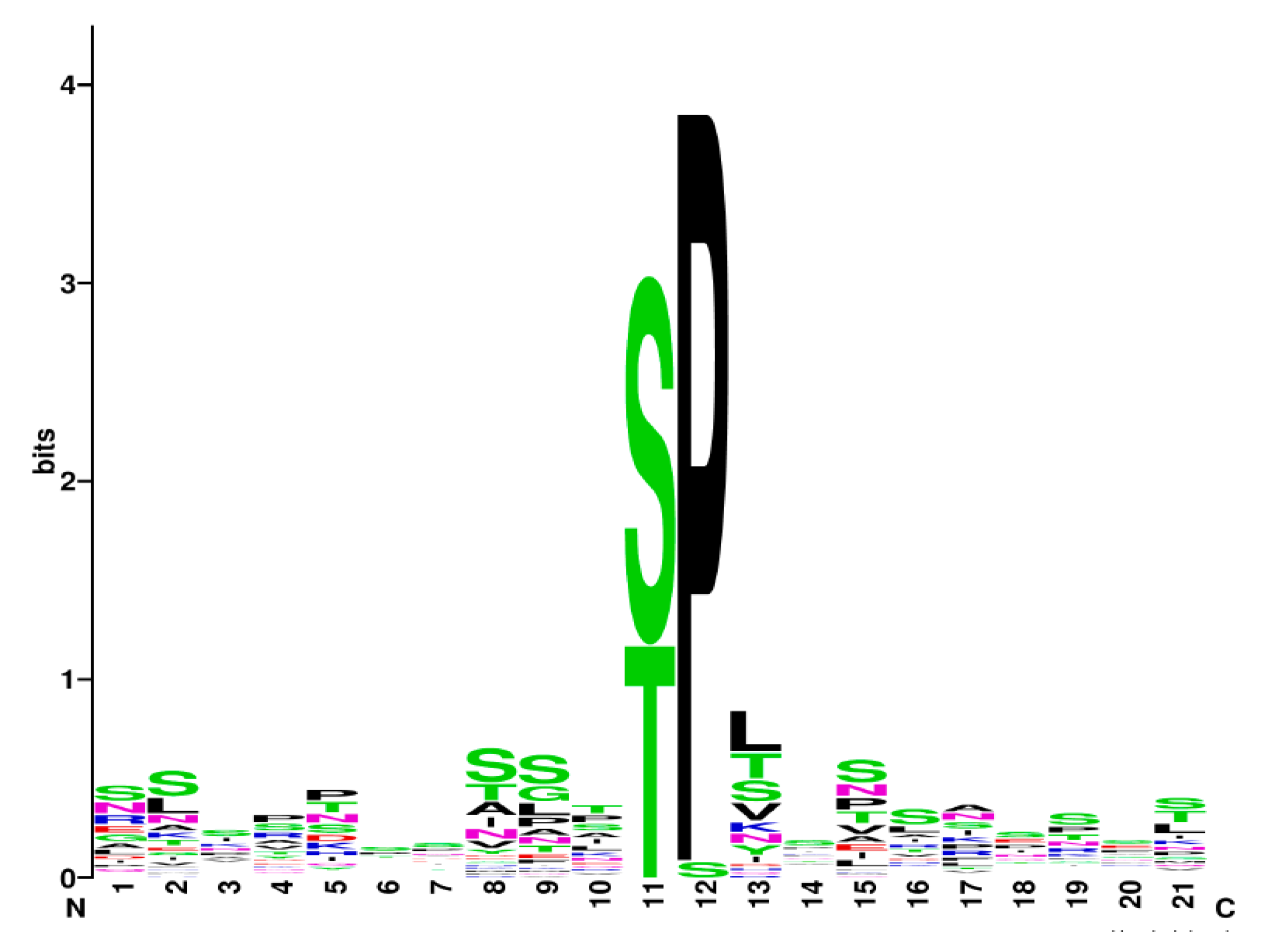
3.9. Is There a Specific Slt2 Phosphorylation Signature?
4. Candidate Slt2 Substrates: A Growing List
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kock, C.; Dufrêne, Y.F.; Heinisch, J.J. Up against the wall: Is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl. Environ. Microbiol. 2015, 81, 806–811. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Chen, R.E.; Thorner, J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1773, 1311–1340. [Google Scholar] [CrossRef]
- Schmelzle, T.; Helliwell, S.B.; Hall, M.N. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell Biol. 2002, 22, 1329–1339. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Rodicio, R. Protein kinase C in fungi-more than just cell wall integrity. FEMS Microbiol. Rev. 2018, 42, fux051. [Google Scholar] [CrossRef]
- Roelants, F.M.; Leskoske, K.L.; Martinez Marshall, M.N.; Locke, M.N.; Thorner, J. The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae. Biomolecules 2017, 7, 66. [Google Scholar] [CrossRef]
- Inagaki, M.; Schmelzle, T.; Yamaguchi, K.; Irie, K.; Hall, M.N.; Matsumoto, K. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol. Cell Biol. 1999, 19, 8344–8352. [Google Scholar] [CrossRef]
- Martín, H.; Flandez, M.; Nombela, C.; Molina, M. Protein phosphatases in MAPK signalling: We keep learning from yeast. Mol. Microbiol. 2005, 58, 6–16. [Google Scholar] [CrossRef]
- Tatjer, L.; Sacristán-Reviriego, A.; Casado, C.; González, A.; Rodríguez-Porrata, B.; Palacios, L.; Canadell, D.; Serra-Cardona, A.; Martín, H.; Molina, M.; et al. Wide-Ranging Effects of the Yeast Ptc1 Protein Phosphatase Acting Through the MAPK Kinase Mkk1. Genetics 2016, 202, 141–156. [Google Scholar] [CrossRef]
- González-Rubio, G.; Fernández-Acero, T.; Martín, H.; Molina, M. Mitogen-Activated Protein Kinase Phosphatases (MKPs) in Fungal Signaling: Conservation, Function, and Regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef]
- Molina, M.; Cid, V.J.; Martín, H. Fine regulation of Saccharomyces cerevisiae MAPK pathways by post-translational modifications. Yeast 2010, 27, 503–511. [Google Scholar] [CrossRef]
- Audhya, A.; Emr, S.D. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2002, 2, 593–605. [Google Scholar] [CrossRef]
- Millson, S.H.; Truman, A.W.; King, V.; Prodromou, C.; Pearl, L.H.; Piper, P.W. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 2005, 4, 849–860. [Google Scholar] [CrossRef]
- Lee, J.; Liu, L.; Levin, D.E. Stressing out or stressing in: Intracellular pathways for SAPK activation. Curr. Genet. 2019, 65, 417–421. [Google Scholar] [CrossRef]
- Jiménez-Gutiérrez, E.; Alegría-Carrasco, E.; Sellers-Moya, A.; Molina, M.; Martín, H. Not just the wall: The other ways to turn the yeast CWI pathway on. Int. Microbiol. 2020, 23, 107–119. [Google Scholar] [CrossRef]
- Martín, H.; Rodríguez-Pachón, J.M.; Ruiz, C.; Nombela, C.; Molina, M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 1511–1519. [Google Scholar] [CrossRef]
- Liu, L.; Levin, D.E. Intracellular mechanism by which genotoxic stress activates yeast SAPK Mpk1. Mol. Biol. Cell 2018, 29, 2898–2909. [Google Scholar] [CrossRef]
- González-Rubio, G.; Sellers-Moya, Á.; Martín, H.; Molina, M. Differential Role of Threonine and Tyrosine Phosphorylation in the Activation and Activity of the Yeast MAPK Slt2. Int. J. Mol. Sci. 2021, 22, 1110. [Google Scholar] [CrossRef]
- Ahmadpour, D.; Maciaszczyk-Dziubinska, E.; Babazadeh, R.; Dahal, S.; Migocka, M.; Andersson, M.; Wysocki, R.; Tamás, M.J.; Hohmann, S. The mitogen-activated protein kinase Slt2 modulates arsenite transport through the aquaglyceroporin Fps1. FEBS Lett. 2016, 590, 3649–3659. [Google Scholar] [CrossRef]
- Pujol-Carrion, N.; Pavón-Vergés, M.; Arroyo, J.; de la Torre-Ruiz, M.A. The MAPK Slt2/Mpk1 plays a role in iron homeostasis through direct regulation of the transcription factor Aft1. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118974. [Google Scholar] [CrossRef]
- Quilis, I.; Gomar-Alba, M.; Igual, J.C. The CWI Pathway: A Versatile Toolbox to Arrest Cell-Cycle Progression. J. Fungi 2021, 7, 1041. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Klionsky, D.J. MAPKs regulate mitophagy in Saccharomyces cerevisiae. Autophagy 2011, 7, 1564–1565. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Walker, L.; Novick, P.; Ferro-Novick, S. Ptc1p regulates cortical ER inheritance via Slt2p. Embo J. 2006, 25, 4413–4422. [Google Scholar] [CrossRef]
- Li, X.; Du, Y.; Siegel, S.; Ferro-Novick, S.; Novick, P. Activation of the mitogen-activated protein kinase, Slt2p, at bud tips blocks a late stage of endoplasmic reticulum inheritance in Saccharomyces cerevisiae. Mol. Biol. Cell 2010, 21, 1772–1782. [Google Scholar] [CrossRef][Green Version]
- Guo, S.; Shen, X.; Yan, G.; Ma, D.; Bai, X.; Li, S.; Jiang, Y. A MAP kinase dependent feedback mechanism controls Rho1 GTPase and actin distribution in yeast. PLoS ONE 2009, 4, e6089. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Sanz, A.B.; Rodríguez-Peña, J.M.; Nombela, C.; Arroyo, J. Rlm1 mediates positive autoregulatory transcriptional feedback that is essential for Slt2-dependent gene expression. J. Cell Sci. 2016, 129, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Gutiérrez, E.; Alegría-Carrasco, E.; Alonso-Rodríguez, E.; Fernández-Acero, T.; Molina, M.; Martín, H. Rewiring the yeast cell wall integrity (CWI) pathway through a synthetic positive feedback circuit unveils a novel role for the MAPKKK Ssk2 in CWI pathway activation. FEBS J. 2020, 287, 4881–4901. [Google Scholar] [CrossRef]
- Soulard, A.; Cremonesi, A.; Moes, S.; Schütz, F.; Jenö, P.; Hall, M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 2010, 21, 3475–3486. [Google Scholar] [CrossRef]
- Leskoske, K.L.; Roelants, F.M.; Emmerstorfer-Augustin, A.; Augustin, C.M.; Si, E.P.; Hill, J.M.; Thorner, J. Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev. 2018, 32, 1576–1590. [Google Scholar] [CrossRef]
- Kim, K.Y.; Levin, D.E. Transcriptional reporters for genes activated by cell wall stress through a non-catalytic mechanism involving Mpk1 and SBF. Yeast 2010, 27, 541–548. [Google Scholar] [CrossRef]
- Kim, K.Y.; Truman, A.W.; Levin, D.E. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell Biol. 2008, 28, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. ERK1/2 MAP kinases: Structure, function, and regulation. Pharm. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef] [PubMed]
- Carlson, S.M.; Chouinard, C.R.; Labadorf, A.; Lam, C.J.; Schmelzle, K.; Fraenkel, E.; White, F.M. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci. Signal. 2011, 4, rs11. [Google Scholar] [CrossRef]
- Mok, J.; Kim, P.M.; Lam, H.Y.; Piccirillo, S.; Zhou, X.; Jeschke, G.R.; Sheridan, D.L.; Parker, S.A.; Desai, V.; Jwa, M.; et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 2010, 3, ra12. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Beltrao, P. Evolution of protein kinase substrate recognition at the active site. PLoS Biol. 2019, 17, e3000341. [Google Scholar] [CrossRef]
- Tanoue, T.; Nishida, E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 2002, 93, 193–202. [Google Scholar] [CrossRef]
- Garai, Á.; Zeke, A.; Gógl, G.; Törő, I.; Fördős, F.; Blankenburg, H.; Bárkai, T.; Varga, J.; Alexa, A.; Emig, D.; et al. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci. Signal. 2012, 5, ra74. [Google Scholar] [CrossRef]
- Tanoue, T.; Adachi, M.; Moriguchi, T.; Nishida, E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000, 2, 110–116. [Google Scholar] [CrossRef]
- Chang, C.I.; Xu, B.E.; Akella, R.; Cobb, M.H.; Goldsmith, E.J. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 2002, 9, 1241–1249. [Google Scholar] [CrossRef]
- Zeke, A.; Bastys, T.; Alexa, A.; Garai, Á.; Mészáros, B.; Kirsch, K.; Dosztányi, Z.; Kalinina, O.V.; Reményi, A. Systematic discovery of linear binding motifs targeting an ancient protein interaction surface on MAP kinases. Mol. Syst. Biol. 2015, 11, 837. [Google Scholar] [CrossRef]
- Akella, R.; Moon, T.M.; Goldsmith, E.J. Unique MAP Kinase binding sites. Biochim. Biophys. Acta 2008, 1784, 48–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- González-Rubio, G.; Sellers-Moya, Á.; Martín, H.; Molina, M. A walk-through MAPK structure and functionality with the 30-year-old yeast MAPK Slt2. Int. Microbiol. 2021, 24, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Irie, K.; Matsumoto, K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell Biol. 1995, 15, 5740–5749. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Takaesu, G.; Hagiwara, M.; Irie, K.; Matsumoto, K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell Biol. 1997, 17, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Quinn, C.M.; Kwak, S.; Talanian, R.V. Current in vitro kinase assay technologies: The quest for a universal format. Curr. Drug Discov. Technol. 2008, 5, 59–69. [Google Scholar] [CrossRef]
- Hastie, C.J.; McLauchlan, H.J.; Cohen, P. Assay of protein kinases using radiolabeled ATP: A protocol. Nat. Protoc. 2006, 1, 968–971. [Google Scholar] [CrossRef]
- Mok, J.; Im, H.; Snyder, M. Global identification of protein kinase substrates by protein microarray analysis. Nat. Protoc. 2009, 4, 1820–1827. [Google Scholar] [CrossRef]
- Shah, K.; Shokat, K.M. A chemical genetic approach for the identification of direct substrates of protein kinases. Methods Mol. Biol. 2003, 233, 253–271. [Google Scholar]
- Hertz, N.T.; Wang, B.T.; Allen, J.J.; Zhang, C.; Dar, A.C.; Burlingame, A.L.; Shokat, K.M. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr. Protoc. Chem. Biol. 2010, 2, 15–36. [Google Scholar] [CrossRef]
- Alonso-Rodríguez, E.; Fernández-Pinar, P.; Sacristán-Reviriego, A.; Molina, M.; Martín, H. An Analog-sensitive Version of the Protein Kinase Slt2 Allows Identification of Novel Targets of the Yeast Cell Wall Integrity Pathway. J. Biol. Chem. 2016, 291, 5461–5472. [Google Scholar] [CrossRef]
- Koch, A.; Hauf, S. Strategies for the identification of kinase substrates using analog-sensitive kinases. Eur J. Cell Biol. 2010, 89, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Uezato, Y. Analysis of protein kinases by Phos-tag SDS-PAGE. J. Proteom. 2022, 255, 104485. [Google Scholar] [CrossRef] [PubMed]
- von Stechow, L.; Francavilla, C.; Olsen, J.V. Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev. Proteom. 2015, 12, 469–487. [Google Scholar] [CrossRef] [PubMed]
- Mascaraque, V.; Hernaez, M.L.; Jimenez-Sanchez, M.; Hansen, R.; Gil, C.; Martin, H.; Cid, V.J.; Molina, M. Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol. Cell Proteom. 2013, 12, 557–574. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.; Tian, R.; Lee, R.E.; Wang, F.; Beauvais, A.; Zou, H.; Megeney, L.A.; Gingras, A.C.; Pawson, T.; Figeys, D.; et al. A novel whole-cell lysate kinase assay identifies substrates of the p38 MAPK in differentiating myoblasts. Skelet Muscle 2012, 2, 5. [Google Scholar] [CrossRef]
- van Drogen, F.; Peter, M. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 2002, 12, 1698–1703. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, M.; Cid, V.J.; Molina, M. Retrophosphorylation of Mkk1 and Mkk2 MAPKKs by the Slt2 MAPK in the yeast cell integrity pathway. J. Biol. Chem. 2007, 282, 31174–31185. [Google Scholar] [CrossRef]
- Flández, M.; Cosano, I.C.; Nombela, C.; Martín, H.; Molina, M. Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 2004, 279, 11027–11034. [Google Scholar] [CrossRef]
- Marín, M.J.; Flández, M.; Bermejo, C.; Arroyo, J.; Martín, H.; Molina, M. Different modulation of the outputs of yeast MAPK-mediated pathways by distinct stimuli and isoforms of the dual-specificity phosphatase Msg5. Mol. Genet. Genom. 2009, 281, 345–359. [Google Scholar] [CrossRef]
- Madden, K.; Sheu, Y.J.; Baetz, K.; Andrews, B.; Snyder, M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 1997, 275, 1781–1784. [Google Scholar] [CrossRef]
- Kim, K.Y.; Truman, A.W.; Caesar, S.; Schlenstedt, G.; Levin, D.E. Yeast Mpk1 cell wall integrity mitogen-activated protein kinase regulates nucleocytoplasmic shuttling of the Swi6 transcriptional regulator. Mol. Biol. Cell 2010, 21, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Strich, R.; Cooper, K.F. Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress. Mol. Biol. Cell 2014, 25, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Krasley, E.; Cooper, K.F.; Mallory, M.J.; Dunbrack, R.; Strich, R. Regulation of the oxidative stress response through Slt2p-dependent destruction of cyclin C in Saccharomyces cerevisiae. Genetics 2006, 172, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Stieg, D.C.; Willis, S.D.; Ganesan, V.; Ong, K.L.; Scuorzo, J.; Song, M.; Grose, J.; Strich, R.; Cooper, K.F. A complex molecular switch directs stress-induced cyclin C nuclear release through SCF(Grr1)-mediated degradation of Med13. Mol. Biol. Cell 2018, 29, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Yurko, N.; Liu, X.; Yamazaki, T.; Hoque, M.; Tian, B.; Manley, J.L. MPK1/SLT2 Links Multiple Stress Responses with Gene Expression in Budding Yeast by Phosphorylating Tyr1 of the RNAP II CTD. Mol. Cell 2017, 68, 913–925.e3. [Google Scholar] [CrossRef]
- Ai, W.; Bertram, P.G.; Tsang, C.K.; Chan, T.F.; Zheng, X.F. Regulation of subtelomeric silencing during stress response. Mol. Cell 2002, 10, 1295–1305. [Google Scholar] [CrossRef]
- Ray, A.; Hector, R.E.; Roy, N.; Song, J.H.; Berkner, K.L.; Runge, K.W. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 2003, 33, 522–526. [Google Scholar] [CrossRef]
- Carmody, S.R.; Tran, E.J.; Apponi, L.H.; Corbett, A.H.; Wente, S.R. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol. Cell Biol. 2010, 30, 5168–5179. [Google Scholar] [CrossRef]
- Duch, A.; Canal, B.; Barroso, S.I.; García-Rubio, M.; Seisenbacher, G.; Aguilera, A.; de Nadal, E.; Posas, F. Multiple signaling kinases target Mrc1 to prevent genomic instability triggered by transcription-replication conflicts. Nat. Commun. 2018, 9, 379. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; De Virgilio, C. TORC1 controls G1-S cell cycle transition in yeast via Mpk1 and the greatwall kinase pathway. Nat. Commun. 2015, 6, 8256. [Google Scholar] [CrossRef]
- Lee, K.S.; Levin, D.E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell Biol. 1992, 12, 172–182. [Google Scholar]
- Ozaki, K.; Tanaka, K.; Imamura, H.; Hihara, T.; Kameyama, T.; Nonaka, H.; Hirano, H.; Matsuura, Y.; Takai, Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996, 15, 2196–2207. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Padmanabha, R.; Snyder, M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 1997, 8, 1829–1844. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Takematsu, H.; Yamaji, T.; Hiramoto, S.; Kozutsumi, Y. Disturbance of sphingolipid biosynthesis abrogates the signaling of Mss4, phosphatidylinositol-4-phosphate 5-kinase, in yeast. J. Biol. Chem. 2005, 280, 18087–18094. [Google Scholar] [CrossRef] [PubMed]
- Yamochi, W.; Tanaka, K.; Nonaka, H.; Maeda, A.; Musha, T.; Takai, Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J. Cell Biol. 1994, 125, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.J.; Tuch, B.B.; Villén, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef] [PubMed]
- Saccharomyces Genome Database. Available online: https://www.yeastgenome.org/ (accessed on 14 February 2022).
- Soler, M.; Plovins, A.; Martín, H.; Molina, M.; Nombela, C. Characterization of domains in the yeast MAP kinase Slt2 (Mpk1) required for functional activity and in vivo interaction with protein kinases Mkk1 and Mkk2. Mol. Microbiol. 1995, 17, 833–842. [Google Scholar] [CrossRef]
- Lee, K.S.; Irie, K.; Gotoh, Y.; Watanabe, Y.; Araki, H.; Nishida, E.; Matsumoto, K.; Levin, D.E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell Biol. 1993, 13, 3067–3075. [Google Scholar]
- Irie, K.; Takase, M.; Lee, K.S.; Levin, D.E.; Araki, H.; Matsumoto, K.; Oshima, Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell Biol. 1993, 13, 3076–3083. [Google Scholar]
- Errede, B.; Gartner, A.; Zhou, Z.; Nasmyth, K.; Ammerer, G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 1993, 362, 261–264. [Google Scholar] [CrossRef]
- Zhou, Z.; Gartner, A.; Cade, R.; Ammerer, G.; Errede, B. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell Biol. 1993, 13, 2069–2080. [Google Scholar] [PubMed]
- Buscà, R.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Front. Cell Dev. Biol. 2016, 4, 53. [Google Scholar] [CrossRef]
- Katagiri, C.; Masuda, K.; Urano, T.; Yamashita, K.; Araki, Y.; Kikuchi, K.; Shima, H. Phosphorylation of Ser-446 determines stability of MKP-7. J. Biol. Chem. 2005, 280, 14716–14722. [Google Scholar] [CrossRef]
- Brondello, J.M.; Pouysségur, J.; McKenzie, F.R. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 1999, 286, 2514–2517. [Google Scholar] [CrossRef] [PubMed]
- Sohaskey, M.L.; Ferrell, J.E., Jr. Activation of p42 mitogen-activated protein kinase (MAPK), but not c-Jun NH(2)-terminal kinase, induces phosphorylation and stabilization of MAPK phosphatase XCL100 in Xenopus oocytes. Mol. Biol. Cell 2002, 13, 454–468. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Portela, P.; Rossi, S. cAMP-PKA signal transduction specificity in Saccharomyces cerevisiae. Curr. Genet. 2020, 66, 1093–1099. [Google Scholar] [CrossRef]
- Santangelo, G.M. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 253–282. [Google Scholar] [CrossRef]
- Johnson, K.E.; Cameron, S.; Toda, T.; Wigler, M.; Zoller, M.J. Expression in Escherichia coli of BCY1, the regulatory subunit of cyclic AMP-dependent protein kinase from Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 1987, 262, 8636–8642. [Google Scholar] [CrossRef]
- Kuret, J.; Johnson, K.E.; Nicolette, C.; Zoller, M.J. Mutagenesis of the regulatory subunit of yeast cAMP-dependent protein kinase. Isolation of site-directed mutants with altered binding affinity for catalytic subunit. J. Biol. Chem. 1988, 263, 9149–9154. [Google Scholar] [CrossRef]
- Griffioen, G.; Branduardi, P.; Ballarini, A.; Anghileri, P.; Norbeck, J.; Baroni, M.D.; Ruis, H. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell Biol. 2001, 21, 511–523. [Google Scholar] [CrossRef]
- Griffioen, G.; Swinnen, S.; Thevelein, J.M. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J. Biol. Chem. 2003, 278, 23460–23471. [Google Scholar] [CrossRef] [PubMed]
- Searle, J.S.; Wood, M.D.; Kaur, M.; Tobin, D.V.; Sanchez, Y. Proteins in the nutrient-sensing and DNA damage checkpoint pathways cooperate to restrain mitotic progression following DNA damage. PLoS Genet. 2011, 7, e1002176. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.N.; Thorner, J. Regulation of TORC2 function and localization by Rab5 GTPases in Saccharomyces cerevisiae. Cell Cycle 2019, 18, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Turjanski, A.G.; Vaqué, J.P.; Gutkind, J.S. MAP kinases and the control of nuclear events. Oncogene 2007, 26, 3240–3253. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Pavón-Vergés, M.; Rodríguez-Peña, J.M.; Arroyo, J. Control of Gene Expression via the Yeast CWI Pathway. Int. J. Mol. Sci. 2022, 23, 1791. [Google Scholar] [CrossRef]
- García, R.; Bermejo, C.; Grau, C.; Pérez, R.; Rodríguez-Peña, J.M.; Francois, J.; Nombela, C.; Arroyo, J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004, 279, 15183–15195. [Google Scholar] [CrossRef]
- Jung, U.S.; Sobering, A.K.; Romeo, M.J.; Levin, D.E. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 2002, 46, 781–789. [Google Scholar] [CrossRef]
- Sanz, A.B.; Garcia, R.; Rodriguez-Pena, J.M.; Nombela, C.; Arroyo, J. Slt2 MAPK association with chromatin is required for transcriptional activation of Rlm1 dependent genes upon cell wall stress. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1029–1039. [Google Scholar] [CrossRef]
- Haase, S.B.; Wittenberg, C. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 2014, 196, 65–90. [Google Scholar] [CrossRef]
- Baetz, K.; Andrews, B. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell Biol. 1999, 19, 6729–6741. [Google Scholar] [CrossRef][Green Version]
- Igual, J.C.; Johnson, A.L.; Johnston, L.H. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996, 15, 5001–5013. [Google Scholar] [CrossRef] [PubMed]
- Truman, A.W.; Kim, K.Y.; Levin, D.E. Mechanism of Mpk1 mitogen-activated protein kinase binding to the Swi4 transcription factor and its regulation by a novel caffeine-induced phosphorylation. Mol. Cell Biol. 2009, 29, 6449–6461. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Mediator and the mechanism of transcriptional activation. Trends BioChem. Sci. 2005, 30, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Harper, T.M.; Taatjes, D.J. The complex structure and function of Mediator. J. Biol. Chem. 2018, 293, 13778–13785. [Google Scholar] [CrossRef] [PubMed]
- Ježek, J.; Smethurst, D.G.J.; Stieg, D.C.; Kiss, Z.A.C.; Hanley, S.E.; Ganesan, V.; Chang, K.T.; Cooper, K.F.; Strich, R. Cyclin C: The Story of a Non-Cycling Cyclin. Biology 2019, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.F.; Mallory, M.J.; Smith, J.B.; Strich, R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 1997, 16, 4665–4675. [Google Scholar] [CrossRef]
- Cooper, K.F.; Mallory, M.J.; Strich, R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell Biol. 1999, 19, 3338–3348. [Google Scholar] [CrossRef]
- Cooper, K.F.; Khakhina, S.; Kim, S.K.; Strich, R. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev. Cell 2014, 28, 161–173. [Google Scholar] [CrossRef]
- Cooper, K.F.; Scarnati, M.S.; Krasley, E.; Mallory, M.J.; Jin, C.; Law, M.J.; Strich, R. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 2012, 125, 1015–1026. [Google Scholar] [CrossRef]
- Levin-Salomon, V.; Maayan, I.; Avrahami-Moyal, L.; Marbach, I.; Livnah, O.; Engelberg, D. When expressed in yeast, mammalian mitogen-activated protein kinases lose proper regulation and become spontaneously phosphorylated. BioChem. J. 2009, 417, 331–340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valencia, A.M.; Kadoch, C. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 2019, 21, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Gartenberg, M.R.; Smith, J.S. The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae. Genetics 2016, 203, 1563–1599. [Google Scholar] [CrossRef] [PubMed]
- Sauty, S.M.; Shaban, K.; Yankulov, K. Gene repression in S. cerevisiae-looking beyond Sir-dependent gene silencing. Curr. Genet. 2021, 67, 3–17. [Google Scholar] [CrossRef]
- Stone, E.M.; Pillus, L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 1996, 135, 571–583. [Google Scholar] [CrossRef]
- Viswanathan, M.; Muthukumar, G.; Cong, Y.S.; Lenard, J. Seripauperins of Saccharomyces cerevisiae: A new multigene family encoding serine-poor relatives of serine-rich proteins. Gene 1994, 148, 149–153. [Google Scholar] [CrossRef]
- Kothiwal, D.; Laloraya, S. A SIR-independent role for cohesin in subtelomeric silencing and organization. Proc. Natl. Acad. Sci. USA 2019, 116, 5659–5664. [Google Scholar] [CrossRef]
- Blasl, A.T.; Schulze, S.; Qin, C.; Graf, L.G.; Vogt, R.; Lammers, M. Post-translational lysine ac(et)ylation in health, ageing and disease. Biol. Chem. 2022, 403, 151–194. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Hecht, A.; Luo, K.; Grunstein, M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997, 11, 83–93. [Google Scholar] [CrossRef]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Krebber, H. Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1. Int. J. Mol. Sci. 2021, 22, 11275. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, C.; Tung, K.S.; Amberg, D.C.; Hopper, A.K.; Cole, C.N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996, 10, 1608–1620. [Google Scholar] [CrossRef] [PubMed]
- Zarnack, K.; Balasubramanian, S.; Gantier, M.P.; Kunetsky, V.; Kracht, M.; Schmitz, M.L.; Sträßer, K. Dynamic mRNP Remodeling in Response to Internal and External Stimuli. Biomolecules 2020, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Alpert, T.; Straube, K.; Carrillo Oesterreich, F.; Herzel, L.; Neugebauer, K.M. Widespread Transcriptional Readthrough Caused by Nab2 Depletion Leads to Chimeric Transcripts with Retained Introns. Cell Rep. 2020, 33, 108324. [Google Scholar] [CrossRef]
- Zinzalla, V.; Graziola, M.; Mastriani, A.; Vanoni, M.; Alberghina, L. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 2007, 63, 1482–1494. [Google Scholar] [CrossRef]
- Escoté, X.; Zapater, M.; Clotet, J.; Posas, F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004, 6, 997–1002. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Jaquenoud, M.; Péli-Gulli, M.P.; Nicastro, R.; De Virgilio, C. TORC1 coordinates the conversion of Sic1 from a target to an inhibitor of cyclin-CDK-Cks1. Cell Discov. 2017, 3, 17012. [Google Scholar] [CrossRef]
- Kono, K.; Al-Zain, A.; Schroeder, L.; Nakanishi, M.; Ikui, A.E. Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2016, 113, 6910–6915. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef]
- Uzunova, S.D.; Zarkov, A.S.; Ivanova, A.M.; Stoynov, S.S.; Nedelcheva-Veleva, M.N. The subunits of the S-phase checkpoint complex Mrc1/Tof1/Csm3: Dynamics and interdependence. Cell Div. 2014, 9, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Voordeckers, K.; Colding, C.; Grasso, L.; Pardo, B.; Hoes, L.; Kominek, J.; Gielens, K.; Dekoster, K.; Gordon, J.; Van der Zande, E.; et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 2020, 11, 3664. [Google Scholar] [CrossRef] [PubMed]
- Duch, A.; Felipe-Abrio, I.; Barroso, S.; Yaakov, G.; García-Rubio, M.; Aguilera, A.; de Nadal, E.; Posas, F. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 2013, 493, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Castelli, L.M.; Talavera, D.; Kershaw, C.J.; Mohammad-Qureshi, S.S.; Costello, J.L.; Rowe, W.; Sims, P.F.; Grant, C.M.; Hubbard, S.J.; Ashe, M.P.; et al. The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation. PLoS Genet. 2015, 11, e1005233. [Google Scholar] [CrossRef]
- Mehta, S.; Li, H.; Hogan, P.G.; Cunningham, K.W. Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol. Cell Biol. 2009, 29, 2777–2793. [Google Scholar] [CrossRef][Green Version]
- MacGilvray, M.E.; Shishkova, E.; Place, M.; Wagner, E.R.; Coon, J.J.; Gasch, A.P. Phosphoproteome Response to Dithiothreitol Reveals Unique Versus Shared Features of Saccharomyces cerevisiae Stress Responses. J. Proteome Res. 2020, 19, 3405–3417. [Google Scholar] [CrossRef]
- Bonilla, M.; Nastase, K.K.; Cunningham, K.W. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002, 21, 2343–2353. [Google Scholar] [CrossRef]
- Chen, Y.; Feldman, D.E.; Deng, C.; Brown, J.A.; De Giacomo, A.F.; Gaw, A.F.; Shi, G.; Le, Q.T.; Brown, J.M.; Koong, A.C. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 2005, 3, 669–677. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global analysis of protein phosphorylation in yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef]
- Shimoji, K.; Jakovljevic, J.; Tsuchihashi, K.; Umeki, Y.; Wan, K.; Kawasaki, S.; Talkish, J.; Woolford, J.L., Jr.; Mizuta, K. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2012, 40, 4574–4588. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fang, T.; Yan, H.; Jiang, L. The protein kinase Cmk2 negatively regulates the calcium/calcineurin signalling pathway and expression of calcium pump genes PMR1 and PMC1 in budding yeast. Cell Commun. Signal. 2019, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Lanze, C.E.; Gandra, R.M.; Foderaro, J.E.; Swenson, K.A.; Douglas, L.M.; Konopka, J.B. Plasma Membrane MCC/Eisosome Domains Promote Stress Resistance in Fungi. Microbiol. Mol. Biol. Rev. 2020, 84, e00063-19. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Gruhler, A.; Liu, Y.; Jensen, O.N.; Dickson, R.C. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 2008, 283, 10433–10444. [Google Scholar] [CrossRef]
- Walther, T.C.; Aguilar, P.S.; Fröhlich, F.; Chu, F.; Moreira, K.; Burlingame, A.L.; Walter, P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007, 26, 4946–4955. [Google Scholar] [CrossRef]
- Pearce, L.R.; Komander, D.; Alessi, D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010, 11, 9–22. [Google Scholar] [CrossRef]
- Foderaro, J.E.; Douglas, L.M.; Konopka, J.B. MCC/Eisosomes Regulate Cell Wall Synthesis and Stress Responses in Fungi. J. Fungi 2017, 3, 61. [Google Scholar] [CrossRef]
- Kozubowski, L.; Panek, H.; Rosenthal, A.; Bloecher, A.; DeMarini, D.J.; Tatchell, K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 2003, 14, 26–39. [Google Scholar] [CrossRef]
- Pérez, J.; Arcones, I.; Gómez, A.; Casquero, V.; Roncero, C. Phosphorylation of Bni4 by MAP kinases contributes to septum assembly during yeast cytokinesis. FEMS Yeast Res. 2016, 16, fow060. [Google Scholar] [CrossRef]
- Dokládal, L.; Stumpe, M.; Hu, Z.; Jaquenoud, M.; Dengjel, J.; De Virgilio, C. Phosphoproteomic responses of TORC1 target kinases reveal discrete and convergent mechanisms that orchestrate the quiescence program in yeast. Cell Rep. 2021, 37, 110149. [Google Scholar] [CrossRef]
- Garrett-Engele, P.; Moilanen, B.; Cyert, M.S. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell Biol. 1995, 15, 4103–4114. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Cosano, I.C.; Levin, D.E.; Molina, M.; Martin, H. Dissecting the transcriptional activation function of the cell wall integrity MAP kinase. Yeast 2007, 24, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Casar, B.; Pinto, A.; Crespo, P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol. Cell 2008, 31, 708–721. [Google Scholar] [CrossRef] [PubMed]
| Substrate | In Vitro Kinase Assay | In Vivo Kinase Assay | Effects on Protein Function/Cell Physiology | P-Site | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Method | Stimulus a | Substrate Expressed in | Method b | Stimulus a | ||||
| Rom2 | Radioactive | Heat stress | Yeast | Mobility shift | Heat stress | Cellular redistribution/ Inactivation of Rho1-mediated CWI signaling | ND | [25] |
| Mkk1 | Radioactive AS kinase | Heat stress | Bacteria | Mobility shift | Vanadate | Downregulation /CWI signaling attenuation | ND | [50,57] |
| Mkk2 | Radioactive | Heat stress | Bacteria | Mobility shift | Vanadate | Downregulation /CWI signaling attenuation | S50 | [57] |
| Msg5 | Radioactive AS kinase | Heat stress | Bacteria c | Mobility shift | Heat stress Cell wall stress (CFW) | Reduced binding to Slt2 /Increased Slt2 activation | ND | [50,58] |
| Bcy1 | Radioactive | Rapa | Bacteria | Anti- p-Bcy1 (T129) | Rapa, SDS, CR, vanadate, Bck1 d | PKA inhibition | T129 | [28] |
| Avo2 | Radioactive | Pkc1 e | Bacteria | Phos-tag | Pkc1 e | Negative regulation of TORC2 activity | (T157, T232, S246, S253, S262, T323, S328, T343, S348) f | [29] |
| Avo3 | Radioactive | Pkc1 e | Bacteria g | Phos-tag h | Pkc1 e | ND | (S12, T25, T29, S51, S85, S88) f | [29] |
| Rlm1 | Radioactive | Heat stress | Bacteria g | Mobility shift | Heat stress | Activation/ Transcriptional induction of cell wall genes | S374, S427, T439 (S234, S261, T276, S299, S518, T646, T654) f | [44,59] |
| Swi6 | Radioactive | Heat stress | Bacteria | Mobility shift | Heat stress | Nuclear exit/Downregulation of transcriptional activity | S238 | [60,61] |
| Swi4 | Radioactive | Heat stress | Insect cells | Epistatic and physical interaction with Slt2 | Heat stress | ND | ND | [60] |
| Ssn8 (Cyclin C) | Radioactive | Oxidative stress (H2O2) | Bacteria | Epistatic and physical interaction with Slt2 | Oxidative stress (H202) | Cytoplasmic release, mitochondrial targeting and degradation/ Mitochondrial fission and transcriptional activation of stress genes | S266 | [62,63] |
| Med13 | Radioactive | Vanadate | Bacteria g | Epistatic interaction with Slt2 | Oxidative stress (H202) | Degradation/ Cytoplasmic release of cyclin C | T835, T837 | [64] |
| Rpb1 | Anti-p-Y | Heat stress | Bacteria g | Anti-p-Y | Cell wall stress (CFW) | Activation/ Transcriptional induction of stress genes | Y1 of heptad repeats YSPTSPS | [65] |
| Sir3 | Radioactive | Rapa, Heat stress | Bacteria, Bacteria g | Mobility shift | Rapa, Non- stimulated cells | Reduced association with subtelomeric sequences/ Derepression of PAU genes, Chronological lifespanshortening | S275, S282 S289, S295 | [66,67] |
| Nab2 | Radioactive | Non- stimulated | Bacteria | Mobility shift | Heat stress | Non-hsp RNA retention in the nucleus/ Recovery of heat-stressed cells | T178, S180 | [68] |
| Mrc1 | Radioactive | NS | Bacteria | Anti- p-S/T | Heat stress | Delay in DNA replication to avoid transcription-replication conflicts | T169, S215, S229 | [69] |
| Sic1 | Anti-p-Sic1 (T163) | Rapa | Bacteria | Anti- p-Sic1 (T163) Phos-tag | Rapa | Stabilization/G1-S arrest | T163 | [70] |
| Rcn2 | AS kinase | Cell wall stress (CR) | Bacteria Yeast | Phospho-peptide increaseh | Pkc1 e | ND | S152, S160, S255 | [50,54] |
| Gga1 | AS kinase | Cell wall stress (CR) | Bacteria Yeast | Phospho-peptide Increaseh | Pkc1 e | ND | ND | [50,54] |
| Caf20 | AS kinase | Cell wall stress (CR) | Bacteria Yeast | Phospho-peptide increaseh | Pkc1 e | ND | T102 | [50,54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Rubio, G.; Sastre-Vergara, L.; Molina, M.; Martín, H.; Fernández-Acero, T. Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity. J. Fungi 2022, 8, 368. https://doi.org/10.3390/jof8040368
González-Rubio G, Sastre-Vergara L, Molina M, Martín H, Fernández-Acero T. Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity. Journal of Fungi. 2022; 8(4):368. https://doi.org/10.3390/jof8040368
Chicago/Turabian StyleGonzález-Rubio, Gema, Lucía Sastre-Vergara, María Molina, Humberto Martín, and Teresa Fernández-Acero. 2022. "Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity" Journal of Fungi 8, no. 4: 368. https://doi.org/10.3390/jof8040368
APA StyleGonzález-Rubio, G., Sastre-Vergara, L., Molina, M., Martín, H., & Fernández-Acero, T. (2022). Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity. Journal of Fungi, 8(4), 368. https://doi.org/10.3390/jof8040368







