Protein Kinase A Regulates Autophagy-Associated Proteins Impacting Growth and Virulence of Aspergillus fumigatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Quantitative LC-MS/MS Analysis
2.2. Construction of Gene Deletions and Mutations in A. fumigatus
2.3. Assessment of Radial Growth, Conidiation, and Antifungal Compound Susceptibility
2.4. Protein Extraction and Purification
2.5. Determination of PKA Enzyme Activity
2.6. Fluorescence Microscopy
2.7. Atg24 Interactome and Bioinformatic Analysis of Protein Functional Cluster Enrichment
2.8. Murine Invasive Aspergillosis Virulence Assays and Histopathological Analysis
3. Results
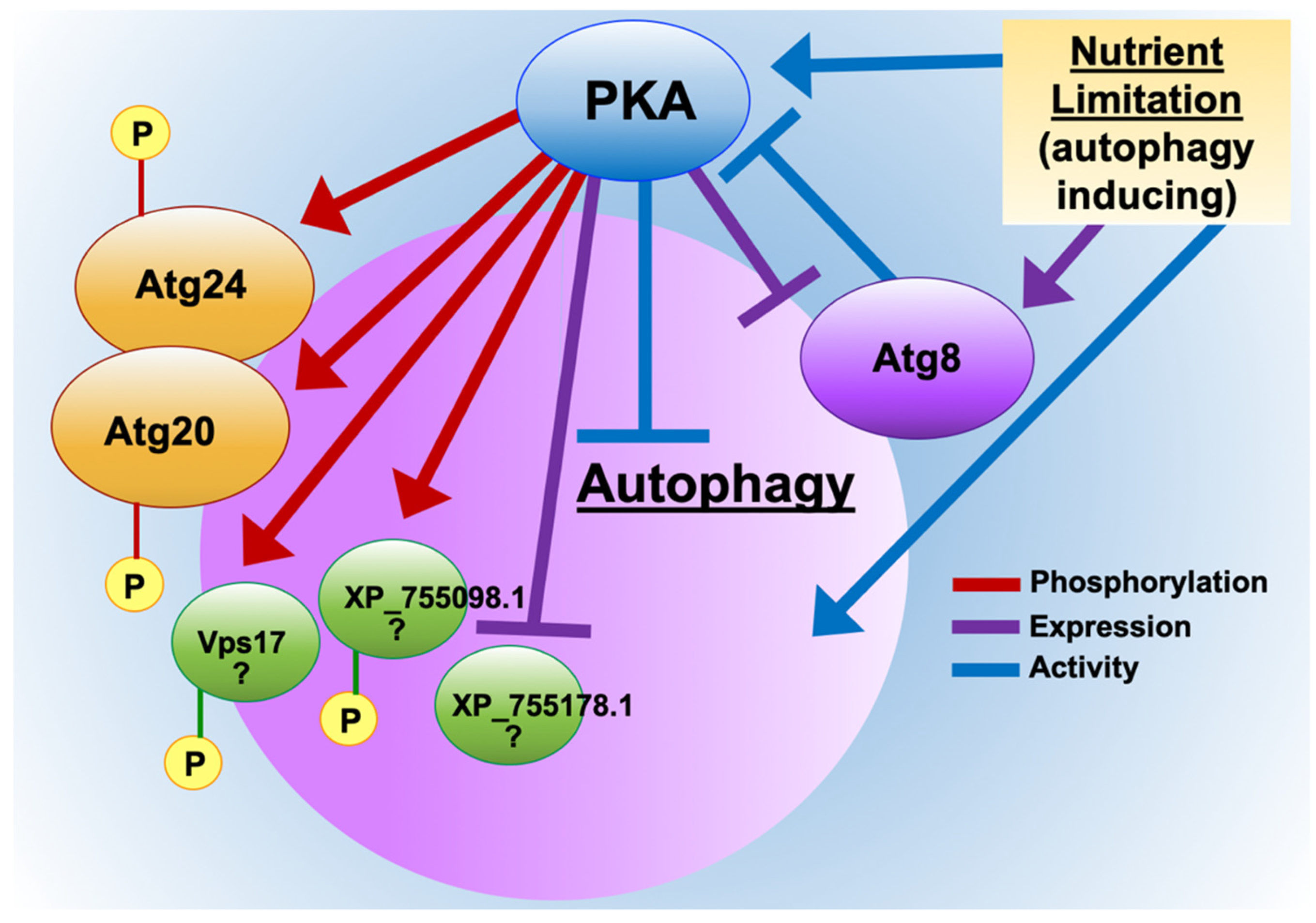
3.1. Advanced Whole-Proteomic and Phosphoproteomic Approaches Identify Autophagy Proteins as Potential PKA-Dependent Effectors
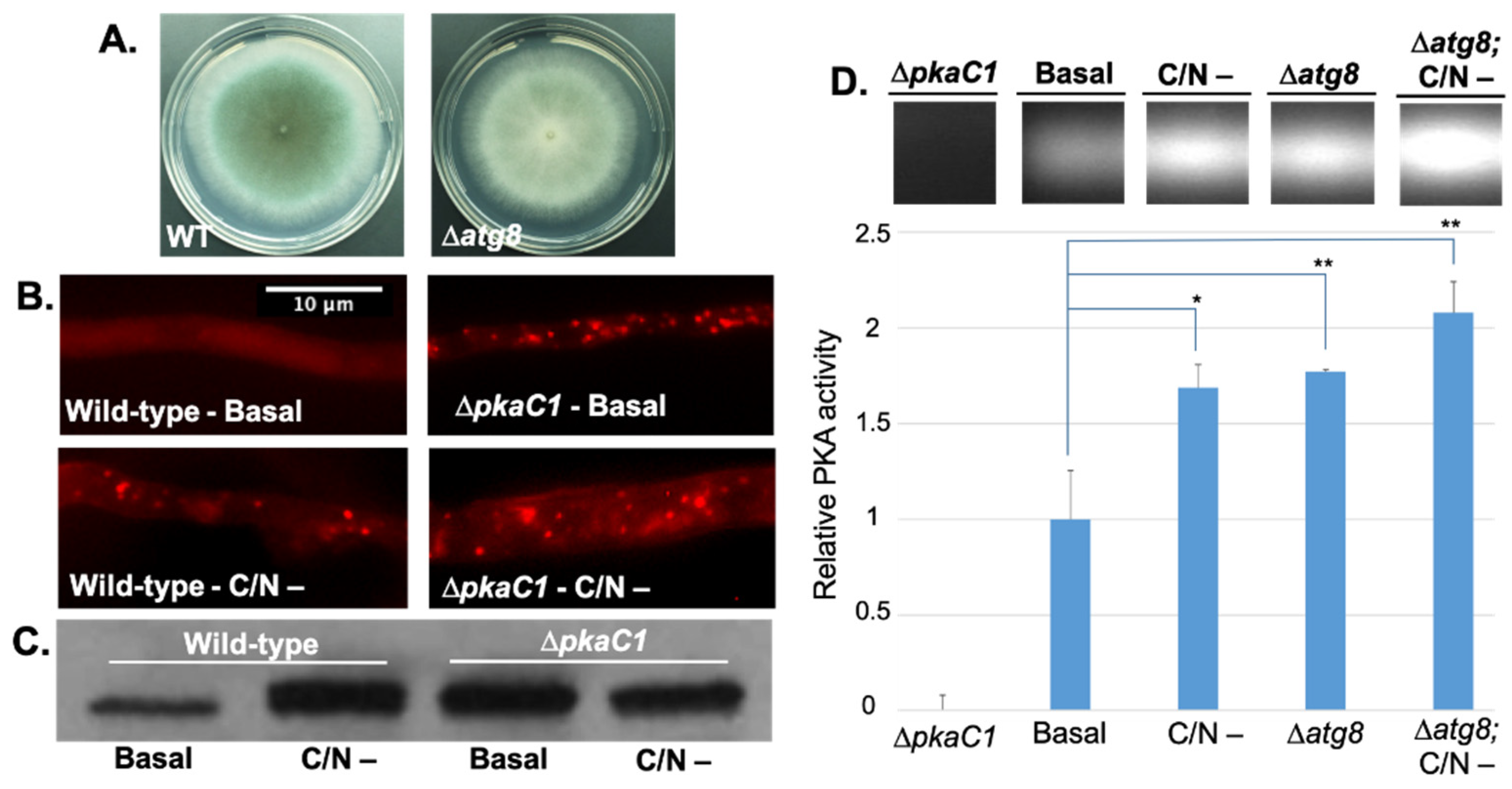
3.2. PKA Regulates Nutrient Sensing and Autophagosome Formation and Exhibits Reciprocal Regulation with the Key Autophagy Marker Protein Atg8
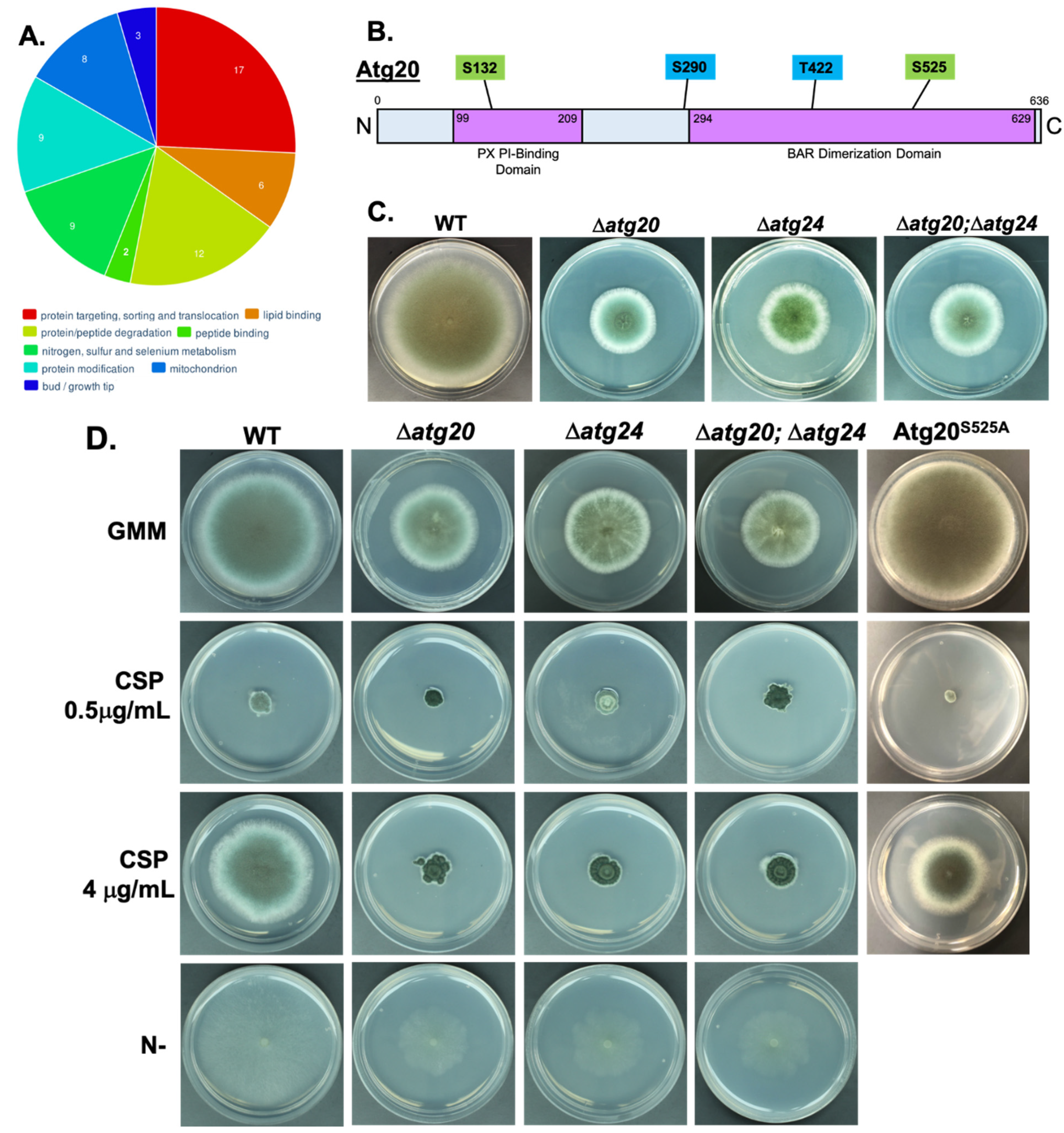
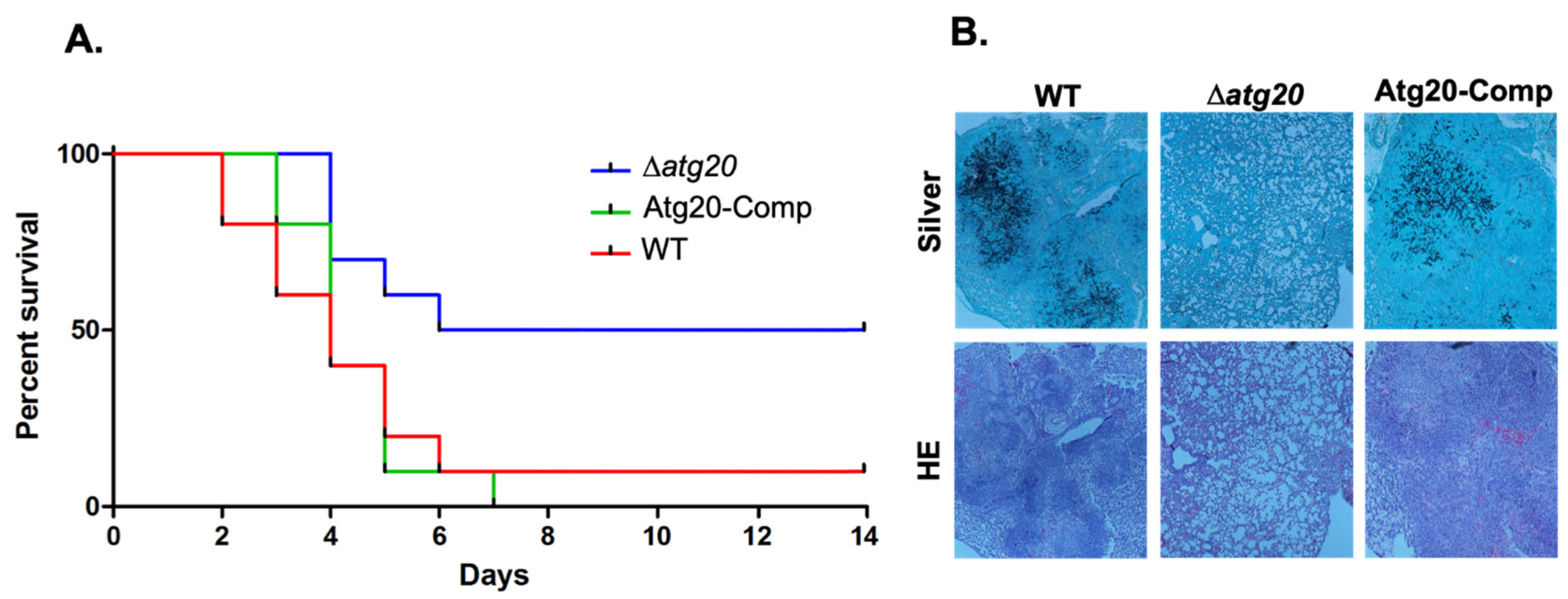
3.3. Atg20 Interacts with Atg24 to Control A. fumigatus Growth and Pathogenesis
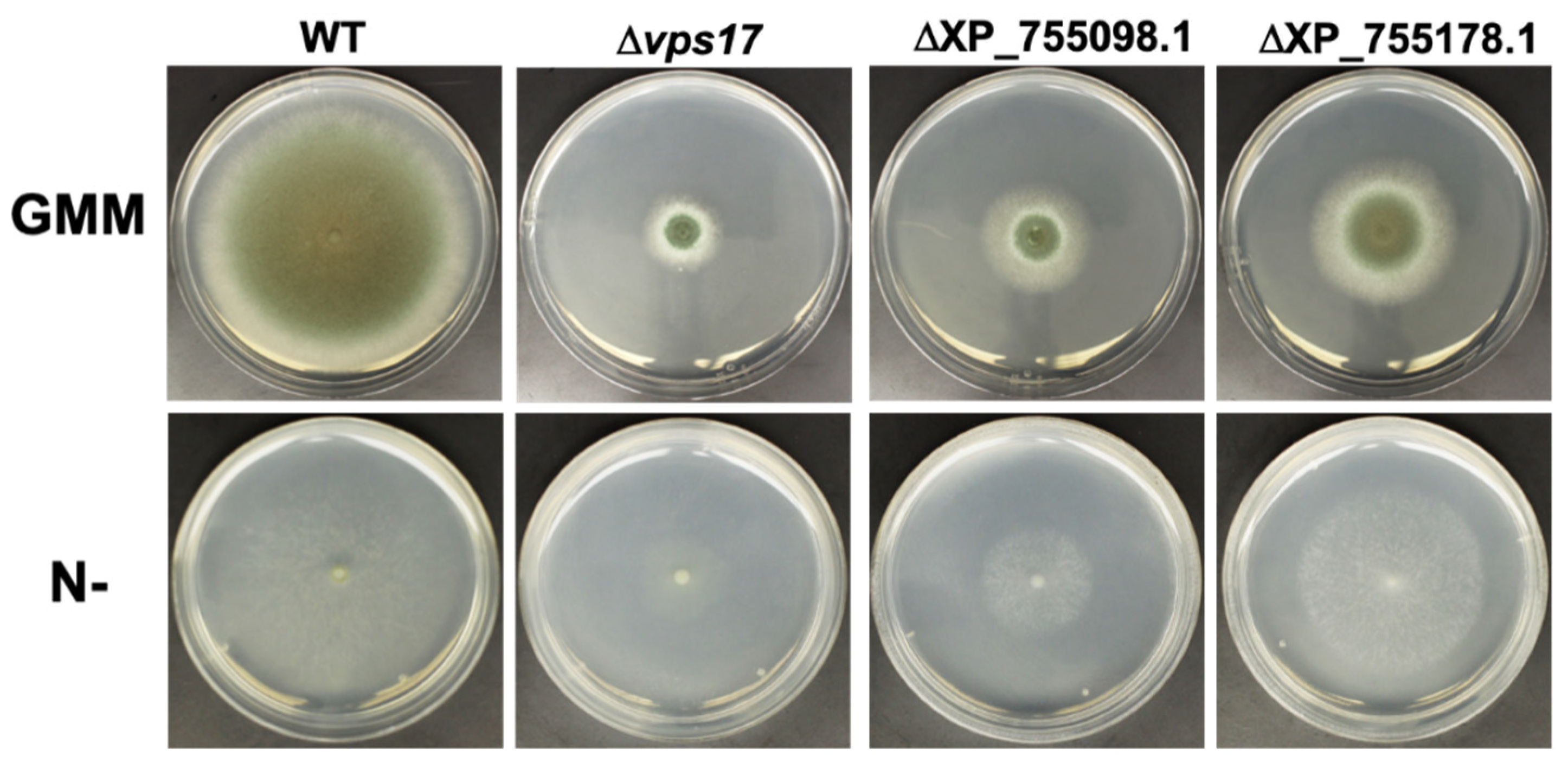
3.4. PKA Regulates Key Cellular Recycling Pathway Proteins Contributing to Fungal Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, W.; Panepinto, J.C.; Fortwendel, J.R.; Fox, L.; Oliver, B.G.; Askew, D.S.; Rhodes, J.C. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 2006, 74, 4865–4874. [Google Scholar] [CrossRef]
- Fuller, K.K.; Richie, D.L.; Feng, X.; Krishnan, K.; Stephens, T.J.; Wikenheiser-Brokamp, K.A.; Askew, D.S.; Rhodes, J.C. Divergent Protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol. Microbiol. 2011, 79, 1045–1062. [Google Scholar] [CrossRef]
- Neofytos, D.; Horn, D.; Anaissie, E.; Steinbach, W.; Olyaei, A.; Fishman, J.; Pfaller, M.; Chang, C.; Webster, K.; Marr, K. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: Analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin. Infect. Dis. 2009, 48, 265–273. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P.; Marr, K.A.; Park, B.J.; Alexander, B.D.; Anaissie, E.J.; Walsh, T.J.; Ito, J.; Andes, D.R.; Baddley, J.W.; Brown, J.M.; et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: Overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 2010, 50, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Caira, M.; Candoni, A.; Offidani, M.; Fianchi, L.; Martino, B.; Pastore, D.; Picardi, M.; Bonini, A.; Chierichini, A.; et al. The epidemiology of fungal infections in patients with hematologic malignancies: The SEIFEM-2004 study. Haematologica 2006, 91, 1068–1075. [Google Scholar]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global analysis of protein phosphorylation in yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Jin, C.; Ren, J.; Yao, X.; Xue, Y. Proteome-wide prediction of PKA phosphorylation sites in eukaryotic kingdom. Genomics 2008, 92, 457–463. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Giansanti, P.; Stokes, M.P.; Silva, J.C.; Scholten, A.; Heck, A.J. Interrogating cAMP-dependent kinase signaling in Jurkat T cells via a protein kinase A targeted immune-precipitation phosphoproteomics approach. Mol. Cell. Proteom. 2013, 12, 3350–3359. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wu, M.; Bing, J.; Tao, L.; Ding, X.; Liu, X.; Huang, G. Global regulatory roles of the cAMP/PKA pathway revealed by phenotypic, transcriptomic and phosphoproteomic analyses in a null mutant of the PKA catalytic subunit in Candida albicans. Mol. Microbiol. 2017, 105, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Franck, W.L.; Gokce, E.; Randall, S.M.; Oh, Y.; Eyre, A.; Muddiman, D.C.; Dean, R.A. Phosphoproteome Analysis Links Protein Phosphorylation to Cellular Remodeling and Metabolic Adaptation during Magnaporthe oryzae Appressorium Development. J. Proteome Res. 2015, 14, 2408–2424. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Juvvadi, P.R.; Waitt, G.; Shaheen, S.; Allen, J., IV; Soderblom, E.J.; Bobay, B.G.; Asfaw, Y.G.; Moseley, M.A.; Steinbach, W.J. The Protein Kinase A-Dependent Phosphoproteome of the Human Pathogen Aspergillus fumigatus Reveals Diverse Virulence-Associated Kinase Targets. Mbio 2020, 11, e02880-20. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Caza, M.; Dong, Y.; Arif, A.A.; Horianopoulos, L.C.; Hu, G.; Johnson, P.; Kronstad, J.W. ATG Genes Influence the Virulence of Cryptococcus neoformans through Contributions beyond Core Autophagy Functions. Infect. Immun. 2018, 86, e00069-18. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Hacham, M.; Waterman, S.R.; Panepinto, J.; Shin, S.; Liu, X.; Gibbons, J.; Valyi-Nagy, T.; Obara, K.; Jaffe, H.A.; et al. PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans. J. Clin. Investig. 2008, 118, 1186–1197. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 2010, 6, 294–295. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Wang, H.; Chen, L.; Zhang, J.; Sun, M.; Xu, J.R.; Wang, C. The PKR regulatory subunit of protein kinase A (PKA) is involved in the regulation of growth, sexual and asexual development, and pathogenesis in Fusarium graminearum. Mol. Plant Pathol. 2018, 19, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Juvvadi, P.R.; Waitt, G.; Soderblom, E.J.; Moseley, M.A.; Nicely, N.I.; Asfaw, Y.G.; Steinbach, W.J. A Novel Phosphoregulatory Switch Controls the Activity and Function of the Major Catalytic Subunit of Protein Kinase A in Aspergillus fumigatus. Mbio 2017, 8, e02319-16. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Belina, D.; Soderblom, E.J.; Moseley, M.A.; Steinbach, W.J. Filamentous fungal-specific septin AspE is phosphorylated in vivo and interacts with actin, tubulin and other septins in the human pathogen Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2013, 431, 547–553. [Google Scholar] [CrossRef] [PubMed]
- da Silva Ferreira, M.E.; Kress, M.R.; Savoldi, M.; Goldman, M.H.; Hartl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.A.J.; Perfect, B.Z.; Pinchai, N.; Park, S.; Perlin, D.S.; Asfaw, Y.G.; Heitman, J.; Perfect, J.R.; Steinbach, W.J. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot. Cell. 2008, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Ma, Y.; Richards, A.D.; Soderblom, E.J.; Moseley, M.A.; Lamoth, F.; Steinbach, W.J. Identification and mutational analyses of phosphorylation sites of the calcineurin-binding protein CbpA and the identification of domains required for calcineurin binding in Aspergillus fumigatus. Front. Microbiol. 2015, 6, 175. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, W.J.; Cramer, R.A.J.; Perfect, B.Z.; Asfaw, Y.G.; Sauer, T.C.; Najvar, L.K.; Kirkpatrick, W.R.; Patterson, T.F.; Benjamin, D.K.J.; Heitman, J.; et al. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Juvvadi, P.R.; Waitt, G.; Soderblom, E.J.; Barrington, B.C.; Asfaw, Y.G.; Moseley, M.A.; Steinbach, W.J. Calcineurin-dependent dephosphorylation of the transcription factor CrzA at specific sites controls conidiation, stress tolerance, and virulence of Aspergillus fumigatus. Mol. Microbiol. 2019, 112, 62–80. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, A.; Fedorova, N.D.; Crabtree, J.; Yu, Y.; Kim, S.; Chen, D.; Loss, O.; Cairns, T.; Goldman, G.; Armstrong-James, D.; et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008, 4, e1000154. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; Carilla-Latorre, S.; Mesquita, A.; Escalante, R. A proteolytic cleavage assay to monitor autophagy in Dictyostelium discoideum. Autophagy 2011, 7, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Deng, Y.Z.; Naqvi, N.I. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2013, 9, 1818–1827. [Google Scholar] [CrossRef]
- Hettema, E.H.; Lewis, M.J.; Black, M.W.; Pelham, H.R. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003, 22, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Gomez, T.S.; Sackey, B.K.; Billadeau, D.D.; Burd, C.G. Rab GTPase regulation of retromer-mediated cargo export during endosome maturation. Mol. Biol. Cell 2012, 23, 2505–2515. [Google Scholar] [CrossRef]
- Debelyy, M.O.; Platta, H.W.; Saffian, D.; Hensel, A.; Thoms, S.; Meyer, H.E.; Warscheid, B.; Girzalsky, W.; Erdmann, R. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 2011, 286, 28223–28234. [Google Scholar] [CrossRef]
- Cunningham, K.W.; Fink, G.R. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+-ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 2226–2237. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos, A.M.; Cyert, M.S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997, 11, 3432–3444. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X.; Qian, H.; Zhang, S.; Lu, L. Calcineurin and Calcium Channel CchA Coordinate the Salt Stress Response by Regulating Cytoplasmic Ca2+ Homeostasis in Aspergillus nidulans. Appl. Environ. Microbiol. 2016, 82, 3420–3430. [Google Scholar] [CrossRef] [PubMed]
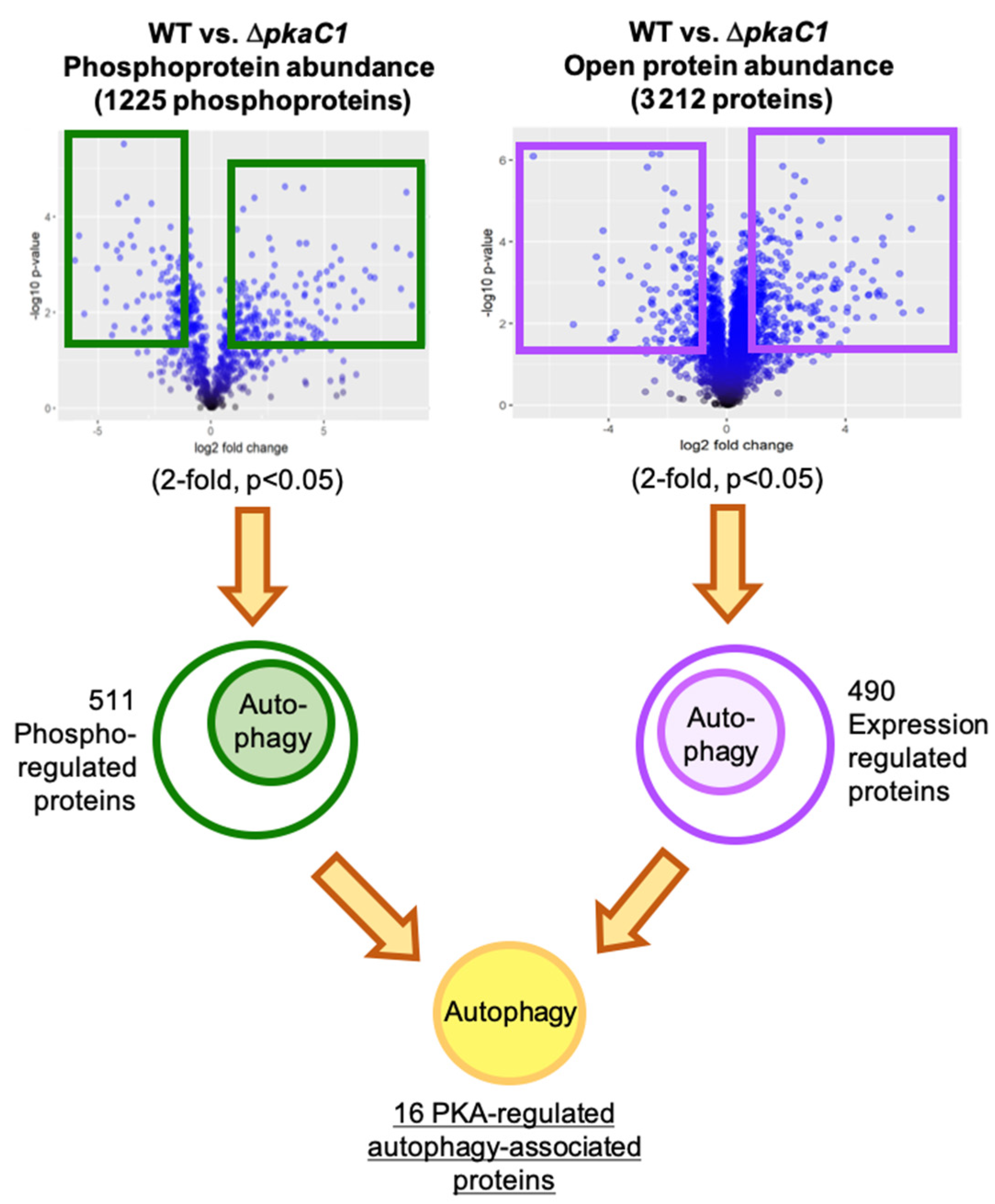
| RefSeq ID | Protein Name | Description | WT/ΔpkaC1 Fold Change | p-Value |
|---|---|---|---|---|
| XP_755360.1 | TorA | TOR pathway phosphatidylinositol 3-kinase | 15.3 | 0.025 |
| XP_753398.1 | NA | N-acetylglucosaminyl-phosphatidylinositol deacetylase | 6.68 | 0.026 |
| XP_752894.1 | Atg27 | autophagy protein | −2.33 | 0.0395 |
| XP_751230.1 | CpyA/Prc1 | carboxypeptidase | −2.79 | 0.002 |
| XP_755098.1 | NA | vacuolar H+/Ca2+ exchanger | −9.47 | 0.049 |
| RefSeq ID | Protein Name | Master Protein Descriptions | WT/ΔpkaC1 Fold Change | p-Value |
|---|---|---|---|---|
| XP_748106.1 | VpsA | vacuolar dynamin-like GTPase | 263.31 | 0.0003 |
| XP_753919.2 | Vps17 | vacuolar protein sorting-associated protein | 63.29 | 0.0093 |
| XP_751536.1 | Atg24 | vacuolar targeting sorting nexin protein | 25.3 | 0.0020 |
| XP_001481667.1 | NA | RING finger protein | 12.78 | 0.0337 |
| XP_755360.1 | TorA | TOR pathway phosphatidylinositol 3-kinase | 7.42 | 0.0156 |
| XP_751003.1 | Vps15 | vacuolar protein kinase | 6.69 | 0.0126 |
| XP_749349.1 | Cue3 | CUE domain protein | 4.1 | 0.0349 |
| XP_755749.2 | Atg20 | autophagy sorting nexin protein | 3.58 | 0.0008 |
| XP_750137.1 | Scn1 | Cut9 interacting protein Scn1 | 3.04 | 0.0132 |
| XP_755178.1 | NA | ubiquitin C-terminal hydrolase | 2.29 | 0.0332 |
| XP_753300.1 | Vac8 | vacuolar armadillo repeat protein | −2 | 0.0200 |
| XP_755238.1 | Vtc4 | vacuolar transporter chaperone | −6.67 | 0.0045 |
| RefSeq ID | Protein Name | Description | Human Similarity | Likely Autophagy Relevance | PKA Motif Phos. | Phos. Fold Change | Expr. Fold Change | p-Value |
|---|---|---|---|---|---|---|---|---|
| XP_753919.2 | Vps17 | Vacuolar protein sorting-associated protein | 0% | High | No | 63.29 | - | 0.0093 |
| XP_755098.1 | NA | Vacuolar H+/Ca2+ exchanger | 0% | High | No | - | −9.47 | 0.049 |
| XP_001481667.1 | NA | RING finger protein | 5% | Moderate | Yes | 12.78 | - | 0.0337 |
| XP_755178.1 | NA | Ubiquitin C-terminal hydrolase | 10% | High | No | 2.29 | - | 0.0332 |
| XP_755238.1 | Vtc4 | Vacuolar transporter chaperone | 7% | High | No | −6.52 | - | 0.0045 |
| XP_750137.1 | Scn1 | Cut9 interacting protein | 7% | Moderate | Yes | 3.04 | - | 0.0132 |
| XP_749349.1 | Cue3 | CUE domain protein | 15% | Moderate | Yes | 4.1 | - | 0.0349 |
| XP_752894.1 | Atg27 | Autophagy protein | 0% | High | No | - | −2.33 | 0.0395 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shwab, E.K.; Juvvadi, P.R.; Shaheen, S.K.; Allen, J., IV; Waitt, G.; Soderblom, E.J.; Asfaw, Y.G.; Moseley, M.A.; Steinbach, W.J. Protein Kinase A Regulates Autophagy-Associated Proteins Impacting Growth and Virulence of Aspergillus fumigatus. J. Fungi 2022, 8, 354. https://doi.org/10.3390/jof8040354
Shwab EK, Juvvadi PR, Shaheen SK, Allen J IV, Waitt G, Soderblom EJ, Asfaw YG, Moseley MA, Steinbach WJ. Protein Kinase A Regulates Autophagy-Associated Proteins Impacting Growth and Virulence of Aspergillus fumigatus. Journal of Fungi. 2022; 8(4):354. https://doi.org/10.3390/jof8040354
Chicago/Turabian StyleShwab, E. Keats, Praveen R. Juvvadi, Shareef K. Shaheen, John Allen, IV, Greg Waitt, Erik J. Soderblom, Yohannes G. Asfaw, M. Arthur Moseley, and William J. Steinbach. 2022. "Protein Kinase A Regulates Autophagy-Associated Proteins Impacting Growth and Virulence of Aspergillus fumigatus" Journal of Fungi 8, no. 4: 354. https://doi.org/10.3390/jof8040354
APA StyleShwab, E. K., Juvvadi, P. R., Shaheen, S. K., Allen, J., IV, Waitt, G., Soderblom, E. J., Asfaw, Y. G., Moseley, M. A., & Steinbach, W. J. (2022). Protein Kinase A Regulates Autophagy-Associated Proteins Impacting Growth and Virulence of Aspergillus fumigatus. Journal of Fungi, 8(4), 354. https://doi.org/10.3390/jof8040354







