Colletotrichum Species Associated with Peaches in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Colletotrichum spp. from Peach Samples
2.2. Morphological Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analyses
2.5. Pathogenicity Test
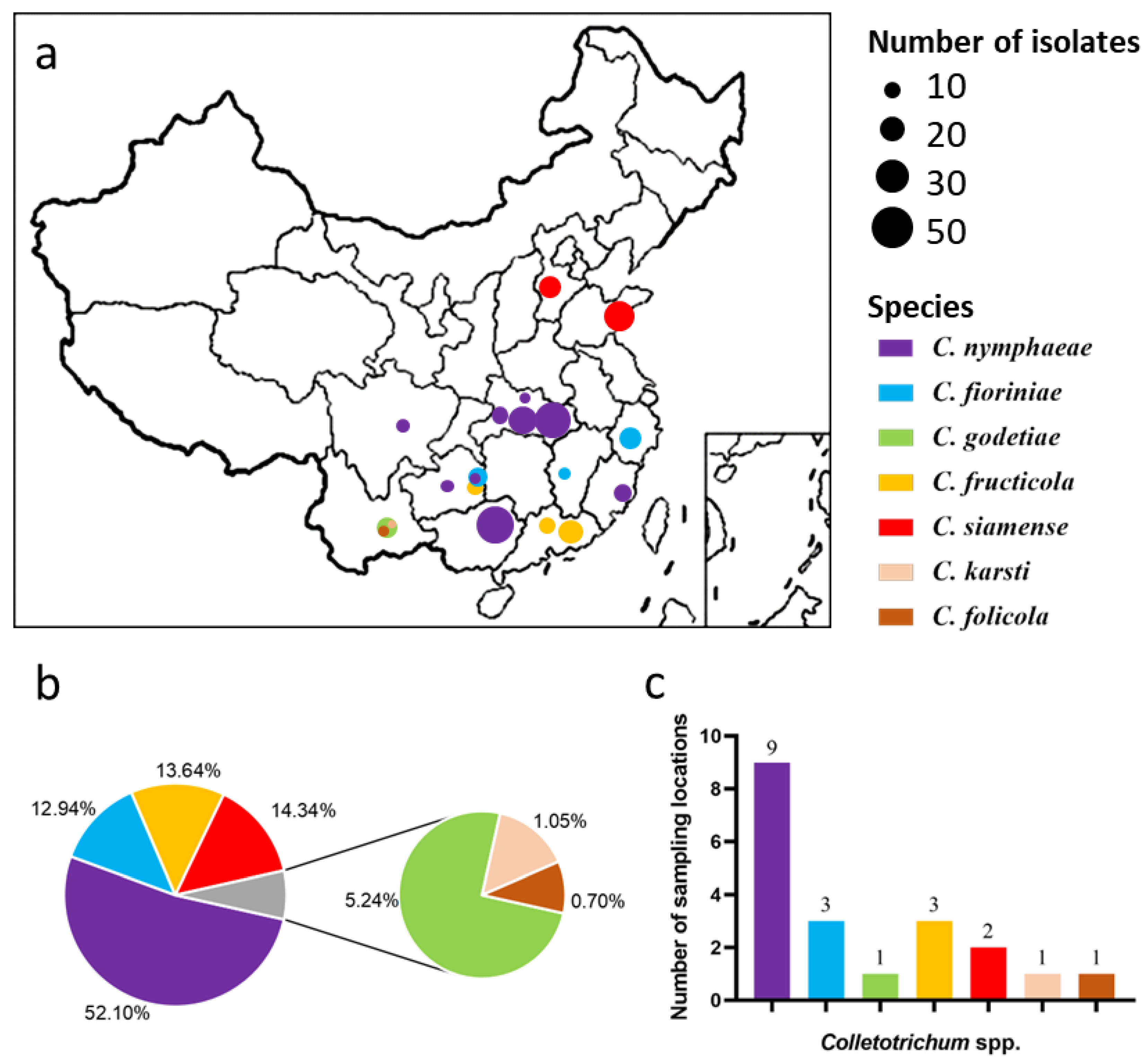
3. Results
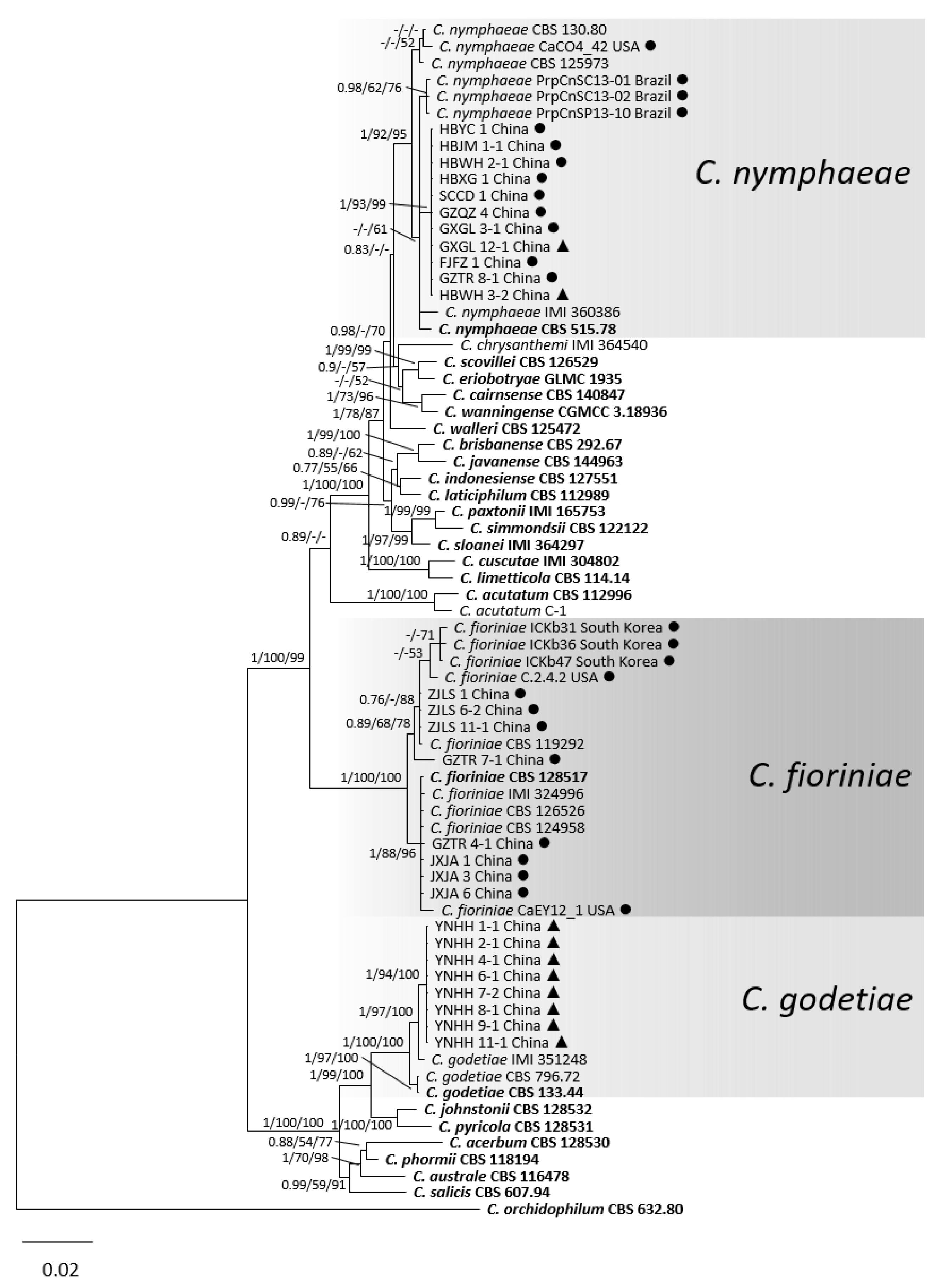
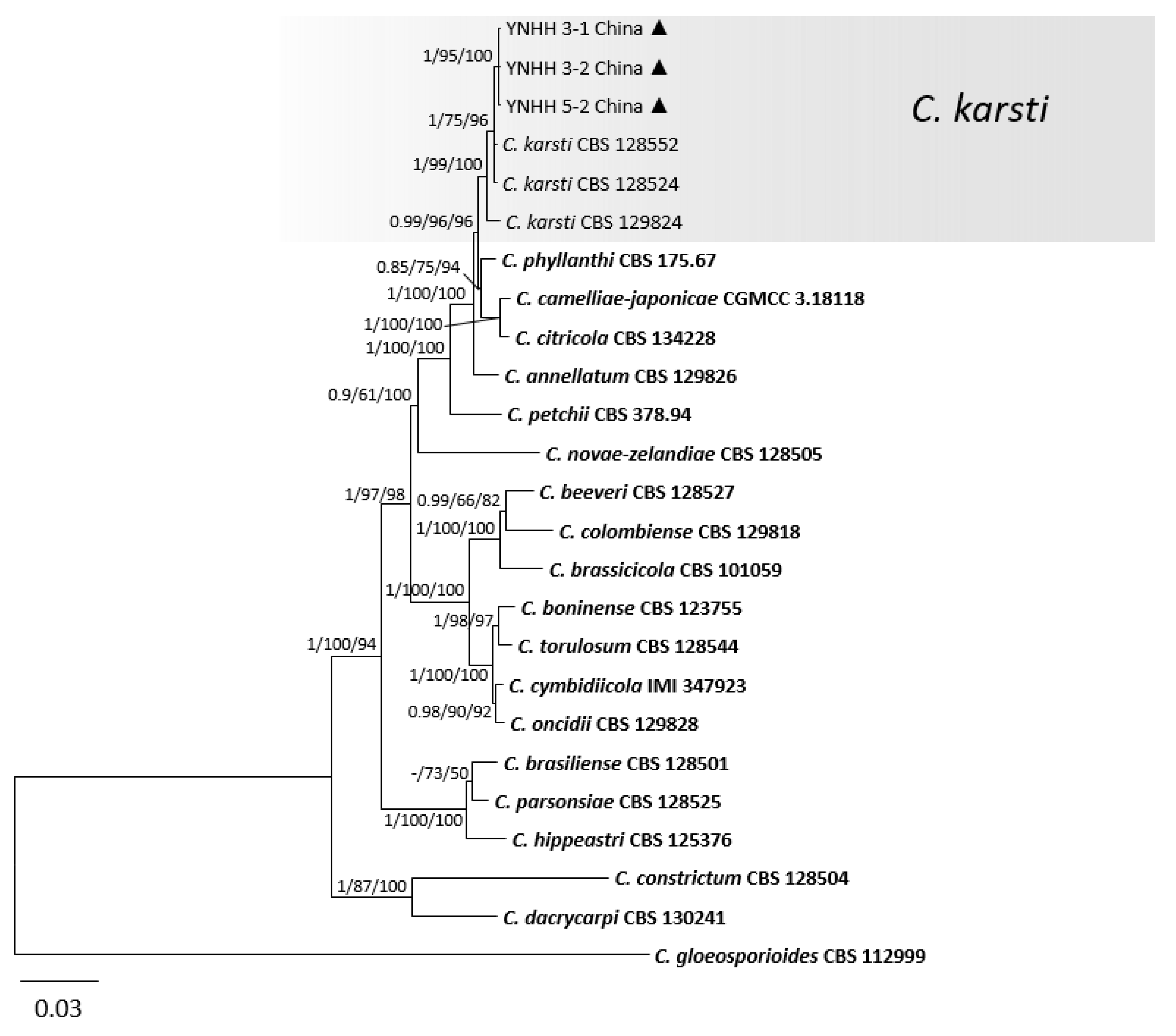
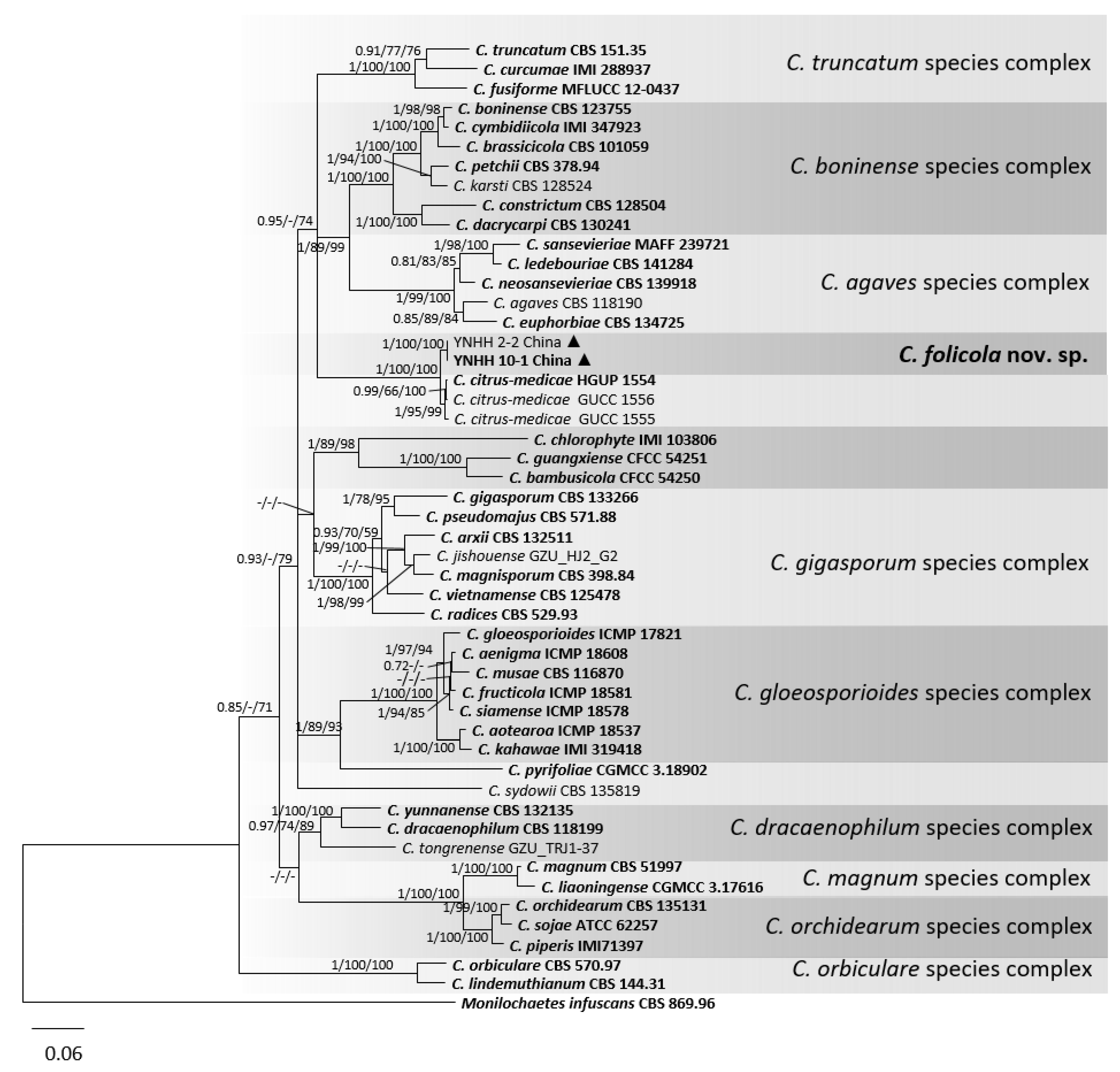
3.1. Phylogenetic Analyses
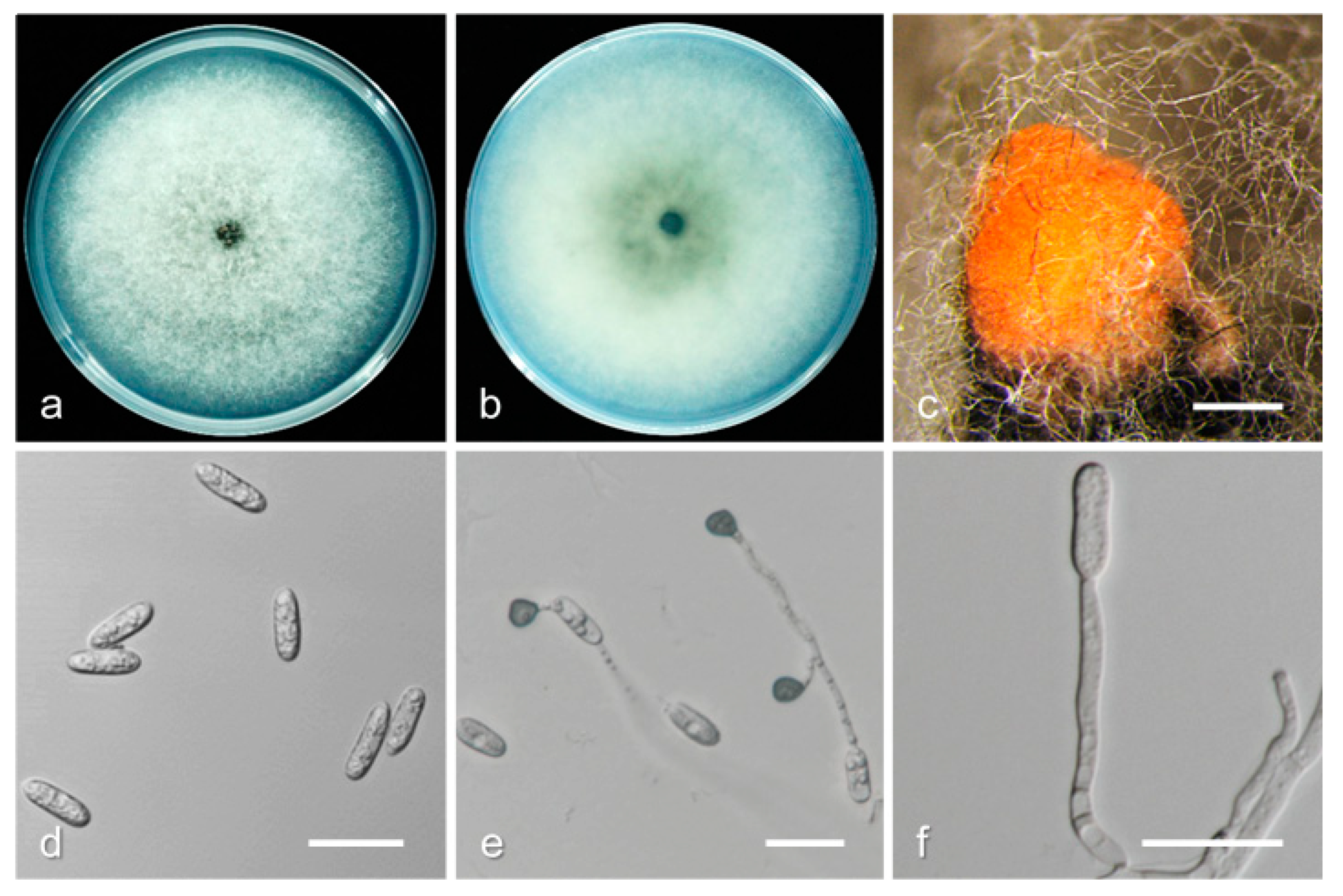
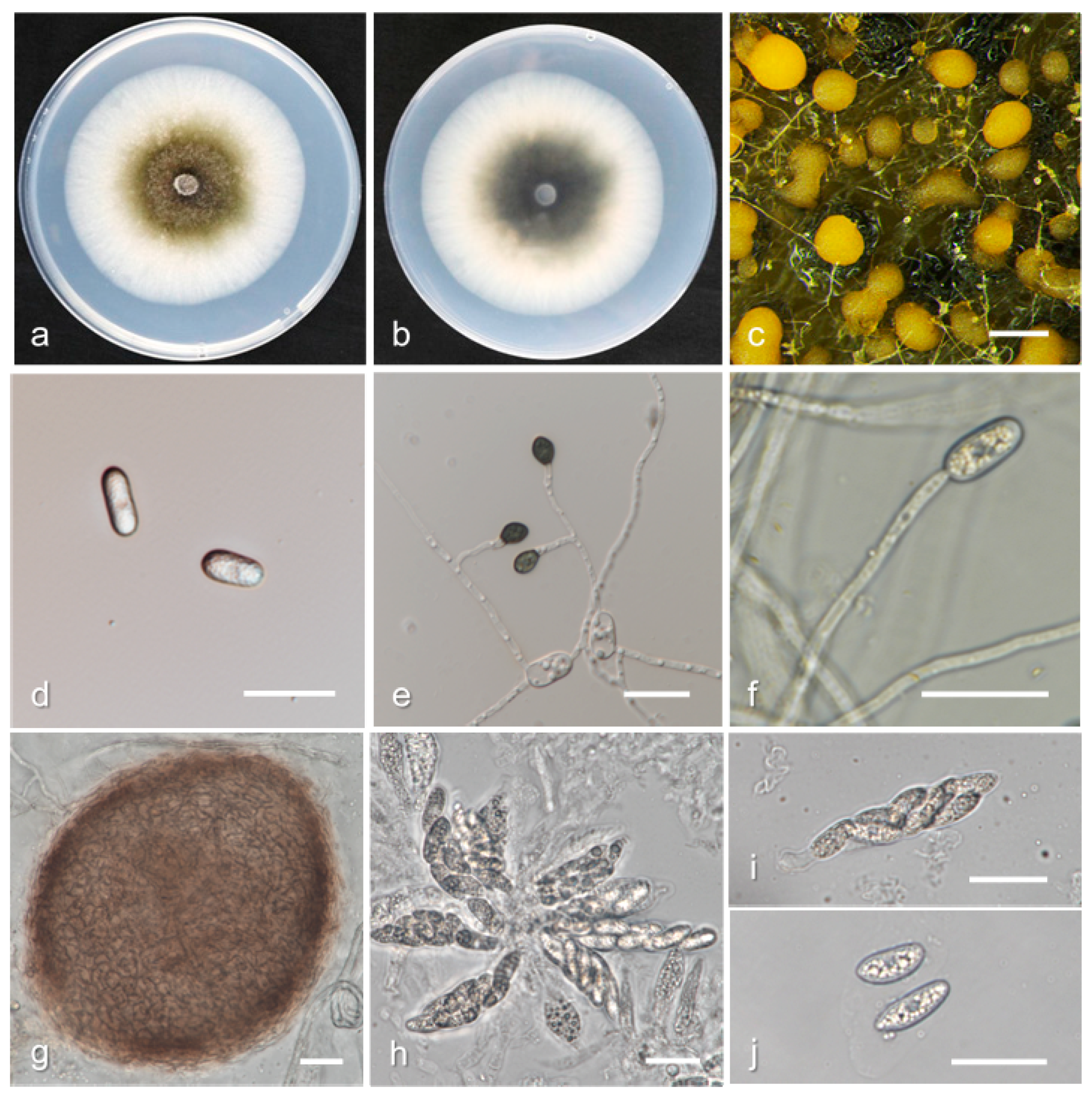
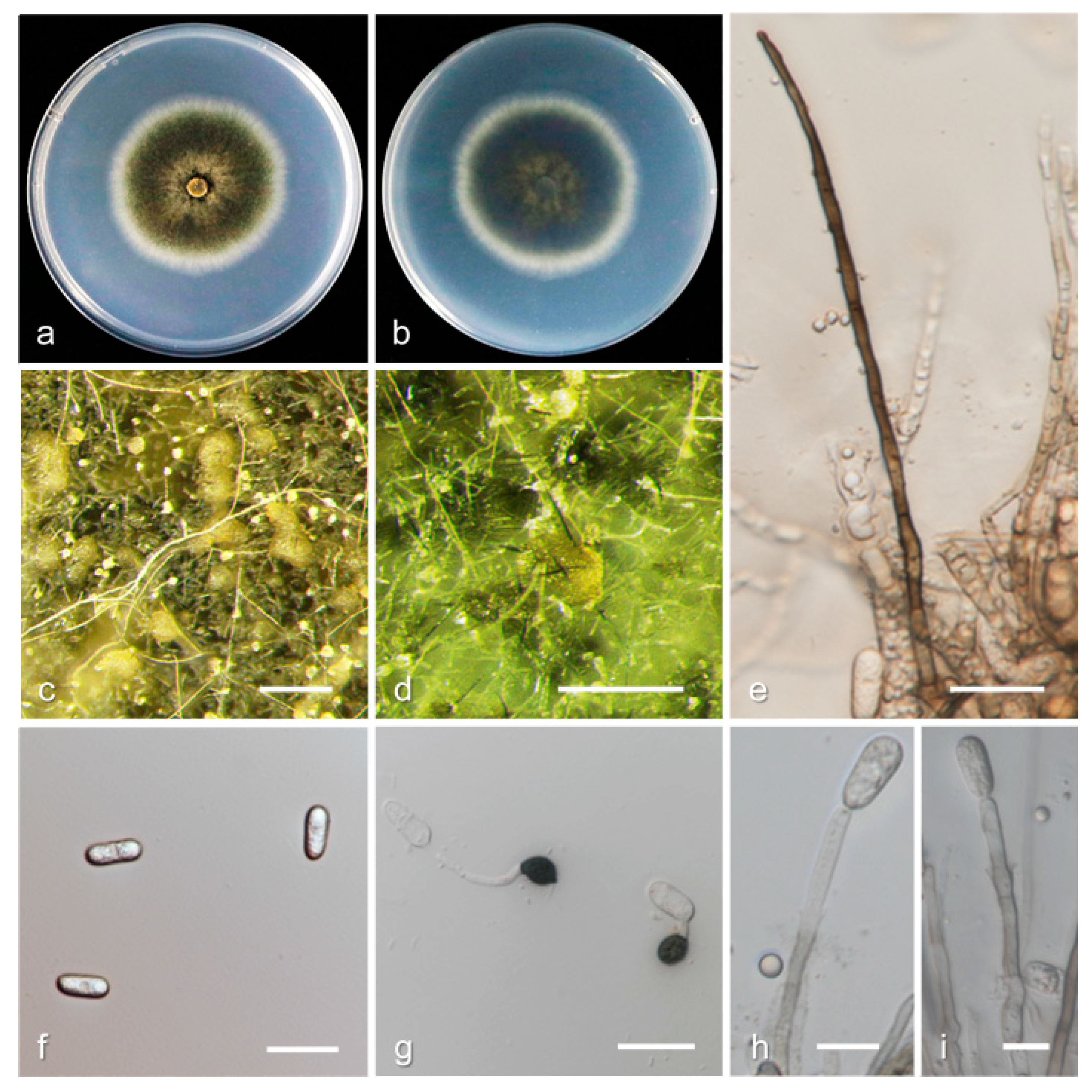
3.2. Taxonomy
3.3. Pathogenicity Tests
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, Y.; Crawford, G.W.; Chen, X. Archaeological evidence for peach (Prunus persica) cultivation and domestication in China. PLoS ONE 2014, 9, e106595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Agricultural Organization of the United Nations (FAOSTAT) Website. 2020. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 14 February 2022).
- Dowling, M.; Peres, N.; Villani, S.; Schnabel, G. Managing Colletotrichum on fruit crops: A “complex” challenge. Plant Dis. 2020, 104, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Dowling, M.; Schnabel, G. Understanding plant diseases using art and technology. Int. J. Fruit Sci. 2020, 20, 959–966. [Google Scholar] [CrossRef]
- Stensvand, A.; Borve, J.; Talgo, V. Overwintering diseased plant parts and newly infected flowers and fruit as sources of inoculum for Colletotrichum acutatum in sour cherry. Plant Dis. 2017, 101, 1207–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, T.B.; Shane, W.W. Epidemiology of the perfect stage of Glomerella cingulata on apples. Phytopathology 1983, 73, 1179–1183. [Google Scholar] [CrossRef]
- De Silva, D.D.; Crous, P.W.; Ades, P.K.; Hyde, K.D.; Taylor, P.W.J. Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol. Rev. 2017, 31, 155–168. [Google Scholar] [CrossRef]
- Fu, M.; Crous, P.W.; Bai, Q.; Zhang, P.F.; Xiang, J.; Guo, Y.S.; Zhao, F.F.; Yang, M.M.; Hong, N.; Xu, W.X.; et al. Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 2019, 42, 1–35. [Google Scholar] [CrossRef] [Green Version]
- Sutton, B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, UK, 1980. [Google Scholar]
- Cacciola, S.O.; Gilardi, G.; Faedda, R.; Schena, L.; Pane, A.; Garibaldi, A.; Gullino, M.L. Characterization of Colletotrichum ocimi population associated with black spot of sweet basil (Ocimum basilicum) in Northern Italy. Plants 2020, 9, 654. [Google Scholar] [CrossRef]
- Riolo, M.; Aloi, F.; Pane, A.; Cara, M.; Cacciola, S.O. Twig and shoot dieback of Citrus, a new disease caused by Colletotrichum species. Cells 2021, 10, 449. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.S.; Waller, J.M.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Liu, Z.Y.; et al. A polyphasic approach for studying Colletotrichum. Fungal Divers 2009, 39, 183–204. [Google Scholar]
- Liu, F.; Wang, M.; Damm, U.; Crous, P.W.; Cai, L. Species boundaries in plant pathogenic fungi: A Colletotrichum case study. BMC Evol. Biol. 2016, 16, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhinhas, P.; Baroncelli, R. Colletotrichum species and complexes: Geographic distribution, host range and conservation status. Fungal Divers. 2021, 110, 109–198. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, X.; Zheng, H.; Zhang, H.; Qiao, M. Fourteen new species of foliar Colletotrichum associated with the invasive plant Ageratina adenophora and surrounding crops. J. Fungi 2022, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yu, Z.; Jiang, X.; Fang, L.; Qiao, M. Endophytic Colletotrichum species from aquatic plants in southwest China. J. Fungi 2022, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.; Zehr, E.I.; Dean, R.A.; Shabi, E. Characteristics of Colletotrichum from peach, apple, pecan, and other hosts. Plant Dis. 1995, 79, 478–482. [Google Scholar] [CrossRef]
- Adaskaveg, J.E.; Hartin, R.J. Characterization of Colletotrichum acutatum isolates causing anthracnose of almond and peach in California. Phytopathology 1997, 87, 979–987. [Google Scholar] [CrossRef] [Green Version]
- Schnabel, G.; Chai, W.; Cox, K.D. Identifying and characterizing summer diseases on ‘Babygold’ peach in South Carolina. Plant Health. Prog. 2006, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.G.; Hong, S.K. Occurrence of anthracnose on peach tree caused by Colletotrichum species. Plant Pathol. J. 2008, 24, 80–83. [Google Scholar] [CrossRef]
- Chen, S.N.; Luo, C.X.; Hu, M.J.; Schnabel, G. Sensitivity of Colletotrichum species, including C. fioriniae and C. nymphaeae, from peach to demethylation inhibitor fungicides. Plant Dis. 2016, 100, 2434–2441. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.J.; Grabke, A.; Schnabel, G. Investigation of the Colletotrichum gloeosporioides species complex causing peach anthracnose in South Carolina. Plant Dis. 2015, 99, 797–805. [Google Scholar] [CrossRef] [Green Version]
- Moreira, R.R.; Silva, G.A.; De Mio, L.L.M. Colletotrichum acutatum complex causing anthracnose on peach in Brazil. Austral. Plant Pathol. 2020, 49, 179–189. [Google Scholar] [CrossRef]
- Lee, D.M.; Hassan, O.; Chang, T. Identification, characterization, and pathogenicity of Colletotrichum species causing anthracnose of peach in Korea. Mycobiology 2020, 48, 210–218. [Google Scholar] [CrossRef]
- Grabke, A.; Williamson, M.; Henderson, G.W.; Schnabel, G. First report of anthracnose on peach fruit caused by Colletotrichum truncatum in South Carolina. Plant Dis. 2014, 98, 1154. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.X.; Ruan, H.C.; Shi, N.N.; Gan, L.; Yang, X.J.; Dai, Y.L.; Chen, F.R. First report of anthracnose on peach fruit caused by Colletotrichum acutatum in China. Plant Dis. 2017, 101, 1678. [Google Scholar] [CrossRef]
- Suzaki, K. Improved method to induce sporulation of Colletotrichum gloeosporioides, causal fungus of grape ripe rot. J. Gen. Plant Pathol. 2011, 77, 81–84. [Google Scholar] [CrossRef]
- Yang, Y.L.; Liu, Z.Y.; Cai, L.; Hyde, K.D.; Yu, Z.N.; McKenzie, E.H.C. Colletotrichum anthracnose of Amaryllidaceae. Fungal Divers. 2009, 39, 123–146. [Google Scholar]
- Chi, M.H.; Park, S.Y.; Lee, Y.H. A quick and safe method for fungal DNA extraction. Plant Pathol. J. 2009, 25, 108–111. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef] [Green Version]
- Diao, Y.Z.; Zhang, C.; Liu, F.; Wang, W.Z.; Liu, L.; Cai, L.; Liu, X.L. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 2017, 38, 20–37. [Google Scholar] [CrossRef] [Green Version]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Johnston, P.R.; Weir, B.S.; Tan, Y.P.; Shivas, R.G.; Crous, P.W. The Colletotrichum boninense species complex. Stud. Mycol. 2012, 59, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin-Felix, Y.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U.; et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModelTest v. 2; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Swofford, D. PAUP 4.0 b10: Phylogenetic Analysis Using Parsimony (*and Other Methods), 4.0 b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Rambaut, A. FigTree v. 1.4.2; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2014; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 January 2016).
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Huson, D.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Van der Aa, H.A. A leaf spot disease of Nymphaea alba in the Netherlands. Neth. J. Plant Pathol. 1978, 84, 109–115. [Google Scholar]
- Marcelino, J.; Giordano, R.; Gouli, S.; Gouli, V.; Parker, B.L.; Skinner, M.; TeBeest, D.; Cesnik, R. Colletotrichum acutatum var. fioriniae (teleomorph: Glomerella acutata var. fioriniae var. nov.) infection of a scale insect. Mycologia 2008, 100, 353–374. [Google Scholar] [CrossRef] [Green Version]
- Marcelino, J.A.P.; Gouli, S.; Parker, B.L.; Skinner, M.; Schwarzberg, L.; Giordano, R. Host plant associations of an entomopathogenic variety of the fungus, Colletotrichum acutatum, recovered from the elongate hemlock scale, Fiorinia externa. J. Insect Sci. 2009, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Shivas, R.G.; Tan, Y.P. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov and C. simmondsii sp nov. Fungal Divers. 2009, 39, 111–122. [Google Scholar]
- Ohlsens Enkes plantepato logiske Laboratorium. 1 April 1942–31 March 1943. In Pest Alerts; CABI: Kopenhagen, Danmark, 1943; p. 21.
- Neergaard, P. Mycological notes. III. 7. Colletotrichum godetiae Neerg. 8. Phoma bellidis Neerg. 9. Zygosporium parasiticum (Grove) Bunting & Mason. 10. Peronospora di-anthicola Barthelet. Friesia 1950, 4, 72–80. [Google Scholar]
- Schena, L.; Abdelfattah, A.; Mosca, S.; Li Destri Nicosia, M.G.; Agosteo, G.E.; Cacciola, S.O. Quantitative detection of Colletotrichum godetiae and C. acutatum sensu stricto in the phyllosphere and carposphere of olive during four phenological phases. Eur. J. Plant Pathol. 2017, 149, 337–347. [Google Scholar] [CrossRef]
- Prihastuti, H.; Cai, L.; Chen, H.; McKenzie, E.H.C.; Hyde, K.D. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009, 39, 89–109. [Google Scholar]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.L.; Cai, L.; Yu, Z.N.; Liu, Z.Y.; Hyde, K.D. Colletotrichum species on Orchidaceae in southwest China. Cryptogam. Mycol. 2011, 32, 229–253. [Google Scholar]
- Wang, W.; de Silva, D.D.; Moslemi, A.; Edwards, J.; Ades, P.K.; Crous, P.W.; Taylor, P.W.J. Colletotrichum species causing anthracnose of citrus in Australia. J. Fungi 2021, 7, 47. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jeewon, R.; Chen, Y.-J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.-B.; Lin, C.-G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020, 103, 219–271. [Google Scholar] [CrossRef]
- Hu, M.J.; Grabke, A.; Dowling, M.E.; Holstein, H.J.; Schnabel, G. Resistance in Colletotrichum siamense from peach and blueberry to thiophanate-methyl and azoxystrobin. Plant Dis. 2015, 99, 806–814. [Google Scholar] [CrossRef] [Green Version]
- Munir, M.; Amsden, B.; Dixon, E.; Vaillancourt, L.; Gauthier, N.A.W. Characterization of Colletotrichum species causing bitter rot of apple in Kentucky orchards. Plant Dis. 2016, 100, 2194–2203. [Google Scholar] [CrossRef] [Green Version]
- Eaton, M.J.; Edwards, S.; Inocencio, H.A.; Machado, F.J.; Nuckles, E.M.; Farman, M.; Gauthier, N.A.; Vaillancourt, L.J. Diversity and cross-infection potential of Colletotrichum causing fruit rots in mixed-fruit orchards in Kentucky. Plant Dis. 2021, 105, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.X.; Liu, Y.; Huang, X.Q.; Zhang, L. First report of anthracnose caused by Colletotrichum nymphaeae on loquat fruit in China. Plant Dis. 2018, 102, 243. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, X.; Khaskheli, M.I.; Sun, X.; Chang, X.; Gong, G. Identification of Colletotrichum species associated with blueberry anthracnose in Sichuan, China. Pathogens 2020, 9, 718. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.C.; Zeng, X.G.; Xiang, F.Y.; Ren, L.; Chen, F.Y.; Gu, Y.C. Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei, China. Plant Dis. 2016, 100, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhang, W.; Zhou, Y.; Liu, Y.; Yan, J.Y.; Li, X.H.; Jayawardena, R.S.; Hyde, K.D. First report of twig anthracnose on grapevine caused by Colletotrichum nymphaeae in China. Plant Dis. 2016, 100, 2530. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Meng, K.; Shu, J.P.; Zhang, W.; Wang, H.J. First report of anthracnose on pecan (Carya illinoensis) caused by Colletotrichum nymphaeae in China. Plant Dis. 2019, 103, 1432–1433. [Google Scholar] [CrossRef]
- Braganca, C.A.D.; Damm, U.; Baroncelli, R.; Massola, N.S.; Crous, P.W. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biol. 2016, 120, 547–561. [Google Scholar] [CrossRef]
- Wang, N.Y.; Forcelini, B.B.; Peres, N.A. Anthracnose fruit and root necrosis of strawberry are caused by a dominant species within the Colletotrichum acutatum species complex in the United States. Phytopathology 2019, 109, 1293–1301. [Google Scholar] [CrossRef]
- Baroncelli, R.; Sreenivasaprasad, S.; Thon, M.R.; Sukno, S.A. First report of apple bitter rot caused by Colletotrichum godetiae in the United Kingdom. Plant Dis. 2014, 98, 1000–1001. [Google Scholar] [CrossRef] [Green Version]
- Munda, A. First report of Colletotrichum fioriniae and C. godetiae causing apple bitter rot in Slovenia. Plant Dis. 2014, 98, 1282. [Google Scholar] [CrossRef]
- Zapparata, A.; Da Lio, D.; Sarrocco, S.; Vannacci, G.; Baroncelli, R. First report of Colletotrichum godetiae causing grape (Vitis vinifera) berry rot in Italy. Plant Dis. 2017, 101, 1051–1052. [Google Scholar] [CrossRef]
- Karimi, K.; Arzanlou, M.; Pertot, I. Weeds as potential inoculum reservoir for Colletotrichum nymphaeae causing strawberry anthracnose in Iran and rep-PCR fingerprinting as useful marker to differentiate C. acutatum complex on strawberry. Front. Microbiol. 2019, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios, J.A.; Pinho, D.B.; Moreira, W.R.; Pereira, O.L.; Rodrigues, F.A. First report of Colletotrichum karstii causing anthracnose on blueberry leaves in Brazil. Plant Dis. 2015, 99, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Velho, A.C.; Stadnik, M.J.; Wallhead, M. Unraveling Colletotrichum species associated with Glomerella leaf spot of apple. Trop. Plant Pathol. 2019, 44, 197–204. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, X. A discussion on structural adjustment of fruit cultivation in Yunnan. Southest China J. Agric. Sci. 2003, 16, 103–106. [Google Scholar]
- Yang, Y.M.; Tian, K.; Hao, J.M.; Pei, S.J.; Yang, Y.X. Biodiversity and biodiversity conservation in Yunnan, China. Biodivers. Conserv. 2004, 13, 813–826. [Google Scholar] [CrossRef]
- Peres, N.A.R.; Souza, N.L.; Peever, T.L.; Timmer, L.W. Benomyl sensitivity of isolates of Colletotrichum acutatum and C. gloeosporioides from citrus. Plant Dis. 2004, 88, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.H.; Ishii, H.; Nishimura, K.; Fukaya, M.; Yano, K.; Kajitani, Y. Fungicide sensitivity and phylogenetic relationship of anthracnose fungi isolated from various fruit crops in Japan. Plant Dis. 2006, 90, 506–512. [Google Scholar] [CrossRef] [Green Version]
- Usman, H.M.; Tan, Q.; Fan, F.; Karim, M.M.; Yin, W.X.; Zhu, F.X.; Luo, C.X. Sensitivity of Colletotrichum nymphaeae to six fungicides and characterization of fludioxonil resistant isolates in China. Plant Dis. 2021, 106, 165–173. [Google Scholar] [CrossRef]
- Schnabel, G.; Tan, Q.; Schneider, V.; Ishii, H. Inherent tolerance of Colletotrichum gloeosporioides to fludioxonil. Pest. Biochem. Physiol. 2021, 172, 6. [Google Scholar] [CrossRef]
- Usman, H.M.; Tan, Q.; Karim, M.M.; Adnan, M.; Yin, W.X.; Zhu, F.X.; Luo, C.X. Sensitivity of C. fructicola and C. siamense of peach in China to multiple classes of fungicides and characterization of pyraclostrobin-resistant isolates. Plant Dis. 2021, 105, 3459–3465. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Guerber, J.C.; Liu, B.; Correll, J.C.; Johnston, P.R. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 2003, 95, 872–895. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Aveskamp, M.M.; de Gruyter, J.; Spiers, A.G.; Crous, P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 2009, 22, 56–62. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Crous, P.W.; Groenewald, J.Z.; Risede, J.M.; Simoneau, P.; Hywel-Jones, N.L. Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Stud. Mycol. 2004, 50, 415–430. [Google Scholar]


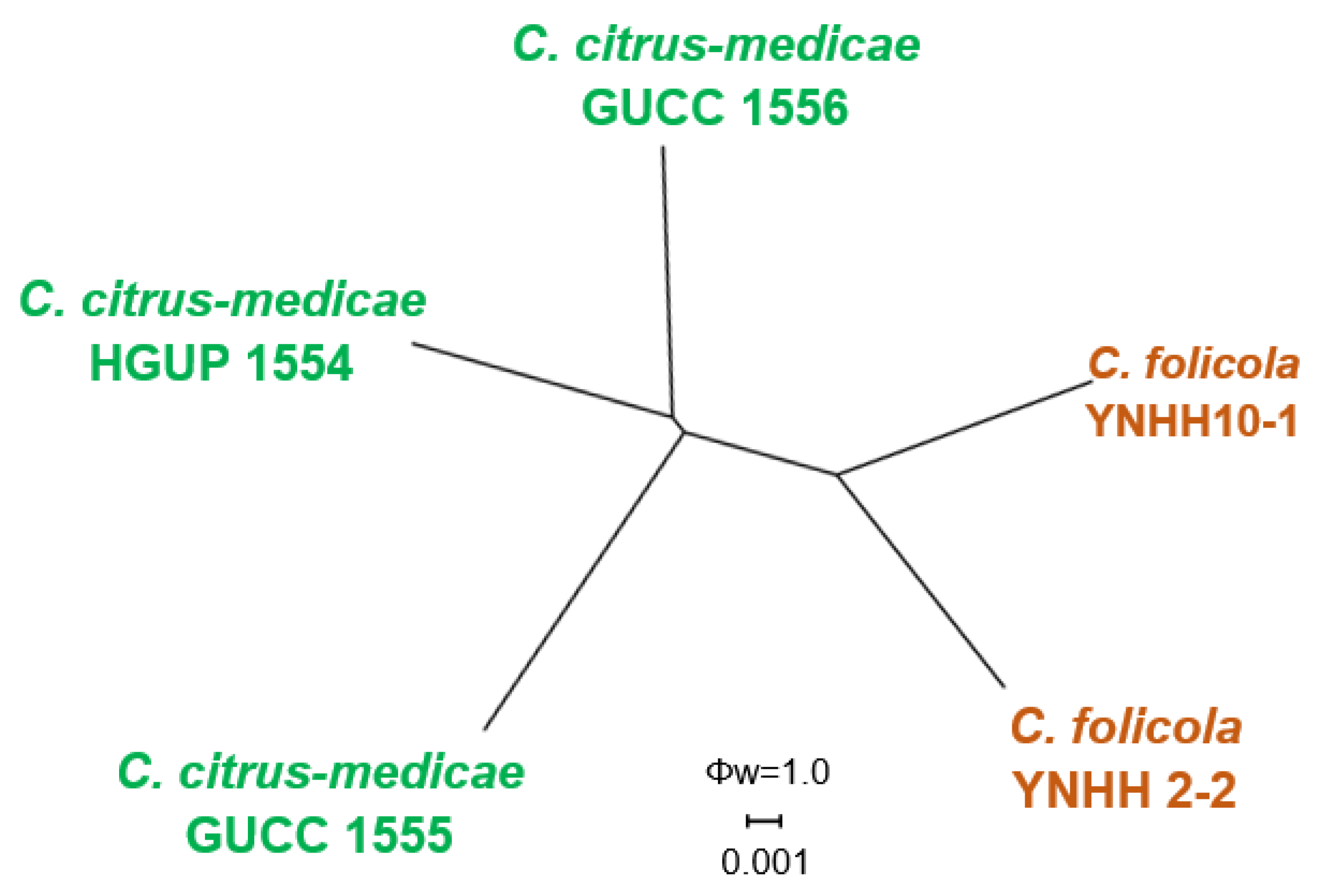

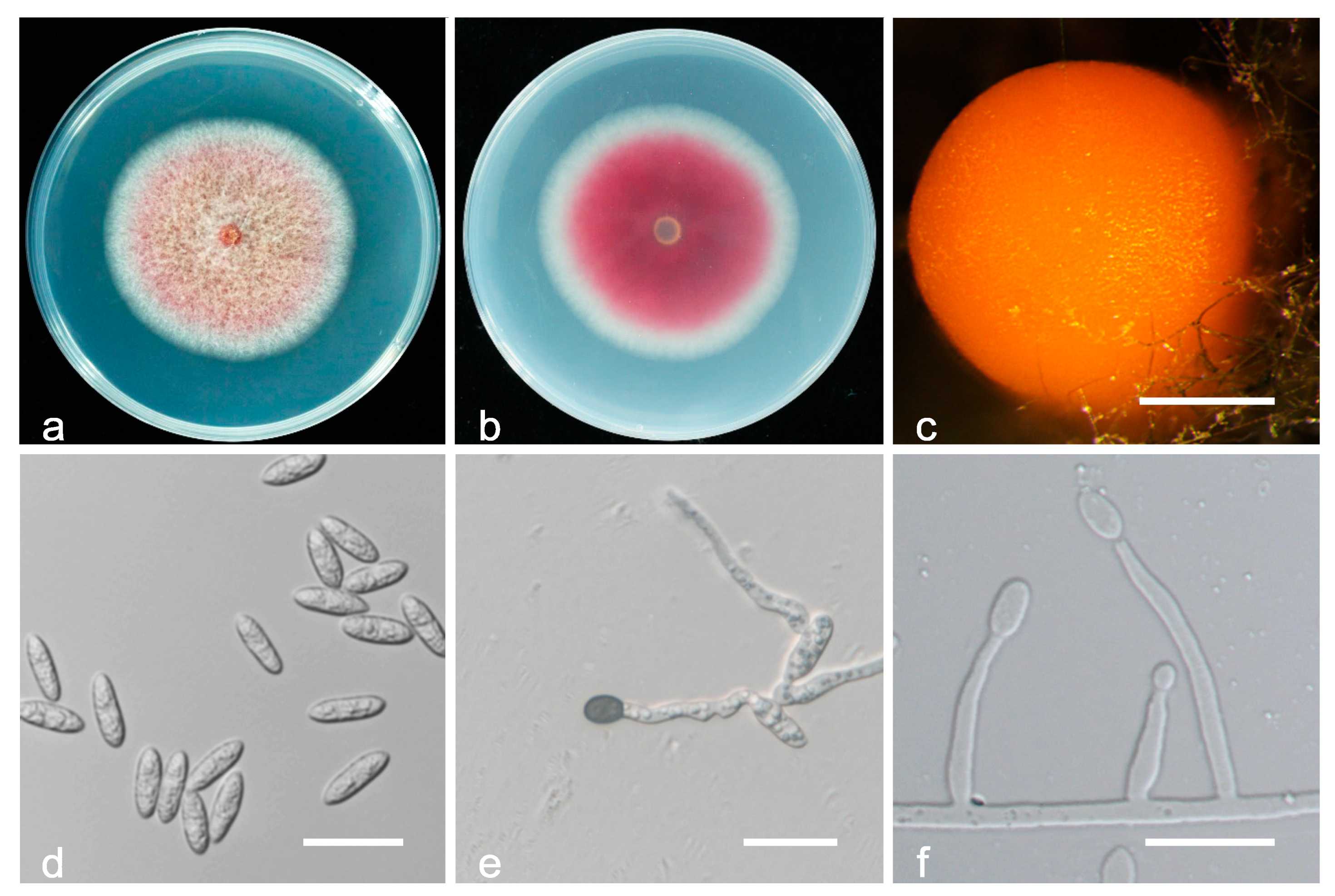
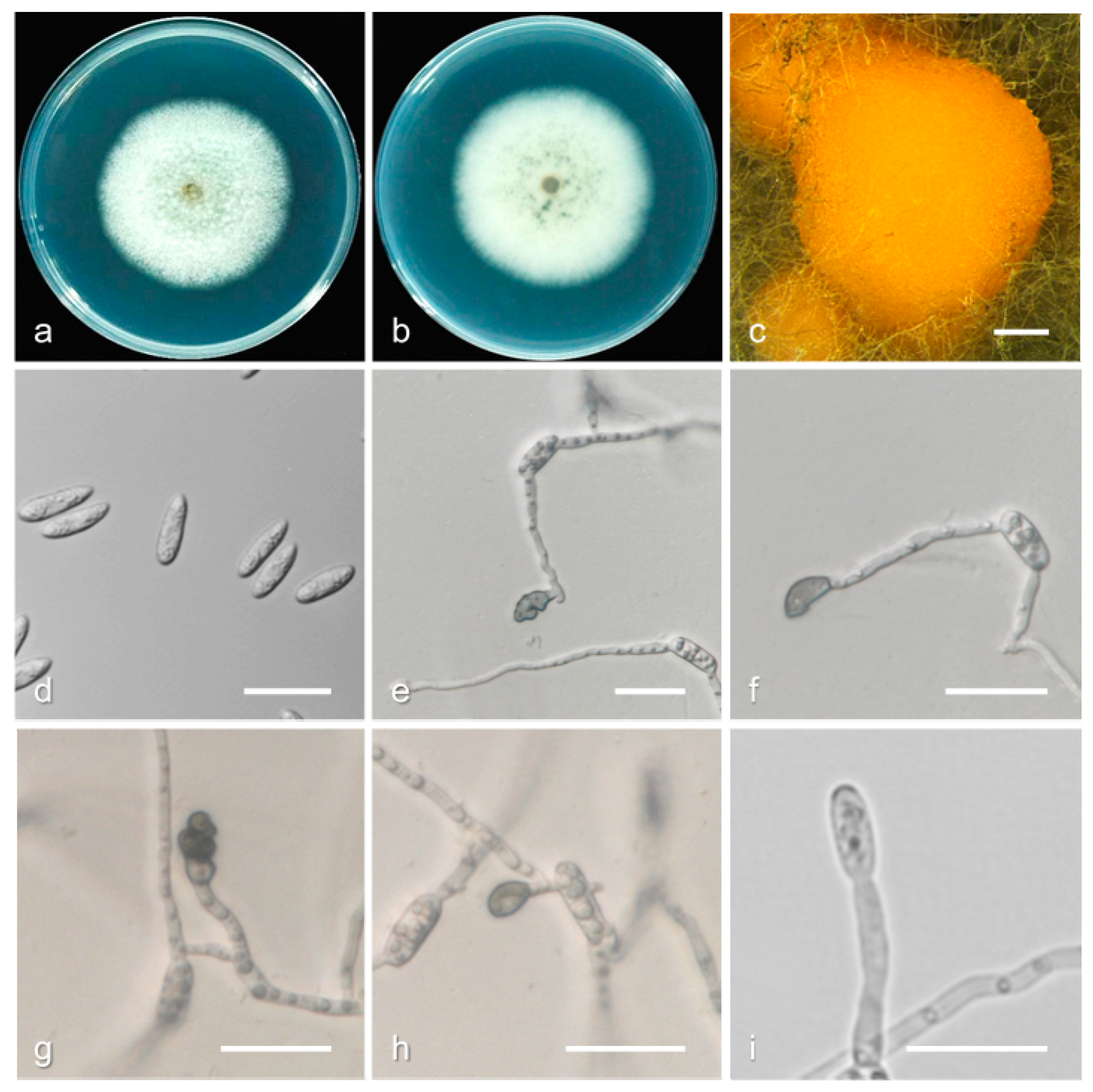
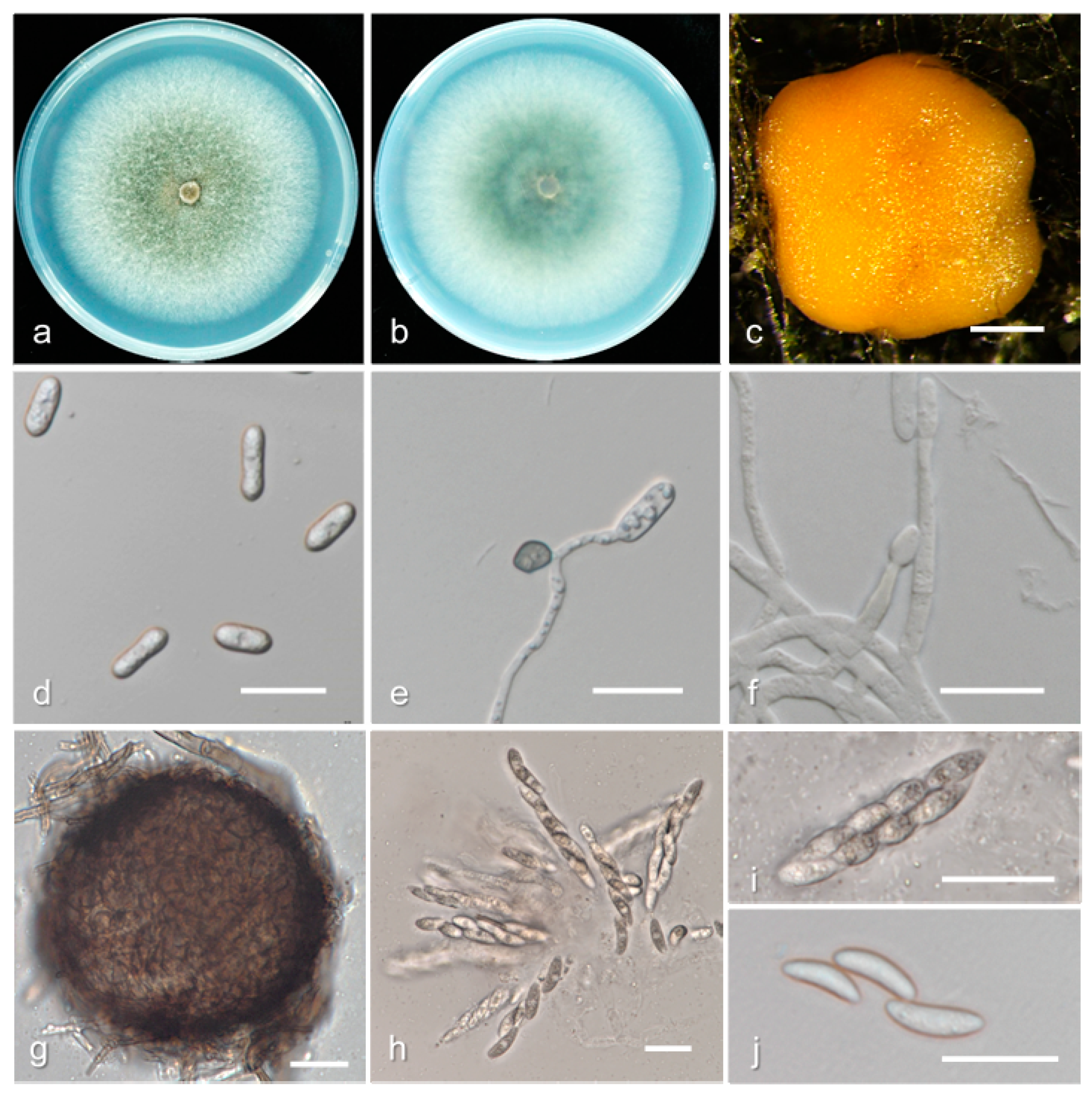


| Species | Culture a | Host | Location | GenBank Accession Number | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | CHS-1 | ACT | HIS3 | TUB2 | CAL | ||||
| C. acerbum | CBS 128530 * | Malus domestica | New Zealand | JQ948459 | JQ948790 | JQ949120 | JQ949780 | JQ949450 | JQ950110 | - |
| C. acutatum | CBS 112996 * | Carica papaya | Australia | JQ005776 | JQ948677 | JQ005797 | JQ005839 | JQ005818 | JQ005860 | - |
| C-1 | Prunus persica | China | KX611163 | KY049983 | - | KY049982 | - | KY049984 | - | |
| C. aenigma | ICMP 18608 * | Persea americana | Israel | JX010244 | JX010044 | JX009774 | JX009443 | - | JX010389 | JX009683 |
| C. aeschynomenes | ICMP 17673 * | Aeschynomene virginica | USA | JX010176 | JX009930 | JX009799 | JX009483 | - | JX010392 | JX009721 |
| C. agaves | CBS 118190 | Agave striate | Mexico | DQ286221 | - | - | - | - | - | - |
| C. alatae | ICMP 17919 * | Dioscorea alata | India | JX010190 | JX009990 | JX009837 | JX009471 | - | JX010383 | JX009738 |
| C. alienum | ICMP 12071 * | Malus domestica | New Zealand | JX010251 | JX010028 | JX009882 | JX009572 | - | JX010411 | JX009654 |
| C. annellatum | CBS 129826 * | Hevea brasiliensis | Colombia | JQ005222 | JQ005309 | JQ005396 | JQ005570 | JQ005483 | JQ005656 | JQ005743 |
| C. aotearoa | ICMP 18537 * | Coprosma sp. | New Zealand | JX010205 | JX010005 | JX009853 | JX009564 | - | JX010420 | JX009611 |
| C. arecicola | CGMCC 3.19667 * | Areca catechu | China | MK914635 | MK935455 | MK935541 | MK935374 | - | MK935498 | - |
| C. artocarpicola | MFLUCC 18-1167 * | Artocarpus heterophyllus | Thailand | MN415991 | MN435568 | MN435569 | MN435570 | - | MN435567 | - |
| C. arxii | CBS 132511 * | Paphiopedilum sp. | Germany | KF687716 | KF687843 | KF687780 | KF687802 | - | KF687881 | - |
| C. asianum | ICMP 18580 * | Coffea arabica | Thailand | FJ972612 | JX010053 | JX009867 | JX009584 | - | JX010406 | FJ917506 |
| C. australe | CBS 116478 * | Trachycarpus fortunei | South Africa | JQ948455 | JQ948786 | JQ949116 | JQ949776 | JQ949446 | JQ950106 | - |
| C.bambusicola | CFCC 54250 * | Phyllostachys edulis | China | MT199632 | MT192844 | MT192871 | MT188638 | - | MT192817 | - |
| C. beeveri | CBS 128527 * | Brachyglottis repanda | New Zealand | JQ005171 | JQ005258 | JQ005345 | JQ005519 | JQ005432 | JQ005605 | JQ005692 |
| C. boninense | CBS 123755 * | Crinum asiaticum var. sinicum | Japan | JQ005153 | JQ005240 | JQ005327 | JQ005501 | JQ005414 | JQ005588 | JQ005674 |
| C. brasiliense | CBS 128501 * | Passiflora edulis | Brazil | JQ005235 | JQ005322 | JQ005409 | JQ005583 | JQ005496 | JQ005669 | JQ005756 |
| C. brassicicola | CBS 101059 * | Brassica oleracea var. gemmifera | New Zealand | JQ005172 | JQ005259 | JQ005346 | JQ005520 | JQ005433 | JQ005606 | JQ005693 |
| C. brisbanense | CBS 292.67 * | Capsicum annuum | Australia | JQ948291 | JQ948621 | JQ948952 | JQ949612 | JQ949282 | JQ949942 | - |
| C. cairnsense | CBS 140847 * | Capsicum annuum | Australia | KU923672 | KU923704 | KU923710 | KU923716 | KU923722 | KU923688 | - |
| C. camelliae-japonicae | CGMCC 3.18118 * | Camellia japonica | Japan | KX853165 | KX893584 | - | KX893576 | - | KX893580 | - |
| C. chlorophyti | IMI 103806 * | Chlorophytum sp. | India | GU227894 | GU228286 | GU228384 | GU227992 | - | GU228188 | - |
| C. chrysanthemi | IMI 364540 | Chrysanthemum coronarium | China | JQ948273 | JQ948603 | JQ948934 | JQ949594 | JQ949264 | JQ949924 | - |
| C. ciggaro | ICMP 18539 * | Olea europaea | Australia | JX010230 | JX009966 | JX009800 | JX009523 | - | JX010434 | JX009635 |
| CBS 237.49 * | Hypericum perforatum | Germany | JX010238 | JX010042 | JX009840 | JX009450 | - | JX010432 | JX009636 | |
| C. citricola | CBS 134228 * | Citrus unshiu | China | KC293576 | KC293736 | - | KC293616 | - | KC293656 | KC293696 |
| C. citrus-medicae | HGUP 1554 *, GUCC 1554 | Citrus medica | China | MN959910 | MT006331 | MT006328 | MT006325 | MT006334 | - | - |
| GUCC 1555 | Citrus medica | China | MN959911 | MT006332 | MT006329 | MT006326 | MT006335 | - | - | |
| GUCC 1556 | Citrus medica | China | MN959912 | MT006333 | MT006330 | MT006327 | MT006336 | - | - | |
| C. clidemiae | ICMP 18658 * | Clidemia hirta | USA | JX010265 | JX009989 | JX009877 | JX009537 | - | JX010438 | JX009645 |
| C. colombiense | CBS 129818 * | Passiflora edulis | Colombia | JQ005174 | JQ005261 | JQ005348 | JQ005522 | JQ005435 | JQ005608 | JQ005695 |
| C. constrictum | CBS 128504 * | Citrus limon | New Zealand | JQ005238 | JQ005325 | JQ005412 | JQ005586 | JQ005499 | JQ005672 | JQ005759 |
| C. cordylinicola | ICMP 18579 * | Cordyline fruticosa | Thailand | JX010226 | JX009975 | JX009864 | HM470235 | - | JX010440 | HM470238 |
| C. curcumae | IMI 288937 * | Curcuma longa | India | GU227893 | GU228285 | GU228383 | GU227991 | - | GU228187 | - |
| C. cuscutae | IMI 304802 * | Cuscuta sp. | Dominica | JQ948195 | JQ948525 | JQ948856 | JQ949516 | JQ949186 | JQ949846 | - |
| C. cymbidiicola | IMI 347923 * | Cymbidium sp. | Australia | JQ005166 | JQ005253 | JQ005340 | JQ005514 | JQ005427 | JQ005600 | JQ005687 |
| C. dacrycarpi | CBS 130241 * | Dacrycarpus dacrydioides | New Zealand | JQ005236 | JQ005323 | JQ005410 | JQ005584 | JQ005497 | JQ005670 | JQ005757 |
| C. dracaenophilum | CBS 118199 * | Dracaena sp. | China | JX519222 | JX546707 | JX519230 | JX519238 | - | JX519247 | - |
| C. eriobotryae | BCRC FU31138 * | Eriobotrya japonica | China | MF772487 | MF795423 | MN191653 | MN191648 | MN19168 | MF795428 | - |
| C. euphorbiae | CBS 134725 * | Euphorbia sp. | South Africa | KF777146 | KF777131 | KF777128 | KF777125 | KF777247 | - | |
| C. fioriniae | CBS 128517 * | Fiorinia externa | USA | JQ948292 | JQ948622 | JQ948953 | JQ949613 | JQ949283 | JQ949943 | - |
| IMI 324996 | Malus pumila | USA | JQ948301 | JQ948631 | JQ948962 | JQ949622 | JQ949292 | JQ949952 | - | |
| CBS 126526 | Primula sp. | Netherlands | JQ948323 | JQ948653 | JQ948984 | JQ949644 | JQ949314 | JQ949974 | - | |
| CBS 124958 | Pyrus sp. | USA | JQ948306 | JQ948636 | JQ948967 | JQ949627 | JQ949297 | JQ949957 | - | |
| CBS 119292 | Vaccinium sp. | New Zealand | JQ948313 | JQ948643 | JQ948974 | JQ949634 | JQ949304 | JQ949964 | - | |
| ICKb31 | Prunus persica | South Korea | LC516639 | LC516653 | LC516660 | - | - | LC516646 | - | |
| ICKb36 | Prunus persica | South Korea | LC516640 | LC516654 | LC516661 | - | - | LC516647 | - | |
| ICKb47 | Prunus persica | South Korea | LC516641 | LC516655 | LC516662 | - | - | LC516648 | - | |
| C.2.4.2 | Prunus persica | USA | KX066091 | KX066094 | - | - | - | KX066088 | - | |
| CaEY12_1 | Prunus persica | USA | KX066093 | KX066096 | - | - | - | KX066090 | - | |
| C. fructicola | ICMP 18581 * | Coffea arabica | Thailand | JX010165 | JX010033 | JX009866 | FJ907426 | - | JX010405 | - |
| ICMP 18613 | Limonium sinuatum | Israel | JX010167 | JX009998 | JX009772 | JX009491 | - | JX010388 | JX009675 | |
| ICMP 18581 * | Coffea arabica | Thailand | JX010165 | JX010033 | JX009866 | FJ907426 | - | JX010405 | FJ917508 | |
| ICMP 18727 | Fragaria × ananassa | USA | JX010179 | JX010035 | JX009812 | JX009565 | - | JX010394 | JX009682 | |
| CBS 125397 * | Tetragastris panamensis | Panama | JX010173 | JX010032 | JX009874 | JX009581 | - | JX010409 | JX009674 | |
| CBS 238.49 * | Ficus edulis | Germany | JX010181 | JX009923 | JX009839 | JX009495 | - | JX010400 | JX009671 | |
| ICKb18 | Prunus persica | South Korea | LC516635 | LC516649 | LC516656 | - | - | LC516642 | LC516663 | |
| ICKb132 | Prunus persica | South Korea | LC516636 | LC516650 | LC516657 | - | - | LC516643 | LC516664 | |
| RR12-3 | Prunus persica | USA | - | KJ769247 | - | - | - | KM245092 | KJ769239 | |
| SE12-1 | Prunus persica | USA | - | KJ769248 | - | - | - | - | KJ769237 | |
| C. fusiforme | MFLUCC 12– 0437 * | unknown | Thailand | KT290266 | KT290255 | KT290253 | KT290251 | - | KT290256 | - |
| C. gigasporum | CBS 133266 * | Centella asiatica | Madagascar | KF687715 | KF687822 | KF687761 | - | - | KF687866 | - |
| C. gloeosporioides | CBS 112999 * | Citrus sinensis | Italy | JQ005152 | JQ005239 | JQ005326 | JQ005500 | JQ005413 | JQ005587 | JQ005673 |
| ICMP 17821 * | Citrus sinensis | Italy | JX010152 | JX010056 | JX009818 | JX009531 | - | JX010445 | JX009731 | |
| C. godetiae | CBS 796.72 | Aeschynomene virginica | USA | JQ948407 | JQ948738 | JQ949068 | JQ949728 | JQ949398 | JQ950058 | - |
| CBS 133.44 * | Clarkia hybrida | Denmark | JQ948402 | JQ948733 | JQ949063 | JQ949723 | JQ949393 | JQ950053 | - | |
| IMI 351248 | Ceanothus sp. | UK | JQ948433 | JQ948764 | JQ949094 | JQ949754 | JQ949424 | JQ950084 | - | |
| C. guangxiense | CFCC 54251 * | Phyllostachys edulis | China | MT199633 | MT192834 | MT192861 | MT188628 | - | MT192805 | - |
| C. hippeastri | CBS 125376 * | Hippeastrum vittatum | China | JQ005231 | JQ005318 | JQ005405 | JQ005579 | JQ005492 | JQ005665 | JQ005752 |
| C. horii | ICMP 10492 * | Diospyros kaki | Japan | GQ329690 | GQ329681 | JX009752 | JX009438 | JX010450 | JX009604 | |
| C. indonesiense | CBS 127551 * | Eucalyptus sp. | Indonesia | JQ948288 | JQ948618 | JQ948949 | JQ949609 | JQ949279 | JQ949939 | - |
| C. javanense | CBS 144963 * | Capsicum annuum | Indonesia | MH846576 | MH846572 | MH846573 | MH846575 | - | MH846574 | - |
| C. jishouense | GZU_HJ2_G2 | Nothapodytes pittosporoides | China | MH482931 | MH681657 | - | MH708134 | - | MH727472 | - |
| C. johnstonii | CBS 128532 * | Solanum lycopersicum | New Zealand | JQ948444 | JQ948775 | JQ949105 | JQ949765 | JQ949435 | JQ950095 | - |
| C. kahawae | IMI 319418 * | Coffea arabica | Kenya | JX010231 | JX010012 | JX009813 | JX009452 | - | JX010444 | - |
| C. karsti | CBS 128524 | Citrullus lanatus | New Zealand | JQ005195 | JQ005282 | JQ005369 | JQ005543 | JQ005456 | JQ005629 | JQ005716 |
| CBS 129824 | Musa AAA | Colombia | JQ005215 | JQ005302 | JQ005389 | JQ005563 | JQ005476 | JQ005649 | JQ005736 | |
| CBS 128552 | Synsepalum dulcificum | Taiwan | JQ005188 | JQ005275 | JQ005362 | JQ005536 | JQ005449 | JQ005622 | JQ005709 | |
| C. laticiphilum | CBS 112989 * | Hevea brasiliensis | India | JQ948289 | JQ948619 | JQ948950 | JQ949610 | JQ949280 | JQ949940 | - |
| C. ledebouriae | CBS 141284 * | Ledebouria floridunda | South Africa | KX228254 | - | - | KX228357 | - | - | - |
| C. liaoningense | CGMCC 3.17616 * | Capsicum sp. | China | KP890104 | KP890135 | KP890127 | KP890097 | - | KP890111 | - |
| C. limetticola | CBS 114.14 * | Citrus aurantifolia | USA | JQ948193 | JQ948523 | JQ948854 | JQ949514 | JQ949184 | JQ949844 | - |
| C. lindemuthianum | CBS 144.31 * | Phaseolus vulgaris | Germany | JQ005779 | JX546712 | JQ005800 | JQ005842 | - | JQ005863 | - |
| C. magnisporum | CBS 398.84 * | unknown | unknown | KF687718 | KF687842 | KF687782 | KF687803 | - | KF687882 | - |
| C. magnum | CBS 519.97 * | Citrullus lanatus | USA | MG600769 | MG600829 | MG600875 | MG600973 | - | MG601036 | - |
| C. makassarense | CBS 143664 * | Capsicum annuum | Indonesia | MH728812 | MH728820 | MH805850 | MH781480 | - | MH846563 | - |
| C. musae | CBS 116870 * | Musa sp. | USA | JX010146 | JX010050 | JX009896 | JX009433 | - | HQ596280 | JX009742 |
| C. neosansevieriae | CBS 139918 * | Sansevieria trifasciata | South Africa | KR476747 | KR476791 | - | KR476790 | - | KR476797 | - |
| C. novae-zelandiae | CBS 128505 * | Capsicum annuum | New Zealand | JQ005228 | JQ005315 | JQ005402 | JQ005576 | JQ005489 | JQ005662 | JQ005749 |
| C. nupharicola | ICMP 18187 * | Nuphar lutea subsp.polysepala | USA | JX010187 | JX009972 | JX009835 | JX009437 | - | JX010398 | JX009663 |
| C. nymphaeae | CBS 515.78 * | Nymphaea alba | Netherlands | JQ948197 | JQ948527 | JQ948858 | JQ949518 | JQ949188 | JQ949848 | - |
| CBS 130.80 | Anemone sp. | Italy | JQ948226 | JQ948556 | JQ948887 | JQ949547 | JQ949217 | JQ949877 | - | |
| IMI 360386 | Pelargonium graveolens | India | JQ948206 | JQ948536 | JQ948867 | JQ949527 | JQ949197 | JQ949857 | - | |
| CBS 125973 | Fragaria × ananassa | UK | JQ948232 | JQ948562 | JQ948893 | JQ949553 | JQ949223 | JQ949883 | - | |
| CaC04_42 | Prunus persica | USA | KX066092 | KX066095 | - | - | - | KX066089 | - | |
| PrpCnSC13–01 | Prunus persica | Brazil | MK761066 | MK770424 | MK770421 | - | - | MK770427 | - | |
| PrpCnSC13–02 | Prunus persica | Brazil | MK765508 | MK770425 | MK770422 | - | - | MK770428 | - | |
| PrpCnSC13–10 | Prunus persica | Brazil | MK765507 | MK770426 | MK770423 | - | - | MK770429 | - | |
| C. oncidii | CBS 129828 * | Oncidium sp. | Germany | JQ005169 | JQ005256 | JQ005343 | JQ005517 | JQ005430 | JQ005603 | JQ005690 |
| C. orbiculare | CBS 570.97 * | Cucumis sativus | Europe | KF178466 | KF178490 | KF178515 | KF178563 | - | KF178587 | - |
| C. orchidearum | CBS 135131 * | Dendrobium nobile | Netherlands | MG600738 | MG600800 | MG600855 | MG600944 | - | MG601005 | - |
| C. orchidophilum | CBS 632.80 * | Dendrobium sp. | USA | JQ948151 | JQ948481 | JQ948812 | JQ949472 | JQ949142 | JQ949802 | - |
| C. parsonsiae | CBS 128525 * | Parsonsia capsularis | New Zealand | JQ005233 | JQ005320 | JQ005407 | JQ005581 | JQ005494 | JQ005667 | JQ005754 |
| C. paxtonii | IMI 165753 * | Musa sp. | Saint Lucia | JQ948285 | JQ948615 | JQ948946 | JQ949606 | JQ949276 | JQ949936 | - |
| C. petchii | CBS 378.94 * | Dracaena marginata | Italy | JQ005223 | JQ005310 | JQ005397 | JQ005571 | JQ005484 | JQ005657 | JQ005744 |
| C. phormii | CBS 118194 * | Phormium sp. | Germany | JQ948446 | JQ948777 | JQ949107 | JQ949767 | JQ949437 | JQ950097 | - |
| C. phyllanthi | CBS 175.67 * | Phyllanthus acidus | India | JQ005221 | JQ005308 | JQ005395 | JQ005569 | JQ005482 | JQ005655 | JQ005742 |
| C. piperis | IMI 71397 * | Piper nigrum | Malaysia | MG600760 | MG600820 | MG600867 | MG600964 | - | MG601027 | - |
| C. pseudomajus | CBS 571.88 * | Camellia sinensis | China | KF687722 | KF687826 | KF687779 | KF687801 | - | KF687883 | - |
| C. psidii | CBS 145.29 * | Psidium sp. | Italy | JX010219 | JX009967 | JX009901 | JX009515 | - | JX010443 | JX009743 |
| C. pyricola | CBS 128531 * | Pyrus communis | New Zealand | JQ948445 | JQ948776 | JQ949106 | JQ949766 | JQ949436 | JQ950096 | - |
| C. pyrifoliae | CGMCC 3.18902 * | Pyrus pyrifolia | China | MG748078 | MG747996 | MG747914 | MG747768 | - | MG748158 | - |
| C. queenslandicum | ICMP 1778 * | Carica papaya | Australia | JX010276 | JX009934 | JX009899 | JX009447 | - | JX010414 | JX009691 |
| C. radicis | CBS 529.93 * | unknown | Costa Rica | KF687719 | KF687825 | KF687762 | KF687785 | - | KF687869 | - |
| C. salicis | CBS 607.94 * | Salix sp. | Netherlands | JQ948460 | JQ948791 | JQ949121 | JQ949781 | JQ949451 | JQ950111 | - |
| C. salsolae | ICMP 19051 * | Salsola tragus | Hungary | JX010242 | JX009916 | JX009863 | JX009562 | - | JX010403 | JX009696 |
| C. sansevieriae | MAFF 239721 * | Sansevieria trifasciata | Japan | AB212991 | - | - | - | - | - | - |
| C. scovillei | CBS 1265299 * | Capsicum sp. | Indonesia | JQ948267 | JQ948597 | JQ948928 | JQ949588 | JQ949258 | JQ949918 | - |
| C. siamense | ICMP 18578 *, MFLU 090230 | Coffea arabica | Thailand | JX010171 | JX009924 | JX009865 | FJ907423 | - | JX010404 | FJ917505 |
| C. siamense (syn. C. hymenocallidis) | CBS 125378 * | Hymenocallis americana | China | JX010278 | JX010019 | GQ856730 | GQ856775 | - | JX010410 | JX009709 |
| C. siamense (syn. C. jasmini-sambac) | CBS 130420 * | Jasminum sambac | Vietnam | HM131511 | HM131497 | JX009895 | HM131507 | - | JX010415 | JX009713 |
| ICKb21 | Prunus persica | South Korea | LC516637 | LC516651 | LC516658 | - | - | LC516644 | LC516665 | |
| ICKb23 | Prunus persica | South Korea | LC516638 | LC516652 | LC516659 | - | - | LC516645 | LC516666 | |
| OD12-1 | Prunus persica | USA | - | KJ769240 | - | - | - | KM245089 | KJ769234 | |
| EY12-1 | Prunus persica | USA | - | KJ769246 | - | - | - | KM245086 | KJ769236 | |
| C. simmondsii | CBS 122122 * | Carica papaya | Australia | JQ948276 | JQ948606 | JQ948937 | JQ949597 | JQ949267 | JQ949927 | - |
| C. sloanei | IMI 364297 * | Theobroma cacao | Malaysia | JQ948287 | JQ948617 | JQ948948 | JQ949608 | JQ949278 | JQ949938 | - |
| C. sojae | ATCC 62257 * | Glycine max | USA | MG600749 | MG600810 | MG600860 | MG600954 | - | MG601016 | - |
| C. sydowii | CBS 135819 | Sambucus sp. | China | KY263783 | KY263785 | KY263787 | KY263791 | - | KY263793 | - |
| C. tainanense | CBS 143666 * | Capsicum annuum | Taiwan | MH728818 | MH728823 | MH805845 | MH781475 | - | MH846558 | - |
| C. theobromicola | CBS 124945 * | Theobroma cacao | Panama | JX010294 | JX010006 | JX009869 | JX009444 | - | JX010447 | JX009591 |
| C. ti | ICMP 4832 * | Cordyline sp. | New Zealand | JX010269 | JX009952 | JX009898 | JX009520 | - | JX010442 | JX009649 |
| C. tongrenense | GZU_TRJ1-37 | Nothapodytes pittosporoides | China | MH482933 | MH705332 | - | MH717074 | - | MH729805 | - |
| C. torulosum | CBS 128544 * | Solanum melongena | New Zealand | JQ005164 | JQ005251 | JQ005338 | JQ005512 | JQ005425 | JQ005598 | JQ005685 |
| C. trichellum | CBS 217.64 * | Hedera helix | UK | GU227812 | GU228204 | GU228302 | GU227910 | - | GU228106 | - |
| C. tropicale | CBS 124949 * | Theobroma cacao | Panama | JX010264 | JX010007 | JX009870 | JX009489 | - | JX010407 | JX009719 |
| C. truncatum | CBS 151.35 * | Phaseolus lunatus | USA | GU227862 | GU228254 | GU228352 | GU227960 | - | GU228156 | - |
| C. vietnamense | CBS 125478 * | Coffea sp. | Vietnam | KF687721 | KF687832 | KF687769 | KF687792 | - | KF687877 | - |
| C. walleri | CBS 125472 * | Coffea sp. | Vietnam | JQ948275 | JQ948605 | JQ948936 | JQ949596 | JQ949266 | JQ949926 | - |
| C. wanningense | CGMCC 3.18936 * | Hevea brasiliensis | China | MG830462 | MG830318 | MG830302 | MG830270 | - | MG830286 | - |
| C. wuxiense | CGMCC 3.17894 * | Camellia sinensis | China | KU251591 | KU252045 | KU251939 | KU251672 | - | KU252200 | KU251833 |
| C. xanthorrhoeae | ICMP 17903 * | Xanthorrhoea preissii | Australia | JX010261 | JX009927 | JX009823 | JX009478 | - | JX010448 | JX009653 |
| C. yunnanense | CBS 132135 * | Buxus sp. | China | JX546804 | JX546706 | JX519231 | JX519239 | - | JX519248 | - |
| Monilochaetes infuscans | CBS 869.96 * | Ipomoea batatas | South Africa | JQ005780 | JX546612 | JQ005801 | JQ005843 | - | JQ005864 | - |
| Gene/Locus | ITS | GAPDH | CHS-1 | HIS3 | ACT | TUB2 | Combined |
|---|---|---|---|---|---|---|---|
| No. of taxa | 72 | 72 | 68 | 60 | 63 | 72 | 72 |
| Aligned length (with gaps) | 546 | 265 | 282 | 387 | 248 | 492 | 2240 |
| Invariable characters | 501 | 152 | 244 | 289 | 170 | 374 | 1750 |
| Uninformative variable characters | 26 | 56 | 13 | 32 | 30 | 60 | 217 |
| Phylogenetically informative characters | 19 | 57 | 25 | 66 | 48 | 58 | 273 |
| Tree length (TL) | 59 | 176 | 64 | 190 | 117 | 165 | 827 |
| Consistency index (CI) | 0.85 | 0.80 | 0.73 | 0.66 | 0.75 | 0.79 | 0.71 |
| Retention index (RI) | 0.97 | 0.95 | 0.94 | 0.93 | 0.94 | 0.94 | 0.93 |
| Rescaled consistency index (RC) | 0.82 | 0.76 | 0.69 | 0.61 | 0.71 | 0.75 | 0.65 |
| Homoplasy index (HI) | 0.15 | 0.20 | 0.27 | 0.34 | 0.25 | 0.21 | 0.30 |
| Nucleotide substitution model | HKY + I | HKY + G | K80 + I | GTR + I + G | GTR + G | GTR + G | GTR + I + G |
| Gene/Locus | ACT | CAL | CHS-1 | GAPDH | ITS | TUB2 | Combined |
|---|---|---|---|---|---|---|---|
| No. of taxa | 54 | 58 | 58 | 62 | 58 | 61 | 62 |
| Aligned length (with gaps) | 314 | 744 | 300 | 307 | 614 | 735 | 3034 |
| Invariable characters | 232 | 520 | 239 | 154 | 555 | 489 | 2209 |
| Uninformative variable characters | 54 | 139 | 22 | 77 | 36 | 156 | 484 |
| Phylogenetically informative characters | 28 | 85 | 39 | 76 | 23 | 90 | 341 |
| Tree length (TL) | 115 | 324 | 102 | 264 | 78 | 349 | 1303 |
| Consistency index (CI) | 0.84 | 0.83 | 0.69 | 0.75 | 0.81 | 0.83 | 0.76 |
| Retention index (RI) | 0.85 | 0.92 | 0.84 | 0.84 | 0.87 | 0.87 | 0.84 |
| Rescaled consistency index (RC) | 0.71 | 0.76 | 0.58 | 0.63 | 0.70 | 0.72 | 0.63 |
| Homoplasy index (HI) | 0.17 | 0.17 | 0.31 | 0.25 | 0.19 | 0.17 | 0.24 |
| Nucleotide substitution model | HKY + G | GTR + G | K80 + G | HKY + I | SYM + I + G | HKY + I | GTR + I + G |
| Gene/Locus | ITS | GAPDH | CHS-1 | HIS3 | ACT | TUB2 | CAL | Combined |
|---|---|---|---|---|---|---|---|---|
| No. of taxa | 25 | 25 | 23 | 23 | 25 | 25 | 24 | 25 |
| Aligned length (with gaps) | 553 | 286 | 280 | 393 | 276 | 502 | 449 | 2763 |
| Invariable characters | 489 | 120 | 224 | 295 | 174 | 348 | 259 | 1932 |
| Uninformative variable characters | 40 | 82 | 25 | 28 | 53 | 75 | 103 | 408 |
| Phylogenetically informative characters | 24 | 84 | 31 | 70 | 49 | 79 | 87 | 423 |
| Tree length (TL) | 87 | 286 | 89 | 210 | 164 | 237 | 300 | 1404 |
| Consistency index (CI) | 0.86 | 0.80 | 0.76 | 0.66 | 0.82 | 0.75 | 0.80 | 0.76 |
| Retention index (RI) | 0.88 | 0.79 | 0.79 | 0.79 | 0.83 | 0.75 | 0.85 | 0.79 |
| Rescaled consistency index (RC) | 0.75 | 0.64 | 0.60 | 0.52 | 0.68 | 0.56 | 0.70 | 0.60 |
| Homoplasy index (HI) | 0.14 | 0.20 | 0.24 | 0.34 | 0.18 | 0.25 | 0.18 | 0.24 |
| Nucleotide substitution model | SYM + I + G | HKY + I | K80 + G | GTR + I + G | GTR + G | HKY + I | HKY + G | GTR + I + G |
| Gene/Locus | ITS | GAPDH | CHS-1 | ACT | TUB2 | combined |
|---|---|---|---|---|---|---|
| No. of taxa | 50 | 47 | 44 | 47 | 44 | 50 |
| Aligned length (with gaps) | 571 | 321 | 265 | 279 | 529 | 1981 |
| Invariable characters | 367 | 63 | 163 | 102 | 223 | 934 |
| Uninformative variable characters | 53 | 21 | 20 | 39 | 50 | 183 |
| Phylogenetically informative characters | 151 | 237 | 82 | 138 | 256 | 864 |
| Tree length (TL) | 630 | 1312 | 389 | 671 | 1300 | 4405 |
| Consistency index (CI) | 0.51 | 0.44 | 0.41 | 0.48 | 0.44 | 0.44 |
| Retention index (RI) | 0.76 | 0.68 | 0.66 | 0.71 | 0.67 | 0.68 |
| Rescaled consistency index (RC) | 0.39 | 0.30 | 0.27 | 0.34 | 0.30 | 0.30 |
| Homoplasy index (HI) | 0.49 | 0.56 | 0.59 | 0.53 | 0.56 | 0.56 |
| Nucleotide substitution model | GTR + I + G | HKY + I + G | GTR + I + G | HKY + I + G | HKY + I + G | GTR + I + G |
| Species | Location | Host | Number of Isolates | Date | Daily Mean Temperature (°C) a |
|---|---|---|---|---|---|
| C. fioriniae | Lishui, Zhejiang | Juicy peach, Yanhong, fruit | 17 | 14 September 2017 | 29 |
| Tongren, Guizhou | Juicy peach, fruit | 14 | 8 August 2018 | 29 | |
| Jian, Jiangxi | Yellow peach, fruit | 6 | 21 August 2018 | 31 | |
| C. folicola | Honghe, Yunnan | Winter peach, Hongxue, leaf | 2 | 17 August 2017 | 26 |
| C. fructicola | Heyuan, Guangdong | Juicy peach, fruit | 19 | 28 June 2017 | 29 |
| Shaoguan, Guangdong | Juicy peach, Yingzui, fruit | 10 | 3 August 2018 | 30 | |
| Tongren, Guizhou | Juicy peach, fruit | 10 | 8 August 2018 | 29 | |
| C. godetiae | Honghe, Yunnan | Winter peach, Hongxue, leaf | 15 | 17 August 2017 | 26 |
| C. karstii | Honghe, Yunnan | Winter peach, Hongxue, leaf | 3 | 17 August 2017 | 26 |
| C. nymphaeae | Yichang, Hubei | Yellow peach, NJC83, fruit | 11 | 30 April 2017 | 19 |
| Jingmen, Hubei | Yellow peach, NJC83, fruit | 14 | 25 April 2017 | 18 | |
| Jingmen, Hubei | Juicy peach, Chunmi, fruit | 11 | 25 April 2017 | 18 | |
| Wuhan, Hubei | Juicy peach, Zaoxianhong, fruit | 17 | 18 April 2017 | 20 | |
| Wuhan, Hubei | Flat peach, Zaoyoupan, fruit | 12 | 18 April 2017 | 20 | |
| Wuhan, Hubei | Juicy peach, leaft | 9 | 14 June 2017 | 25 | |
| Xiaogan, Hubei | Juicy peach, Chunmei, fruit | 4 | 10 May 2017 | 20 | |
| Qingzhen, Guizhou | Juicy peach, Yingqing, fruit | 8 | 21 August 2017 | 24 | |
| Tongren, Guizhou | Juicy peach, fruit | 2 | 08 August 2018 | 29 | |
| Guilin, Guangxi | Juicy peach, Chunmi, fruit | 38 | 18 May 2018 | 25 | |
| Guilin, Guangxi | Juicy peach, Chunmi, leaf | 4 | 18 May 2018 | 25 | |
| Fuzhou, Fujian | Yellow peach, huangjinmi, fruit | 12 | 27 July 2018 | 31 | |
| Chengdu, Sichuan | Yellow peach, Zhongtaojinmi, fruit | 7 | 28 June 2018 | 26 | |
| C. siamense | Qingdao, Shandong | Juicy peach, Yangjiaomi, fruit | 27 | 22 August 2017 | 27 |
| Shijiazhuang, Hebei | Juicy peach, Dajiubao, fruit | 14 | 3 August 2018 | 30 | |
| Total | 286 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Q.; Schnabel, G.; Chaisiri, C.; Yin, L.-F.; Yin, W.-X.; Luo, C.-X. Colletotrichum Species Associated with Peaches in China. J. Fungi 2022, 8, 313. https://doi.org/10.3390/jof8030313
Tan Q, Schnabel G, Chaisiri C, Yin L-F, Yin W-X, Luo C-X. Colletotrichum Species Associated with Peaches in China. Journal of Fungi. 2022; 8(3):313. https://doi.org/10.3390/jof8030313
Chicago/Turabian StyleTan, Qin, Guido Schnabel, Chingchai Chaisiri, Liang-Fen Yin, Wei-Xiao Yin, and Chao-Xi Luo. 2022. "Colletotrichum Species Associated with Peaches in China" Journal of Fungi 8, no. 3: 313. https://doi.org/10.3390/jof8030313
APA StyleTan, Q., Schnabel, G., Chaisiri, C., Yin, L.-F., Yin, W.-X., & Luo, C.-X. (2022). Colletotrichum Species Associated with Peaches in China. Journal of Fungi, 8(3), 313. https://doi.org/10.3390/jof8030313







