Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Plant Material Collection and Decomposition
2.3. Metagenomics
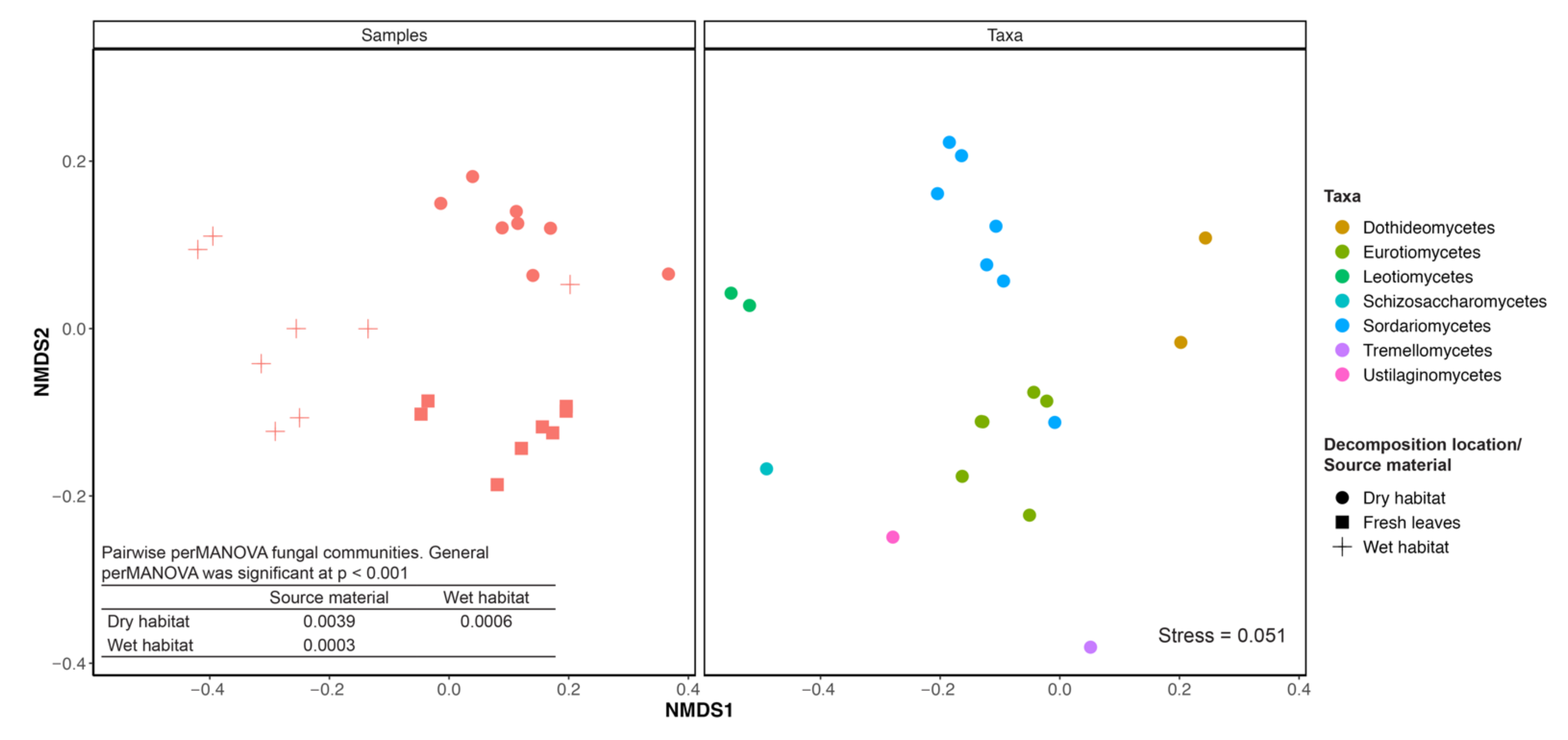
2.4. Effects of Habitat on Fungal Communities
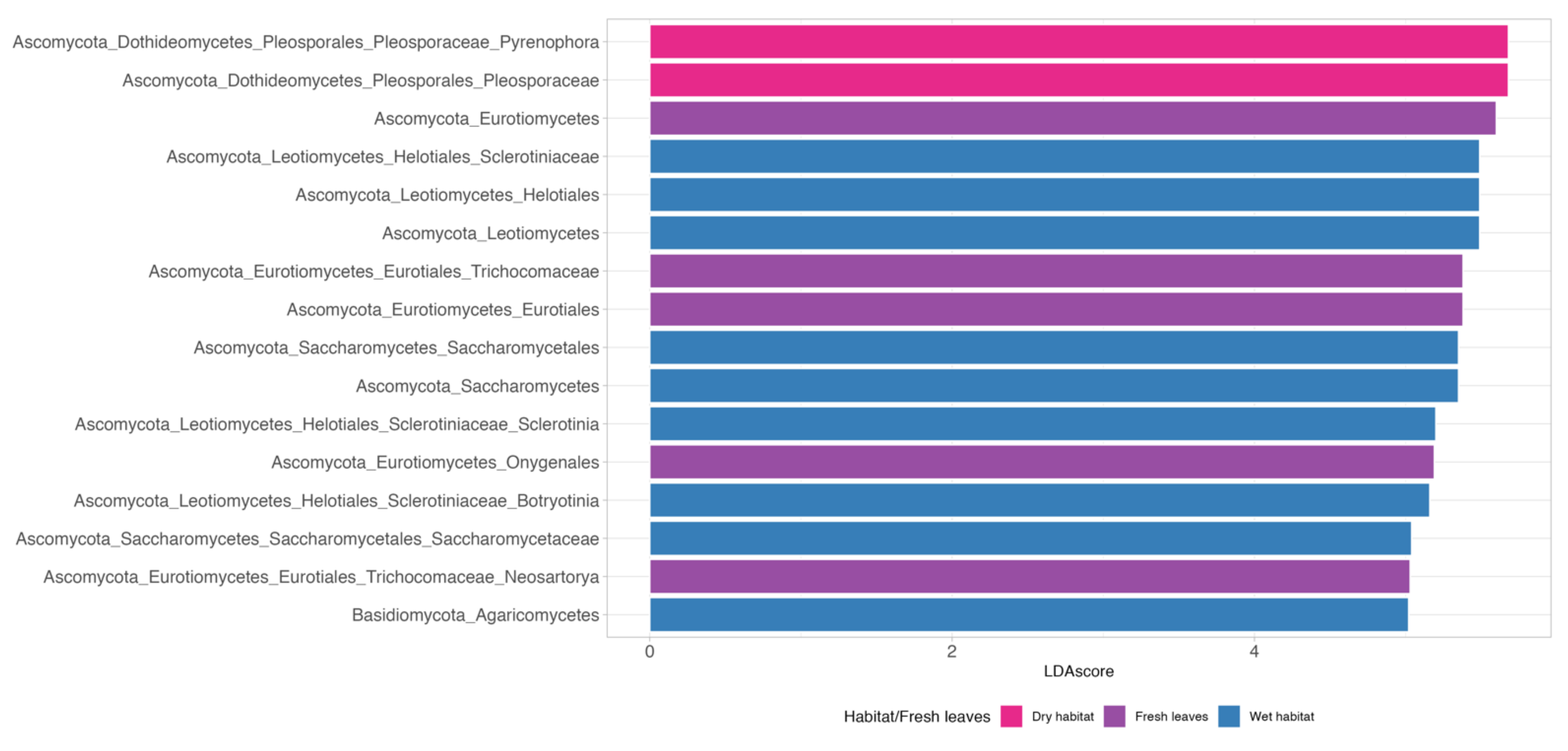
2.5. Indicator Species
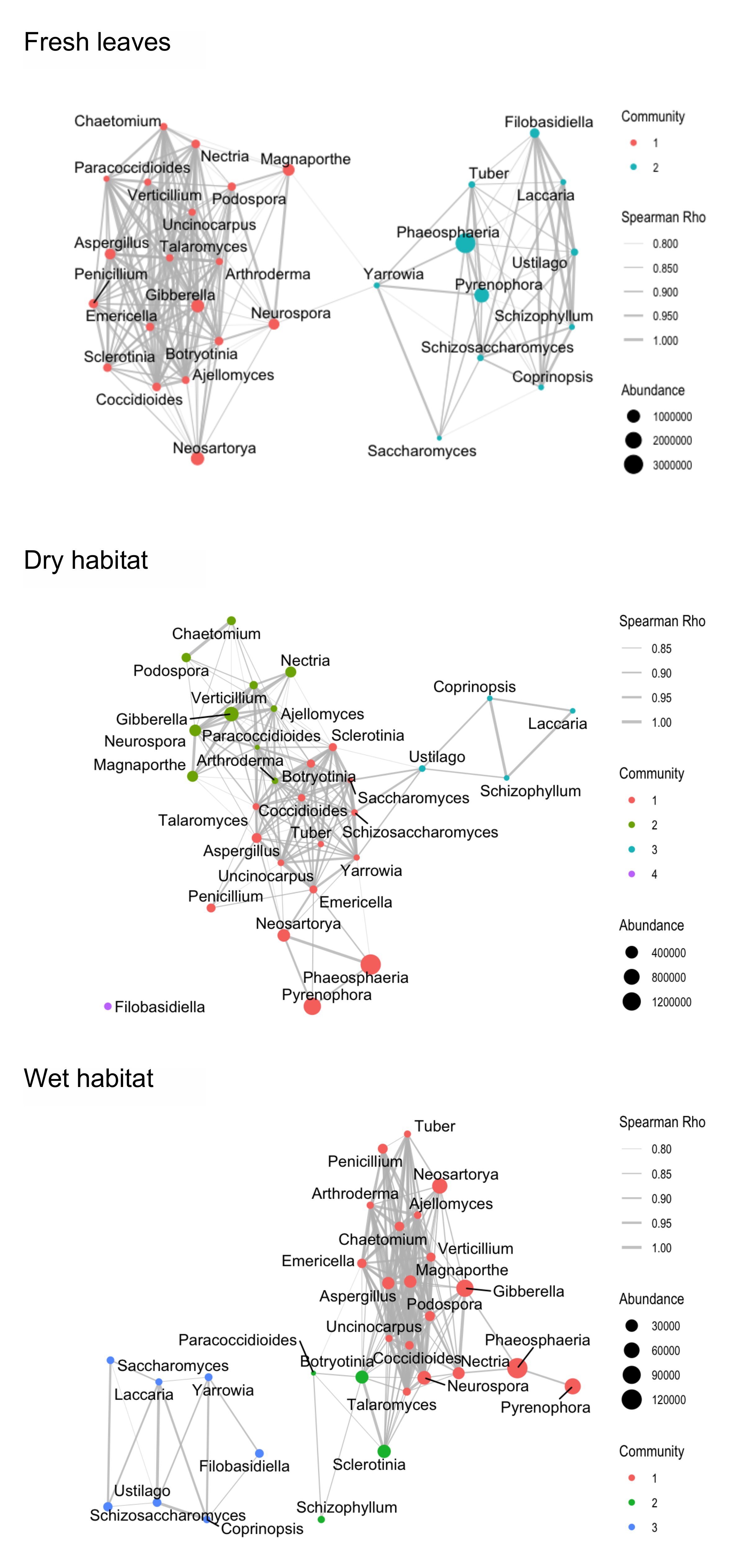
2.6. Co-Occurrence Networks
3. Results
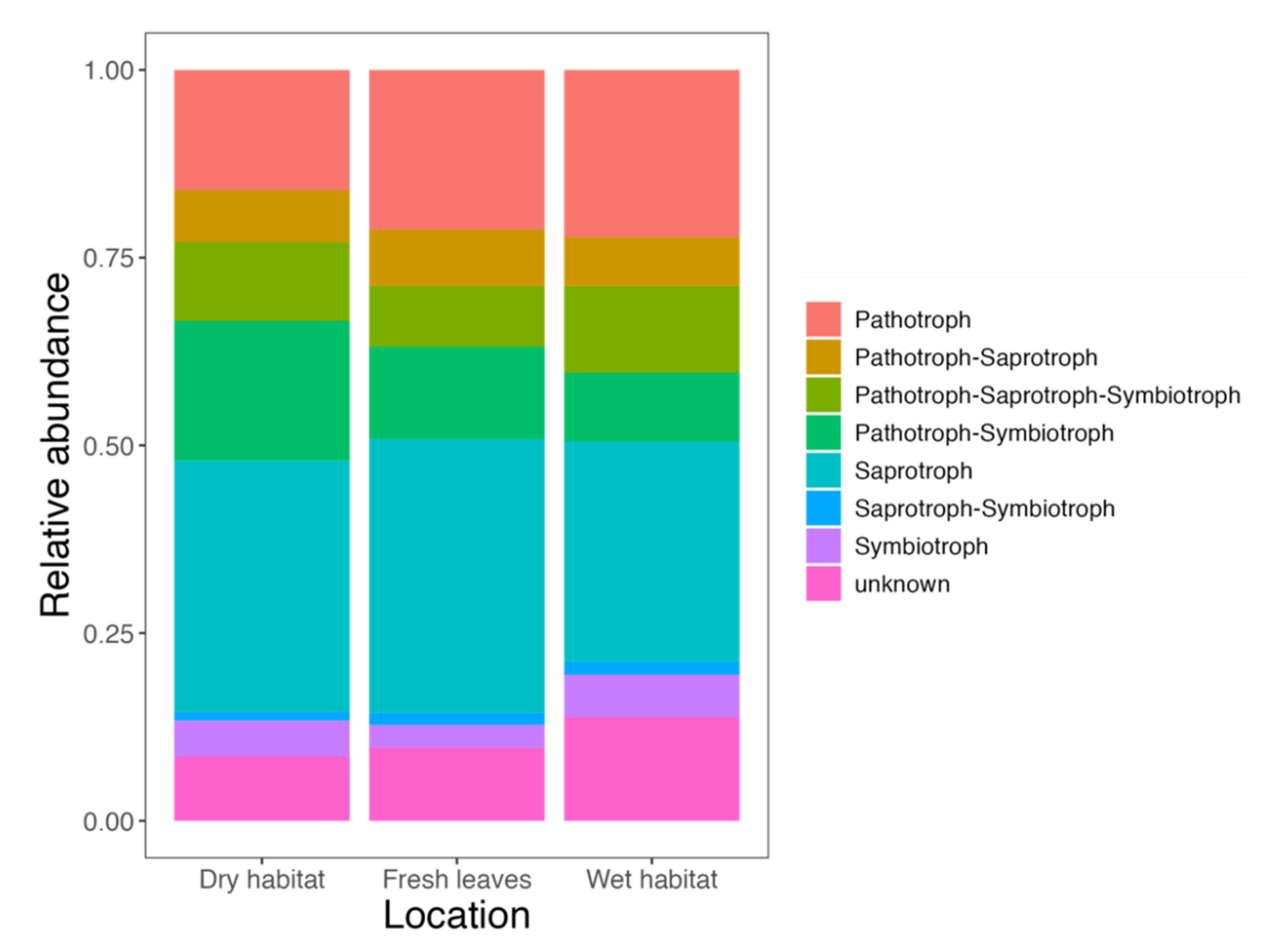
3.1. Diversity and Community Composition
3.2. Indicator Species
3.3. Co-Occurrence Networks
4. Discussion
4.1. Diversity and Community Composition
4.2. Indicator Species
4.3. Co-Occurrence Networks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cronk, J.; Fennesy, S. Wetland Plants: Biology and Ecology; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Longhi, D.; Bartoli, M.; Viaroli, P. Decomposition of four macrophytes in wetland sediments: Organic matter and nutrient decay and associated benthic processes. Aquat. Bot. 2008, 89, 303–310. [Google Scholar] [CrossRef]
- Dolinar, N.; Rudolf, M.; Šraj, N.; Gaberščik, A. Environmental changes affect ecosystem services of the intermittent Lake Cerknica. Ecol. Complex. 2010, 7, 403–409. [Google Scholar] [CrossRef]
- Gaberščik, A.; Grašič, M.; Abram, D.; Zelnik, I. Water level fluctuations and air temperatures affect common reed habitus and productivity in an intermittent wetland ecosystem. Water 2020, 12, 2806. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Gentekaki, E.; Jeewon, R.; Kuo, C.H.; Promputtha, I.; Hyde, K.D. Life in leaf litter: Fungal community succession during decomposition. Mycosphere 2021, 12, 406–429. [Google Scholar] [CrossRef]
- Van Ryckegem, G.; Verbeken, A. Fungal ecology and succession on Phragmites australis in a brackish tidal marsh. I. Leaf sheaths. Fungal Divers. 2005, 19, 157–187. [Google Scholar]
- Van Ryckegem, G.; Verbeken, A. Fungal diversity and community structure on Phragmites australis (Poaceae) along a salinity gradient in the Scheldt estuary (Belgium). Nova Hedwig. 2005, 80, 173–197. [Google Scholar] [CrossRef]
- Komínková, D.; Kuehn, K.A.; Büsing, N.; Steiner, D.; Gessner, M.O. Microbial biomass, growth, and respiration associated with submerged litter of Phragmites australis decomposing in a littoral reed stand of a large lake. Aquat. Microb. Ecol. 2000, 22, 271–282. [Google Scholar] [CrossRef]
- Yarwood, S.A. The role of wetland microorganisms in plant-litter decomposition and soil organic matter formation: A critical review. FEMS Microbiol. Ecol. 2018, 94, fiy175. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, V.; Chauvet, E. Synergistic effects of water temperature and dissolved nutrients on litter decomposition and associated fungi. Glob. Chang. Biol. 2011, 17, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Bedford, A.P. Decomposition of Phragmites australis litter in seasonally flooded and exposed areas of a managed reedbed. Wetlands 2005, 25, 713–720. [Google Scholar] [CrossRef]
- Dolinar, N.; Regvar, M.; Abram, D.; Gaberščik, A. Water-level fluctuations as a driver of Phragmites australis primary productivity, litter decomposition, and fungal root colonisation in an intermittent wetland. Hydrobiologia 2016, 774, 69–80. [Google Scholar] [CrossRef]
- Zhao, B.; Xing, P.; Wu, Q.L. Interactions between bacteria and fungi in macrophyte leaf litter decomposition. Environ. Microbiol. 2021, 23, 1130–1144. [Google Scholar] [CrossRef]
- Langhans, S.D.; Tockner, K. The role of timing, duration, and frequency of inundation in controlling leaf litter decomposition in a river-floodplain ecosystem (Tagliamento, northeastern Italy). Oecologia 2006, 147, 501–509. [Google Scholar] [CrossRef]
- Likar, M.; Dolinar, N.; Vogel-Mikuš, K.; Gaberščik, A.; Regvar, M. Elemental composition and fungal colonisation of decomposing Phragmites australis (Cav.) Trin. ex Steud. litter at different water regimes. Acta Biol. Slov. 2018, 61, 71–84. [Google Scholar]
- Chiba De Castro, W.A.; Almeida, R.V.; Xavier, R.O.; Bianchini, I.; Moya, H.; Silva Matos, D.M. Litter accumulation and biomass dynamics in riparian zones in tropical South America of the Asian invasive plant Hedychium coronarium J. König (Zingiberaceae). Plant. Ecol. Divers. 2020, 13, 47–59. [Google Scholar] [CrossRef]
- Tant, C.J.; Rosemond, A.D.; Helton, A.M.; First, M.R. Nutrient enrichment alters the magnitude and timing of fungal, bacterial, and detritivore contributions to litter breakdown. Freshw. Sci. 2015, 34, 1259–1271. [Google Scholar] [CrossRef]
- Siuda, W.; Chróst, R.J. Decomposition and Utilization of Particulate Organic Matter by Bacteria in Lakes of Different Trophic Status. Pol. J. Environ. Stud. 2002, 11, 53–65. [Google Scholar]
- De Boer, W.; Folman, L.B.; Klein Gunnewiek, P.J.A.; Svensson, T.; Bastviken, D.; Öberg, G.; del Rio, J.C.; Boddy, L. Mechanism of antibacterial activity of the white-rot fungus Hypholoma fasciculare colonizing wood. Can. J. Microbiol. 2010, 56, 380–388. [Google Scholar] [CrossRef]
- Suding, K.N.; Collins, S.L.; Gough, L.; Clark, C.; Cleland, E.E.; Gross, K.L.; Milchunas, D.G.; Pennings, S. Functional-and abundance-based mechanisms explain diversity loss due to N fertilization. Proc. Natl. Acad. Sci. USA 2005, 102, 4387–4392. [Google Scholar] [CrossRef] [Green Version]
- Loreau, M. Linking biodiversity and ecosystems: Towards a unifying ecological theory. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J. BLAT—The BLAST-like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 March 2022).
- De Cáceres, M.; Jansen, F.; Dell, N. indicspecies: Relationship Between Species and Groups of Sites. Available online: https://cran.r-project.org/web/packages/indicspecies/index.html (accessed on 1 March 2022).
- Dufrene, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Sapkota, R.; Knorr, K.; Jørgensen, L.N.; O’Hanlon, K.A.; Nicolaisen, M. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol. 2015, 207, 1134–1144. [Google Scholar] [CrossRef]
- Eusemann, P.; Schnittler, M.; Nilsson, R.H.; Jumpponen, A.; Dahl, M.B.; Würth, D.G.; Buras, A.; Wilmking, M.; Unterseher, M. Habitat conditions and phenological tree traits overrule the influence of tree genotype in the needle mycobiome-Picea glauca system at an arctic treeline ecotone. New Phytol. 2016, 211, 1221–1231. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. qvalue: Q-Value Estimation for False Discovery Rate Control. Available online: http://github.com/jdstorey/qvalue (accessed on 1 March 2022).
- Kai, G. microbial: Do 16s Data Analysis and Generate Figures. Available online: https://cran.r-project.org/web/packages/microbial/index.html (accessed on 1 March 2022).
- Nepusz, T. igraph: Network Analysis and Visualization. Available online: https://cran.r-project.org/web/packages/igraph/index.html (accessed on 1 March 2022).
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research. Available online: https://cran.r-project.org/web/packages/psych/index.html (accessed on 1 March 2022).
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. Available online: https://cran.r-project.org/web/packages/ggraph/index.html (accessed on 1 March 2022).
- Pedersen, T.L. tidygraph: A Tidy API for Graph Manipulation. Available online: https://cran.r-project.org/web/packages/tidygraph/index.html (accessed on 1 March 2022).
- Faust, K.; Raes, J. Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Montoya, D.; Yallop, M.L.; Memmott, J. Functional group diversity increases with modularity in complex food webs. Nat. Commun. 2015, 6, 7379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Mosqueira, M.J.; Fusi, M.; Ramond, J.B.; Merlino, G.; Booth, J.M.; Maggs-Kölling, G.; Cowan, D.A.; Daffonchio, D. Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome 2018, 6, 215. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Hoshino, T.; Inoue, H.; Yano, S. Taxonomic revision of the cellulose-degrading fungus Acremonium cellulolyticus nomen nudum to talaromyces based on phylogenetic analysis. FEMS Microbiol. Lett. 2014, 351, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Alfredsson, H.; Clymans, W.; Stadmark, J.; Conley, D.; Rousk, J. Bacterial and fungal colonization and decomposition of submerged plant litter: Consequences for biogenic silica dissolution. FEMS Microbiol. Ecol. 2016, 92, fiw011. [Google Scholar] [CrossRef] [Green Version]
- Purahong, W.; Wubet, T.; Lentendu, G.; Schloter, M.; Pecyna, M.J.; Kapturska, D.; Hofrichter, M.; Krüger, D.; Buscot, F. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol. Ecol. 2016, 25, 4059–4074. [Google Scholar] [CrossRef]
- Wallis, E.; Raulings, E. Relationship between water regime and hummock-building by Melaleuca ericifolia and Phragmites australis in a brackish wetland. Aquat. Bot. 2011, 95, 182–188. [Google Scholar] [CrossRef]
- Klančnik, K.; Vogel-Mikuš, K.; Kelemen, M.; Vavpetič, P.; Pelicon, P.; Kump, P.; Jezeršek, D.; Gianoncelli, A.; Gaberščik, A. Leaf optical properties are affected by the location and type of deposited biominerals. J. Photochem. Photobiol. B Biol. 2014, 140, 276–285. [Google Scholar] [CrossRef]
- Siddique, A.B.; Unterseher, M. A cost-effective and efficient strategy for Illumina sequencing of fungal communities: A case study of beech endophytes identified elevation as main explanatory factor for diversity and community composition. Fungal Ecol. 2016, 20, 175–185. [Google Scholar] [CrossRef]
- Vincent, J.B.; Weiblen, G.D.; May, G. Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol. Ecol. 2016, 25, 825–841. [Google Scholar] [CrossRef]
- Yang, T.; Sun, H.; Shen, C.; Chu, H. Fungal assemblages in different habitats in an Erman’s birch forest. Front. Microbiol. 2016, 7, 1368. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zheng, Y.; Xu, G.; Guo, Q.; Tan, J.; Ding, G. Fungal diversity within the phyllosphere of Pinus massoniana and the possible involvement of phyllospheric fungi in litter decomposition. Fungal Biol. 2021, 125, 785–795. [Google Scholar] [CrossRef]
- Davey, M.L.; Heegaard, E.; Halvorsen, R.; Ohlson, M. Amplicon-pyrosequencing-based detection of compositional shifts in bryophyte-associated fungal communities along an elevation gradient. Mol. Ecol. 2013, 22, 368–383. [Google Scholar] [CrossRef]
- Peršoh, D. Plant-associated fungal communities in the light of meta’omics. Fungal Divers. 2015, 75, 1–25. [Google Scholar] [CrossRef]
- Angelini, P.; Rubini, A.; Gigante, D.; Reale, L.; Pagiotti, R.; Venanzoni, R. The endophytic fungal communities associated with the leaves and roots of the common reed (Phragmites australis) in Lake Trasimeno (Perugia, Italy) in declining and healthy stands. Fungal Ecol. 2012, 5, 683–693. [Google Scholar] [CrossRef]
- Findley, K.; Rodriguez-Carres, M.; Metin, B.; Kroiss, J.; Fonseca, Á.; Vilgalys, R.; Heitman, J. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the tremellales. Eukaryot. Cell 2009, 8, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Voriskova, J.; Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013, 7, 477–486. [Google Scholar] [CrossRef]
- Vivelo, S.; Bhatnagar, J.M. An evolutionary signal to fungal succession during plant litter decay. FEMS Microbiol. Ecol. 2019, 95, fiz145. [Google Scholar] [CrossRef]
- Baldrian, P.; Valášková, V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 2008, 32, 501–521. [Google Scholar] [CrossRef] [Green Version]
- Duong, L.M.; Mckenzie, E.H.C.; Lumyong, S.; Hyde, K.D. Fungal succession on senescent leaves of Castanopsis diversifolia in Doi Suthep-Pui National Park, Thailand. Fungal Divers. 2008, 30, 23–36. [Google Scholar]
- Vacher, C.; Hampe, A.; Porté, A.J.; Sauer, U.; Compant, S.; Morris, C.E. The Phyllosphere: Microbial Jungle at the Plant–Climate Interface. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Bukovská, P.; Jelínková, M.; Hršelová, H.; Sỳkorová, Z.; Gryndler, M. Terminal restriction fragment length measurement errors are affected mainly by fragment length, G+C nucleotide content and secondary structure melting point. J. Microbiol. Methods 2010, 82, 223–228. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Vicente, V.A.; Najafzadeh, M.J.; Harrak, M.J.; Badali, H.; Seyedmousavi, S. Waterborne Exophiala species causing disease in cold-blooded animals. Pers. Mol. Phylogeny Evol. Fungi 2011, 27, 46–72. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, B.C.; Zhang, C.; van Dorst, J. Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-antarctic soil through cultivation using both a high and a low nutrient media approach. Front. Microbiol. 2011, 2, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koivusaari, P.; Tejesvi, M.V.; Tolkkinen, M.; Markkola, A.; Mykrä, H.; Pirttilä, A.M. Fungi originating from tree leaves contribute to fungal diversity of litter in streams. Front. Microbiol. 2019, 10, 651. [Google Scholar] [CrossRef] [PubMed]
- Wirsel, S.G.R.; Leibinger, W.; Ernst, M.; Mendgen, K. Genetic diversity of fungi closely associated with common reed. New Phytol. 2001, 149, 589–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef]
- Häffner, E.; Konietzki, S.; Diederichsen, E. Keeping control: The role of senescence and development in plant pathogenesis and defense. Plants 2015, 4, 449–488. [Google Scholar] [CrossRef]
- Qian, X.; Chen, L.; Guo, X.; He, D.; Shi, M.; Zhang, D. Shifts in community composition and co-occurrence patterns of phyllosphere fungi inhabiting Mussaenda shikokiana along an elevation gradient. PeerJ 2018, 6, e5767. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Aschenbrenner, I.A.; Cernava, T.; Erlacher, A.; Berg, G.; Grube, M. Differential sharing and distinct co-occurrence networks among spatially close bacterial microbiota of bark, mosses and lichens. Mol. Ecol. 2017, 26, 2826–2838. [Google Scholar] [CrossRef]
- Chow, C.E.T.; Kim, D.Y.; Sachdeva, R.; Caron, D.A.; Fuhrman, J.A. Top-down controls on bacterial community structure: Microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014, 8, 816–829. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, N.; López-Quintero, C.A.; Vasco-Palacios, A.M.; Frisvad, J.C.; Theelen, B.; Boekhout, T.; Samson, R.A.; Houbraken, J. Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycol. Prog. 2016, 15, 1041–1056. [Google Scholar] [CrossRef] [Green Version]
| Observed | Chao1 | Shannon | Simpson | |
|---|---|---|---|---|
| Fresh leaves | 117.0 ± 3.0 | 132.4 ± 11.0 | 2.706 ± 0.08 | 0.876 ± 0.02 |
| Dry habitat | 110.1 ± 6.4 | 122.9 ± 7.0 | 2.521 ± 0.14 | 0.859 ± 0.03 |
| Wet habitat | 85.25 ± 9.3 | 85.25 ± 9.3 | 2.926 ± 0.26 | 0.910 ± 0.04 |
| Network Parameter | Fresh Leaves | Dry Habitat | Wet Habitat |
|---|---|---|---|
| No. nodes | 30 | 30 | 30 |
| No. edges | 229 | 187 | 171 |
| Average degree | 15.3 | 12.5 | 11.4 |
| Clustering coefficient | 0.90 | 0.72 | 0.87 |
| Network diameter | 4.24 | 4.43 | 4.31 |
| Network centralization | 0.16 | 0.29 | 0.23 |
| Network density | 0.53 | 0.43 | 0.39 |
| Average path length | 2.25 | 1.96 | 1.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Likar, M.; Grašič, M.; Stres, B.; Regvar, M.; Gaberščik, A. Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats. J. Fungi 2022, 8, 284. https://doi.org/10.3390/jof8030284
Likar M, Grašič M, Stres B, Regvar M, Gaberščik A. Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats. Journal of Fungi. 2022; 8(3):284. https://doi.org/10.3390/jof8030284
Chicago/Turabian StyleLikar, Matevž, Mateja Grašič, Blaž Stres, Marjana Regvar, and Alenka Gaberščik. 2022. "Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats" Journal of Fungi 8, no. 3: 284. https://doi.org/10.3390/jof8030284
APA StyleLikar, M., Grašič, M., Stres, B., Regvar, M., & Gaberščik, A. (2022). Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats. Journal of Fungi, 8(3), 284. https://doi.org/10.3390/jof8030284








