Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Fungal Isolation and Cultivation
2.3. DNA Extraction and Amplification
2.4. Phylogenetic Analysis
2.5. Morphological Study
3. Results
3.1. Isolated Fungi
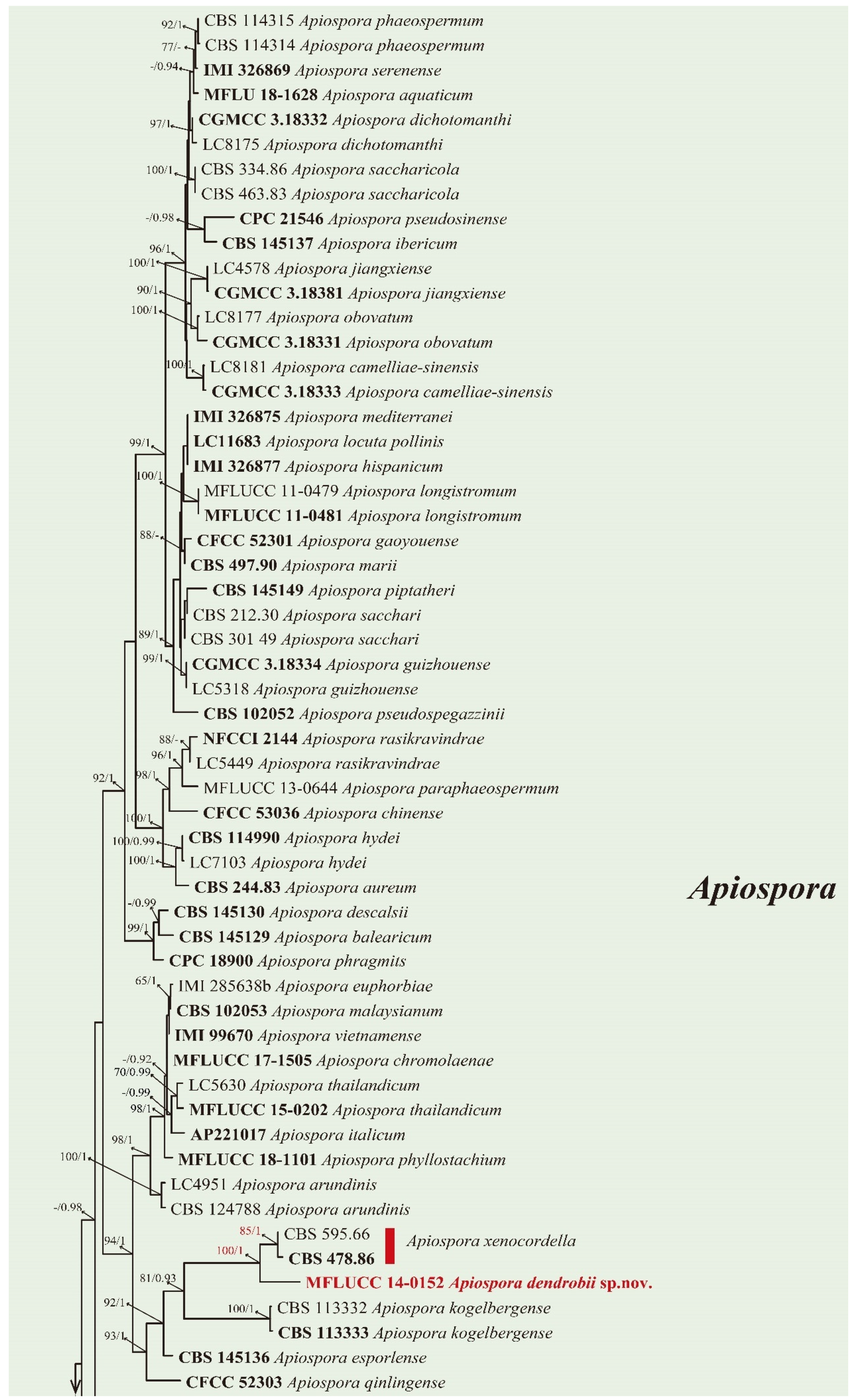
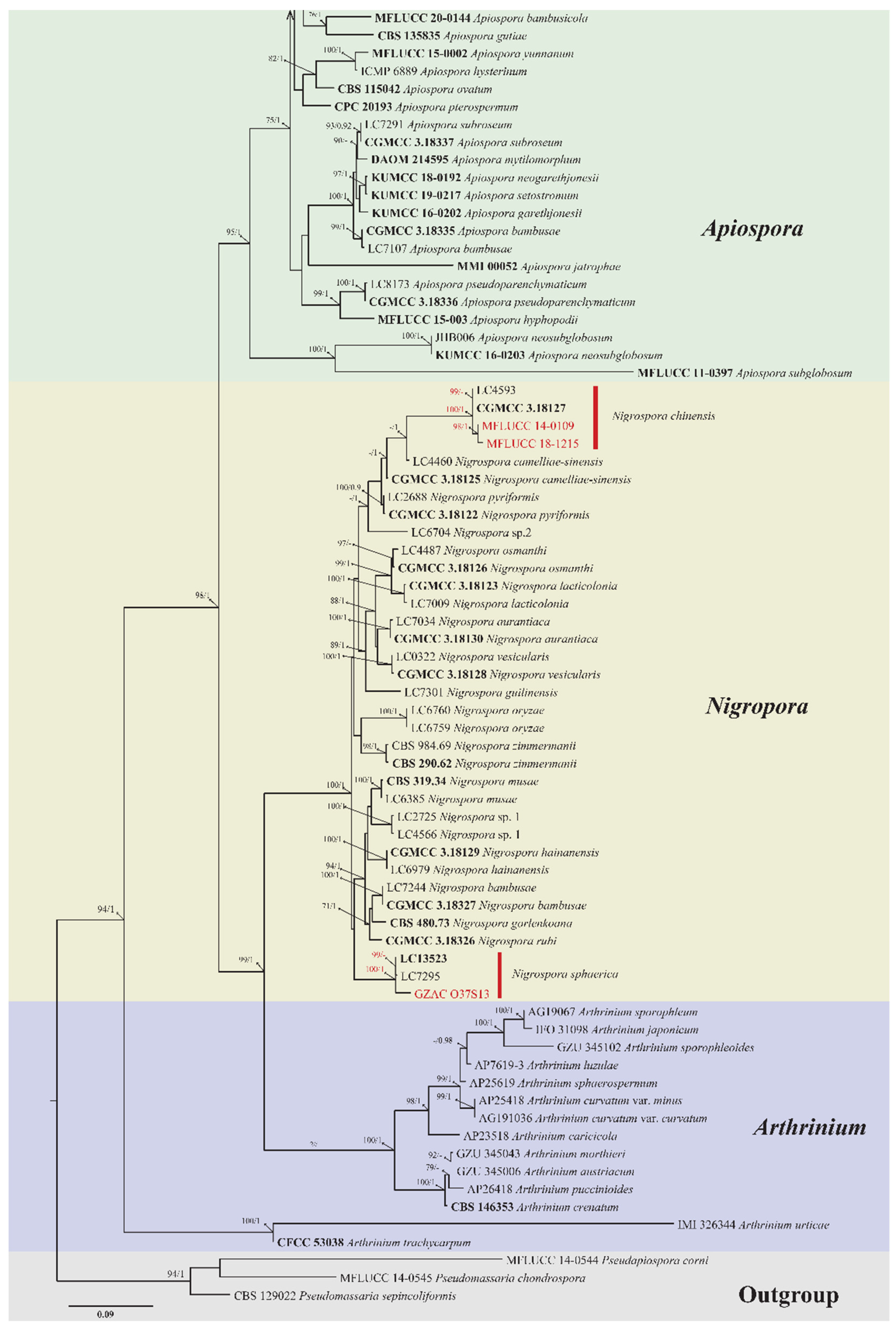
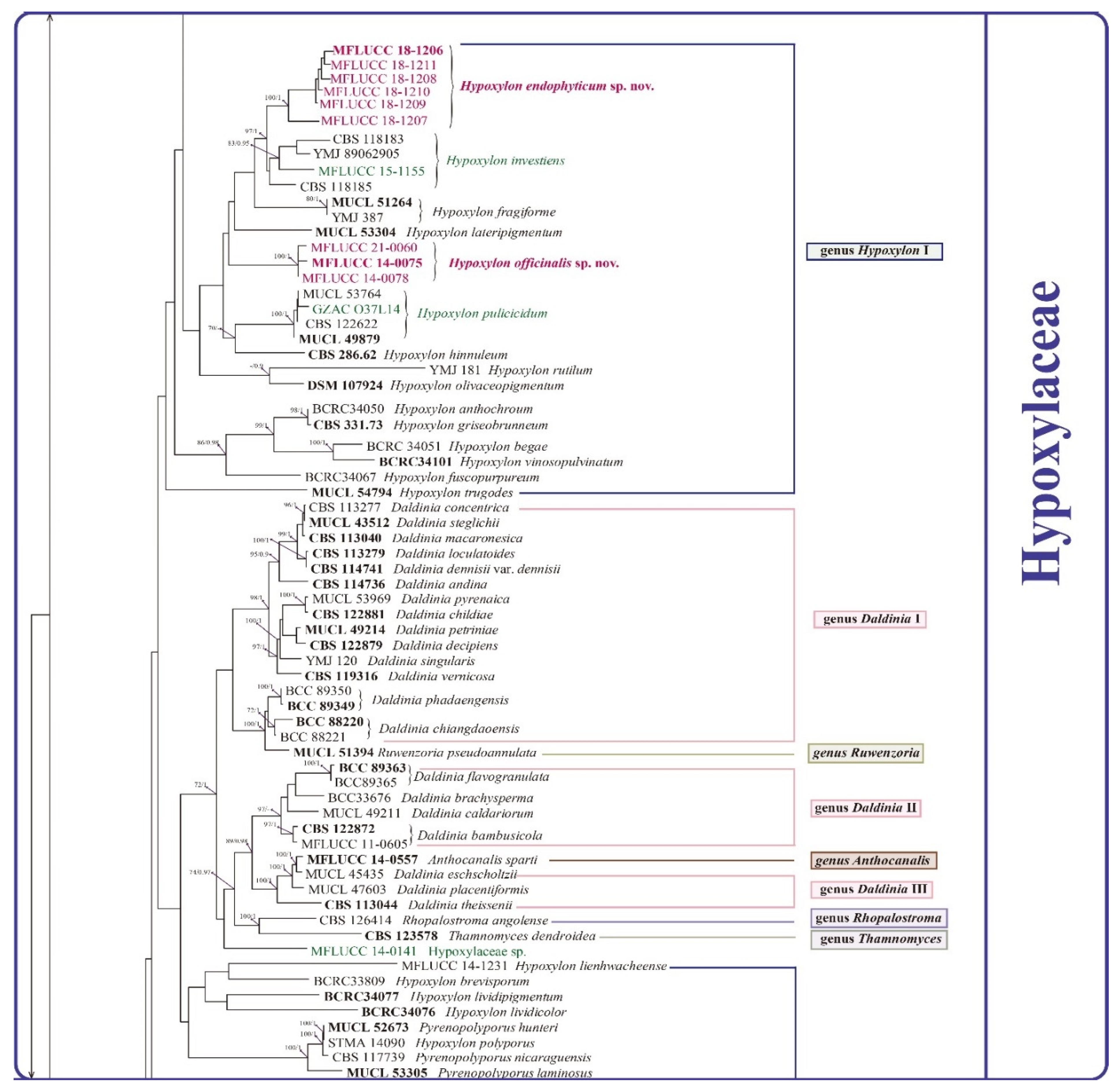
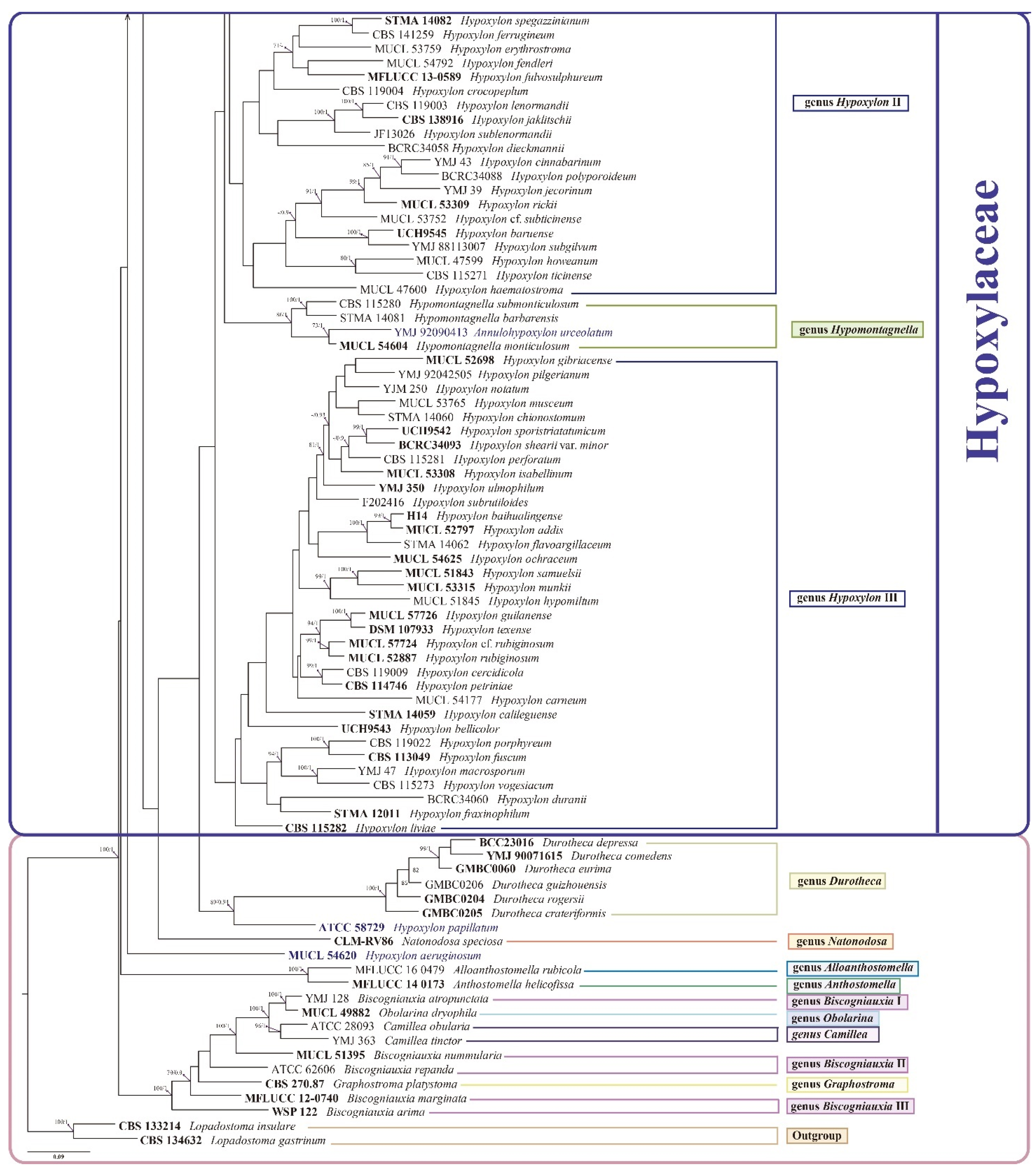
3.2. Phylogenetic Analysis
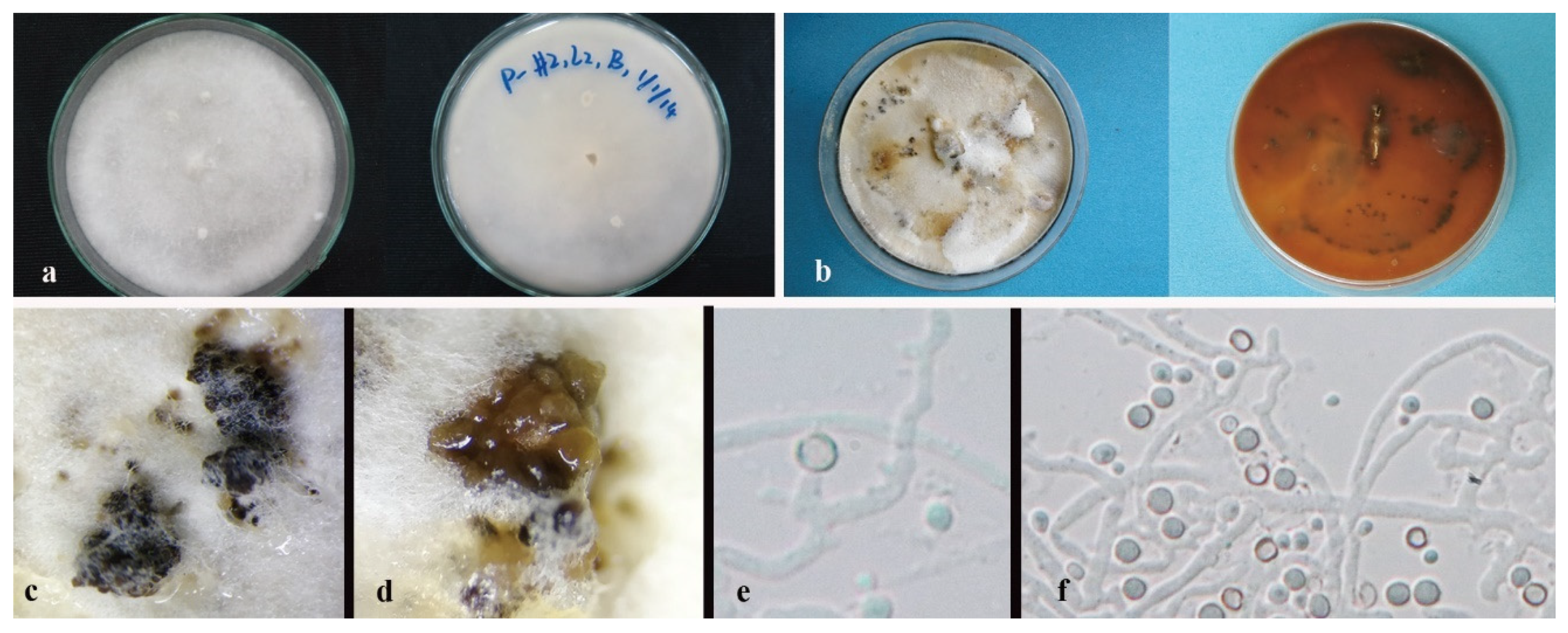
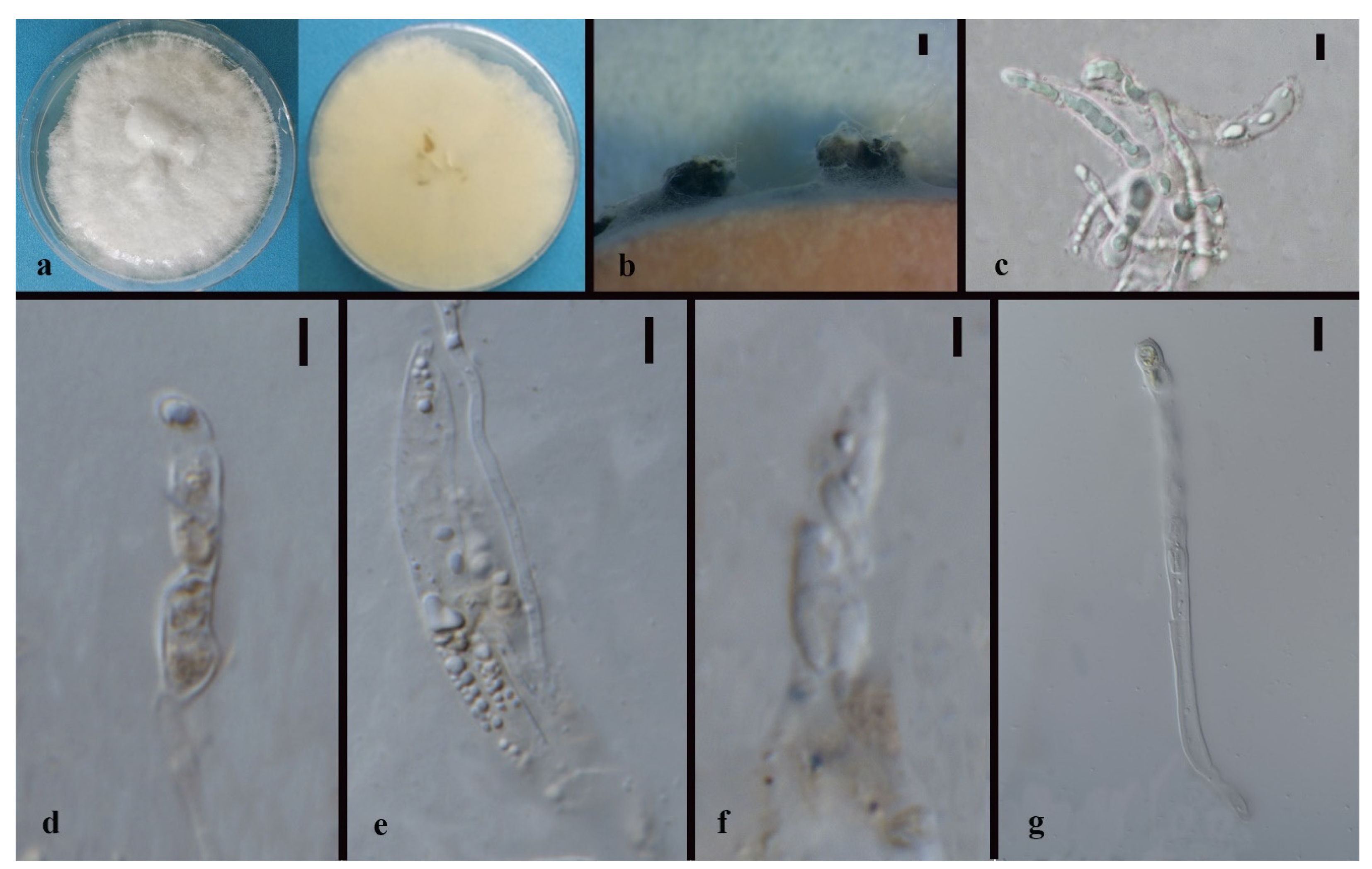
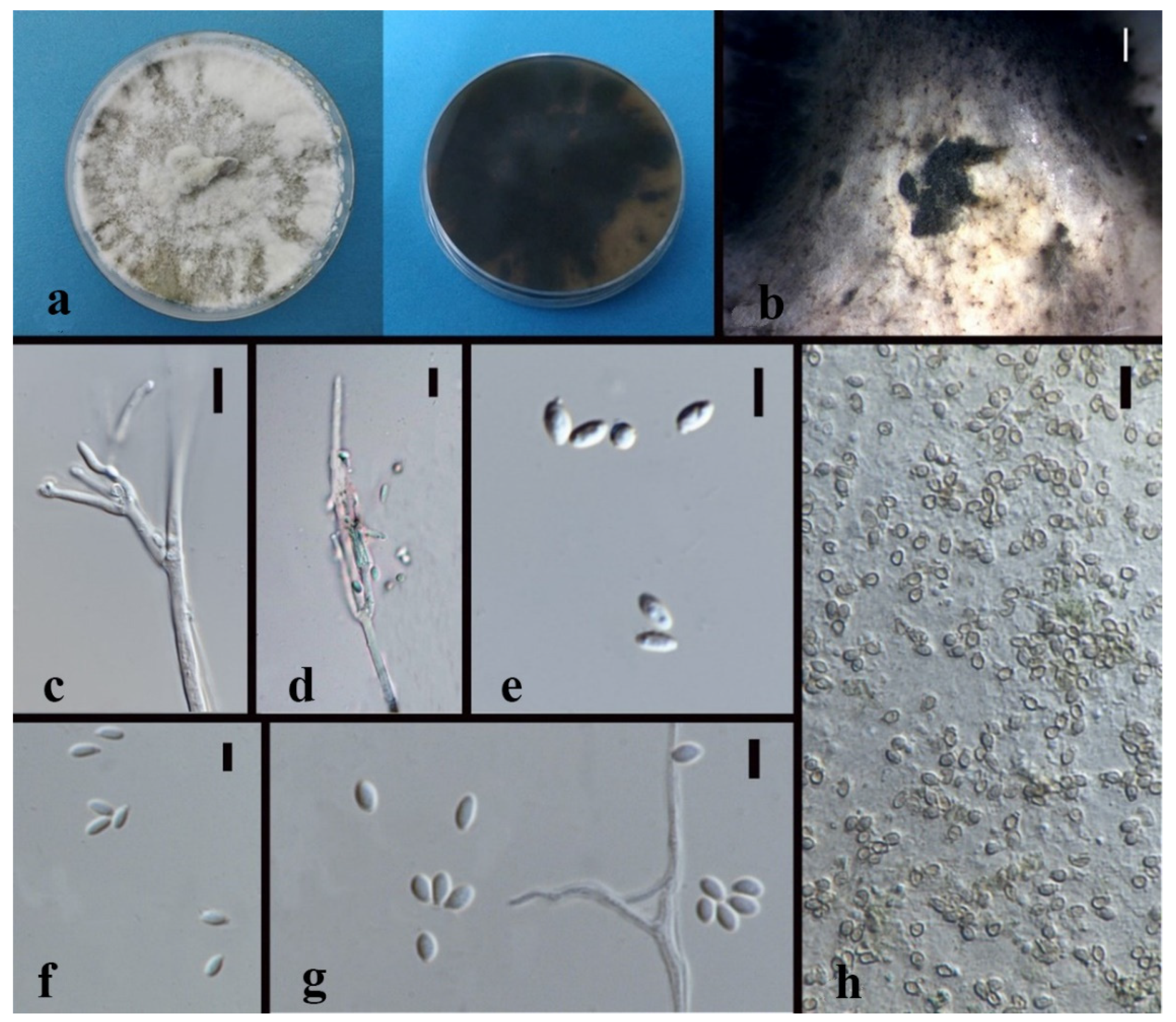
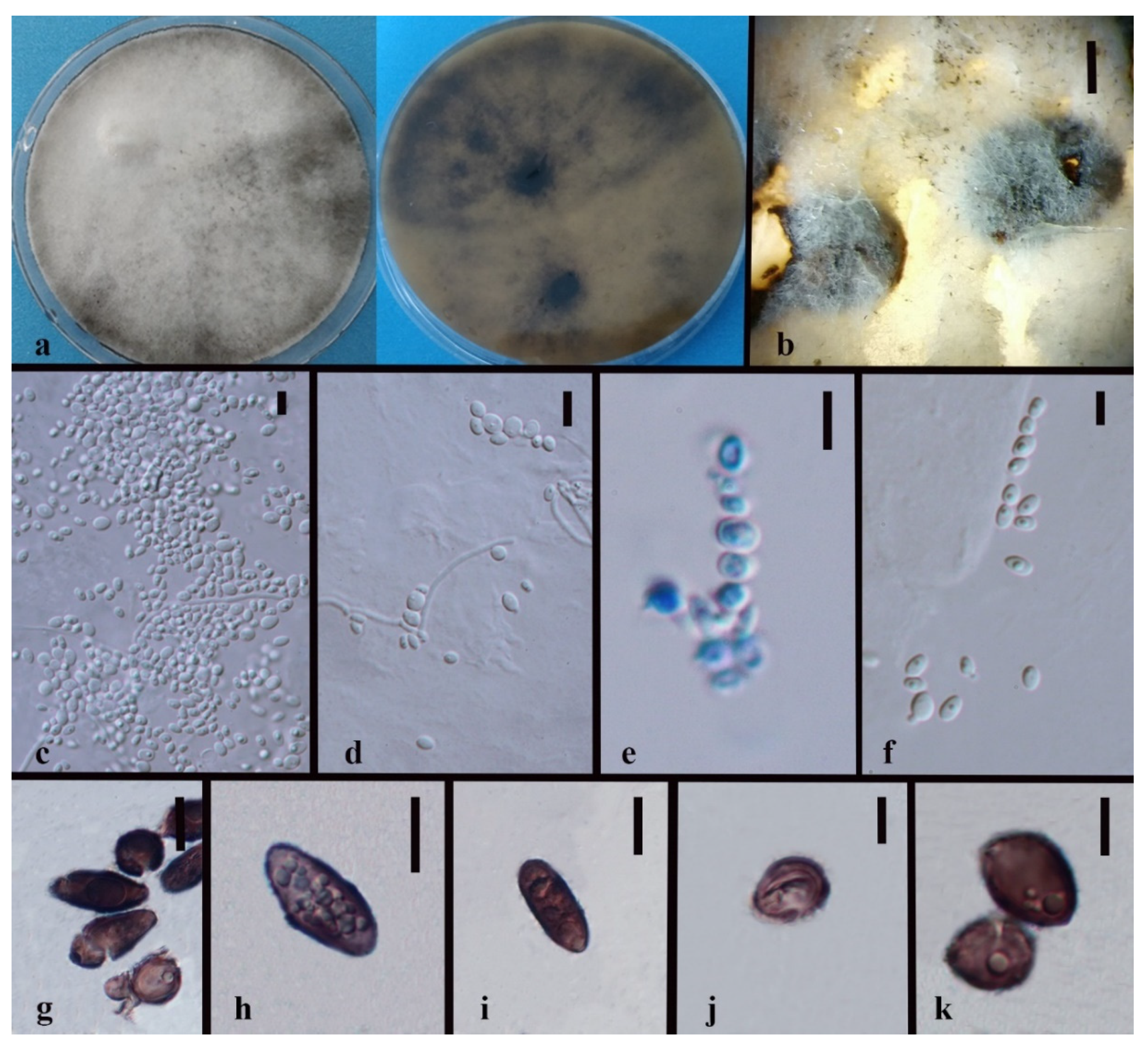
3.3. Taxonomy
4. Discussion
4.1. Xylariaceous Endophyte Associated with Dendrobium
4.2. The Dilemma for Xylaria Endophyte Verification
4.3. HTS Application for Fungal Endophyte
4.4. Potential Roles of Xylariaceous Endophyte
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Moudi, M.; Go, R. Morphological study of four sections of genus Dendrobium SW.(Orchidaceae) in Peninsular Malaysia. Pak. J. Bot. 2017, 49, 569–577. [Google Scholar]
- Wu, M.; Shu, Y.; Song, L.; Liu, B.; Zhang, L.; Wang, L.; Liu, Y.; Bi, J.; Xiong, C.; Cao, Z.; et al. Prenatal exposure to thallium is associated with decreased mitochondrial DNA copy number in newborns: Evidence from a birth cohort study. Environ. Int. 2019, 129, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.; Rahman, M.; Jamal, A.; Rahman, M.; Khalekuzzaman, M. Multiple shoot regeneration in Dendrobium fimbriatum Hook an ornamental orchid. J. Anim. Plant Sci. 2013, 23, 1140–1145. [Google Scholar]
- Arnold, A.E.; Mejia, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fracchia, S.; Aranda-Rickert, A.; Rothen, C.; Sede, S. Associated fungi, symbiotic germination and in vitro seedling development of the rare Andean terrestrial orchid Chloraea riojana. Flora 2016, 224, 106–111. [Google Scholar] [CrossRef]
- Rashmi, M.; Kushveer, J.S.; Sarma, V.V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 2019, 10, 798–1079. [Google Scholar] [CrossRef]
- Xie, X.G.; Zhang, F.M.; Yang, T.; Chen, Y.; Li, X.G.; Dai, C.C. Endophytic fungus drives nodulation and N2 fixation attributable to specific root exudates. MBio 2019, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, E.; Peterson, L. Effect of a dark septate fungal endophyte on seed germination and protocorm development in a terrestrial orchid. Symbiosis 2014, 43, 45–52. [Google Scholar]
- Meng, Y.Y.; Shao, S.C.; Liu, S.J.; Gao, J.Y. Do the fungi associated with roots of adult plants support seed germination? A case study on Dendrobium exile (Orchidaceae). Glob. Ecol. Conserv. 2019, 17, 1–20. [Google Scholar] [CrossRef]
- Sarsaiya, S.; Shi, J.; Chen, J. A comprehensive review on fungal endophytes and its dynamics on Orchidaceae plants: Current research, challenges, and future possibilities. Bioengineered 2019, 10, 316–334. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Han, Q.B.; Li, S.L.; Chen, X.J.; Wang, X.N.; Zhao, Z.Z.; Chen, H.B. Chemistry, bioactivity and quality control of Dendrobium, a commonly used tonic herb in traditional Chinese medicine. Phytochem. Rev. 2013, 12, 341–367. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, H.; Liu, Y. Chemical constituents from Dendrobium brymerianum Rchb. f. Biochem. Syst. Ecol. 2014, 57, 175–177. [Google Scholar] [CrossRef]
- Ramesh, V.; Santosh, K.; Pavunraj, M.; Karunakaran, C.; Rajendran, A. In-vitro antifungal and anticancer potential of Xylaria curta fruiting body fractions against human fungal pathogen and cancer cell lines. Curr. Res. Environ. Appl. Mycol. 2015, 5, 20–26. [Google Scholar] [CrossRef]
- Divate, R.D.; Chung, Y.C. In vitro and in vivo assessment of anti-inflammatory and immunomodulatory activities of Xylaria nigripes mycelium. J. Funct. Foods 2017, 35, 81–89. [Google Scholar] [CrossRef]
- Becker, K.; Wongkanoun, S.; Wessel, A.C.; Bills, G.F.; Stadler, M.; Luangsa-Ard, J.J. Phylogenetic and chemotaxonomic studies confirm the affinities of Stromatoneurospora phoenix to the coprophilous Xylariaceae. J. Fungi 2020, 6, 144. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Pourmoghaddam, M.J.; Cedeño-Sanchez, M.; Surup, F.; Khodaparast, S.A.; Krisai-Greilhuber, I.; Voglmayr, H.; Stradal, T.E.; Stadler, M. Resolution of the complex (Hypoxylaceae, Xylariales) and discovery and biological characterization of two of its prominent secondary metabolites. J. Fungi 2021, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.D.; Liew, E.C.Y.; Hyde, K.D. The Xylariales: A monophyletic order containing 7 families. Fungal Divers. 2003, 13, 175–208. [Google Scholar]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.G.; McKenzie, E.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H. Families of sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Rajeshkumar, K.C.; Hawksworth, D.L.; Madrid, H.; Kirk, P.M.; Braun, U.; Singh, R.V.; Crous, P.W.; Kukwa, M.; et al. Notes for genera: Ascomycota. Fungal Divers. 2017, 86, 1–594. [Google Scholar]
- Jaklitsch, W.M.; Voglmayr, H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Divers. 2012, 52, 75–98. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Hyde, K.D.; Sir, E.B.; Thambugala, K.M.; Tian, Q.; Samarakoon, M.C.; McKenzie, E.H.C.; Jayasiri, S.C.; Tibpromma, S.; Bhat, J.D.; et al. Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers. 2018, 88, 1–165. [Google Scholar] [CrossRef]
- Konta, S.; Hyde, K.D.; Phookamsak, R.; Xu, J.C.; Maharachchikumbura, S.S.N.; Daranagama, D.A.; McKenzie, E.H.; Boonmee, S.; Tibpromma, S.; Eungwanichayapant, P.D.; et al. Polyphyletic genera in Xylariaceae (Xylariales): Neoxylaria gen. nov. and Stilbohypoxylon. Mycosphere 2020, 11, 2629–2651. [Google Scholar] [CrossRef]
- Whalley, A. The xylariaceous way of life. Mycol. Res. 1996, 100, 897–922. [Google Scholar] [CrossRef]
- Hyde, K.D.; Fröhlich, J.; Taylor, J.E. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 1998, 50, 21–80. [Google Scholar]
- Osono, T.; To-Anun, C.; Hagiwara, Y.; Hirose, D. Decomposition of wood, petiole and leaf litter by Xylaria species from northern Thailand. Fungal Ecol. 2011, 4, 210–218. [Google Scholar] [CrossRef] [Green Version]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.G.; McKenzie, E.H.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Stadler, M.; Hellwig, V. Chemotaxonomy of the Xylariaceae and remarkable bioactive compounds from Xylariales and their associated asexual stages. Recent Res. Dev. Phytochem. 2005, 9, 41–93. [Google Scholar]
- Xu, F.; Zhang, Y.; Wang, J.J.; Pang, J.Y.; Huang, C.H.; Wu, X.Y.; She, Z.G.; Vrijmoed, L.L.P.; Jones, E.B.G.; Lin, Y.H. Benzofuran derivatives from the mangrove endophytic fungus Xylaria sp. (#2508). J. Nat. Prod. 2008, 71, 1251–1253. [Google Scholar]
- Pažoutová, S.; Follert, S.; Bitzer, J.; Keck, M.; Surup, F.; Šrůtka, P.; Holuša, J.; Stadler, M. A new endophytic insect-associated Daldinia species, recognised from a comparison of secondary metabolite profiles and molecular phylogeny. Fungal Divers. 2013, 60, 107–123. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Kjer, J.; Proksch, P. Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 2010, 41, 1–16. [Google Scholar] [CrossRef]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021, 74, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Yuan, Z.L.; Chen, Y.C.; Yang, Y. Diverse non-mycorrhizal fungal endophytes inhabiting an epiphytic, medicinal orchid (Dendrobium nobile): Estimation and characterization. World J. Microbiol. Biotechnol. 2009, 25, 295–303. [Google Scholar] [CrossRef]
- Chen, J.; Hu, K.X.; Hou, X.Q.; Guo, S.X. Endophytic fungi assemblages from 10 Dendrobium medicinal plants (Orchidaceae). World J. Microbiol. Biotechnol. 2011, 27, 1009–1016. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.C.; Xing, Y.M.; Wang, Y.Q.; Xing, X.K.; Zhang, D.W.; Liang, H.Q.; Guo, S.X. Diversity and taxonomy of endophytic xylariaceous fungi from medicinal plants of Dendrobium (Orchidaceae). PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Parthibhan, S.; Rao, M.V.; Kumar, T.S. Culturable fungal endophytes in shoots of Dendrobium aqueum Lindley—An imperiled orchid. Ecol. Genet. Genom. 2017, 3, 18–24. [Google Scholar] [CrossRef]
- Sarsaiya, S.; Jain, A.; Jia, Q.; Fan, X.; Shu, F.; Chen, Z.; Zhou, Q.; Shi, J.; Chen, J. Molecular identification of endophytic fungi and their pathogenicity evaluation against Dendrobium nobile and Dendrobium officinale. Int. J. Mol. Sci. 2020, 21, 316. [Google Scholar] [CrossRef] [Green Version]
- Kasmir, J.; Senthilkumar, S.; Britto, S.; Raj, J. Identification of fungal endophytes from Orchidaceae members based on nrITS (internal transcribed spacer) region. Int. Res. J. Biotechnol. 2011, 2, 139–144. [Google Scholar]
- Ma, X.Y.; Kang, J.C.; Nontachaiyapoom, S.; Wen, T.; Hyde, K.D. Non-mycorrhizal endophytic fungi from orchids. Curr. Sci. 2015, 109, 72–87. [Google Scholar]
- Doilom, M.; Manawasinghe, I.S.; Jeewon, R.; Jayawardena, R.S.; Tibpromma, S.; Hongsanan, S.; Meepol, W.; Lumyong, S.; Jones, E.B.; Hyde, K.D. Can ITS sequence data identify fungal endophytes from cultures? A case study from Rhizophora apiculata. Mycosphere 2017, 8, 1869–1892. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; de Cock, A.; Dissanayake, A.J.; Glockling, S.L.; Goonasekara, I.D.; et al. One stop shop: Backbones trees for important phytopathogenic genera: I. Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef] [Green Version]
- Rogers, J.D.; Ju, Y.M.; Hemmes, D.E. Hypoxylon rectangulosporum sp. nov., Xylaria psidii sp. nov., and comments on taxa of Podosordaria and Stromatoneurospora. Mycologia 1992, 84, 166–172. [Google Scholar] [CrossRef]
- Rodrigues, K.F.; Leuchtmann, A.; Petrini, O. Endophytic species of Xylaria: Cultural and isozymic studies. Sydowia 1993, 45, 116–138. [Google Scholar]
- Wang, Y.U.; Guo, L.D.; Hyde, K.D. Taxonomic placement of sterile morphotypes of endophytic fungi from Pinus tabulaeformis (Pinaceae) in northeast China based on rDNA sequences. Fungal Divers. 2005, 20, 235–260. [Google Scholar]
- Tejesvi, M.V.; Mahesh, B.; Nalini, M.S.; Prakash, H.S.; Kini, K.R.; Subbiah, V.; Shetty, H.S. Fungal endophyte assemblages from ethnopharmaceutically important medicinal trees. Can. J. Microbiol. 2006, 427–535. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.M.; Lin, C.R.; Fang, M.J.; Rogers, J.D.; Fournier, J.; Lechat, C.; Ju, Y.M. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenet. Evol. 2010, 54, 957–969. [Google Scholar] [CrossRef]
- Vemireddy, B.; Madasi, A.; Ajmeera, A.; Vanteru, K.R. Distribution and diversity of endophytic fungi associated with three medicinal tree species from Eturnagaram Wildlife Sanctuary, TS, India. J. Appl. Biol. 2020, 8, 7–12. [Google Scholar]
- Ding, G.; Zhang, D.; Ding, X.; Zhou, Q.; Zhang, W.; Li, X. Genetic variation and conservation of the endangered Chinese endemic herb Dendrobium officinale based on SRAP analysis. Plant Syst. Evol. 2008, 276, 149–156. [Google Scholar] [CrossRef]
- Nontachaiyapoom, S.; Sasirat, S.; Manoch, L. Isolation and identification of Rhizoctonia-like fungi from roots of three orchid genera, Paphiopedilum, Dendrobium, and Cymbidium, collected in Chiang Rai and Chiang Mai provinces of Thailand. Mycorrhiza 2010, 20, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.T.; Ackerman, J.D.; Bayman, P. Diversity and host specificity of endophytic Rhizoctonia-like fungi from tropical orchids. Am. J. Bot. 2002, 89, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Petrini, O. Fungal endophytes of tree leaves. In Microbial Ecology of Leaves; Springer: Berlin/Heidelberg, Germany, 1991; pp. 179–197. [Google Scholar]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Wang, M.; Liu, F.; Crous, P.W.; Cai, L. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 2017, 39, 118–142. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Nannf., Nova Acta R. Soc. Scient. upsal., Ser. 1932, 4, p. 66. Available online: https://www.facesoffungi.org/diaporthales-facesoffungi-number-fof/ (accessed on 21 January 2022).
- Zhou, H.; Abuduaini, A.; Xie, H.; Kang, R.; Suo, F.; Huang, L. The complete mitochondrial genome of wood-rotting fungus Xylaria hypoxylon. Mitochondrial DNA B Resour. 2019, 4, 3848–3849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crous, P.W.; Groenewald, J.Z. A phylogenetic re-evaluation of Arthrinium. IMA Fungus 2013, 4, 133–154. [Google Scholar] [CrossRef] [Green Version]
- Sacc., Atti della Società veneto-trentina di scienze naturali residente in Padova. 1875; 4, 85.
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.G.; McKenzie, E.H.; Huang, S.K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Hughes, S.J. Conidiophores, conidia and classification. Can. J. Bot. 1953, 31, 577–659. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Crous, P.W.; Redhead, S.A.; Reynolds, D.R.; Samson, R.A.; Seifert, K.A.; Taylor, J.W.; Wingfield, M.J.; Abaci, Ö.; Aime, C.; et al. The Amsterdam declaration on fungal nomenclature. IMA Fungus 2011, 2, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Dai, D.Q.; Jiang, H.B.; Tang, L.Z.; Bhat, D.J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 2016, 7, 1332–1345. [Google Scholar] [CrossRef]
- Pintos, Á.; Alvarado, P.; Planas, J.; Jarling, R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys 2019, 49, 15–48. [Google Scholar] [CrossRef] [Green Version]
- Pintos, Á.; Alvarado, P. Phylogenetic delimitation of Apiospora and Arthrinium. J. Syst. Evol. 2021, 7, 197–221. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tang, Y.; Fang, X.; Qiao, T.; Han, S.; Zhu, T. Whole-genome sequence of Arthrinium phaeospermum, a globally distributed pathogenic fungus. Genomics 2020, 112, 919–929. [Google Scholar] [CrossRef]
- Zimmerman, A. Ueber einige an tropischen Kulturpflanzen beobachtete Pilze III. Zentralblatt Fur Bakteriol Parasitenkd. Centbl. Bakt. ParasitKde Abt. I 1902, 8, 220. [Google Scholar]
- Seifert, K.A.; Gams, W. The genera of Hyphomycetes—2011 update. Pers. Mol. Phylogeny Evol. Fungi 2011, 27, 119–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meepagala, K.M.; Becnel, J.J.; Estep, A.S. Phomalactone as the active constituent against mosquitoes from Nigrospora spherica. J. Agric. Sci. 2015, 6, 1195–1200. [Google Scholar]
- Huang, R.; Wang, T.; Xie, X.S.; Ma, K.X.; Fang, X.W.; Wu, S.H. Secondary metabolites from an endophytic fungus Nigrospora sp. Chem. Nat. Compd. 2016, 52, 697–699. [Google Scholar] [CrossRef]
- Mason, E.W.; Brit, T. On species of the genus Nigrospora Zimmermann recorded on monocotyledons. Trans. Brit. Mycol. Soc. 1927, 12, 152–165. [Google Scholar] [CrossRef]
- De Lamarck, J.B.; De Candolle, A.P. Flore française; Chez H. Agasse: Paris, France, 1805; Volume 2, pp. 1–600. [Google Scholar]
- Hsieh, H.M.; Ju, Y.M.; Rogers, J.D. Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 2005, 97, 844–865. [Google Scholar] [CrossRef]
- Fournier, J.; Stadler, M.; Hyde, K.D.; Duong, M.L. The new genus Rostrohypoxylon and two new Annulohypoxylon species from Northern Thailand. Fungal Divers. 2010, 40, 23–36. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Liu, X.; Chamyuang, S.; Stadler, M.; Hyde, K. A multiple gene genealogy reveals phylogenetic placement of Rhopalostroma lekae. Phytotaxa 2014, 186, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Bulliard, P. Histoire des Champignons de la France; Harvard University Herbaria & Libraries: Cambridge, MA, USA, 1791; p. 168. [Google Scholar]
- Ju, Y.M.; Rogers, J.D. A Revision of the Genus Hypoxylon, Illustrated, 1st ed.; APS Press, The American Phytopathological Society: Taiwan, China, 1996. [Google Scholar]
- Tomsheck, A.R.; Strobel, G.A.; Booth, E.; Geary, B.; Spakowicz, D.; Knighton, B.; Floerchinger, C.; Sears, J.; Liarzi, O.; Ezra, D. Hypoxylon sp., an endophyte of Persea indica, producing 1, 8-cineole and other bioactive volatiles with fuel potential. Microb. Ecol. 2010, 60, 903–914. [Google Scholar] [CrossRef]
- Curtis, M.A. North Carolina Geological and Natural History Survey Papers; N.C. Institution for the Deaf and Dumb and Blind: Raleigh, NC, USA, 1867; Volume 3, p. 140. [Google Scholar]
- Bills, G.F.; Gonzalez-Menendez, V.; Martin, J.; Platas, G.; Fournier, J.; Persoh, D.; Stadler, M. Hypoxylon pulicicidum sp. nov (Ascomycota, Xylariales), a Pantropical Insecticide-Producing Endophyte. PLoS ONE 2012, 7, e46687. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, M.C.; Thongbai, B.; Hyde, K.D.; Brönstrup, M.; Beutling, U.; Lambert, C.; Miller, A.N.; Liu, J.K.; Promputtha, I.; Stadler, M. Elucidation of the life cycle of the endophytic genus Muscodor and its transfer to Induratia in Induratiaceae fam. nov., based on a polyphasic taxonomic approach. Fungal Divers. 2020, 101, 177–210. [Google Scholar] [CrossRef]
- Chen, J.; Wang, H.; Guo, S.X. Isolation and identification of endophytic and mycorrhizal fungi from seeds and roots of Dendrobium (Orchidaceae). Mycorrhiza 2012, 22, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.F. In Nat. Arr. Brit. Pl. (London). 1821, 1, p. 516. Available online: https://www.facesoffungi.org/nemania-facesoffungi-number-fof03055/ (accessed on 21 January 2022).
- Pouzar, Z. Reassessment of the Hypoxylon serpens-complex. Ceská Mykol. 1985, 39, 15–25. [Google Scholar]
- Schrank; von Paula, F. Baierische Flora. 1789; 1, pp. 1–753. Available online: https://library.awe.gov.au/cgi-bin/koha/opac-detail.pl?biblionumber=3317&shelfbrowse_itemnumber=182013 (accessed on 21 January 2022).
- Ju, Y.M.; Rogers, J.D. In N. Amer. Fung. 2012, 7, p. 18. Available online: http://www.speciesfungorum.org/Names/SynSpecies.asp?RecordID=546721 (accessed on 21 January 2022).
- Fries, E.M. Novae symbolae mycologicae, in peregrinis terris a botanicis danicis collectae. Nova Acta Regiae Societatis Scientiarum Upsaliensis. Ser. 3 1851, 1, 17–136. [Google Scholar]
- Chen, K.L.; Kirschner, R. Fungi from leaves of lotus (Nelumbo nucifera). Mycol Pro. 2018, 17, 275–293. [Google Scholar] [CrossRef]
- Lloyd, C.G. Xylaria notes. 1918, 7, pp. 1–15. Available online: http://www.cybertruffle.org.uk/cgi-bin/bibquery.pl?author=lloyd,%20c.g (accessed on 21 January 2022).
- Montagne, J.P.F.C. Cryptogamia Guyanensis seu plantarum cellularium in Guyana gallica annis 1835-1849 a cl. Leprieur collectarum enumeration universalis. Annales des Sciences Naturelles Botanique. Sér. 4 1855, 3, 91–144. [Google Scholar]
- Lloyd, C.G. Mycological Notes 65. Mycol. Writ. 1921, 6, 1029–1101. [Google Scholar]
- Spegazzini, C. Fungi Puiggariani. Pugillus 1. Boletín De La Acad. Nac. De Cienc. En Córdoba 1889, 11, 381–622. [Google Scholar]
- Lee, D.J.; Lee, J.S.; Lee, H.B.; Choi, Y.J. Four endophytic ascomycetes new to Korea: Cladosporium anthropophilum, C. pseudocladosporioides, Daldinia eschscholtzii, and Nigrospora chinensis. Kor. J. Mycol. 2019, 47, 187–197. [Google Scholar]
- Davis, E.C.; Franklin, J.B.; Shaw, A.J.; Vilgalys, R. Endophytic Xylaria (Xylariaceae) among liverworts and angiosperms: Phylogenetics, distribution, and symbiosis. Am. J. Bot. 2003, 90, 1661–1667. [Google Scholar] [CrossRef] [PubMed]
- Ezra, D.; Hess, W.; Strobel, G.A. New endophytic isolates of Muscodor albus, a volatile-antibiotic-producing fungus. Microbiology 2004, 150, 4023–4031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, A.; Matsuoka, S.; Masuya, H.; Mori, A.S.; Hirose, D.; Osono, T. Comparison of the diversity, composition, and host recurrence of xylariaceous endophytes in subtropical, cool temperate, and subboreal regions in Japan. Popul. Ecol. 2014, 56, 289–300. [Google Scholar] [CrossRef]
- Bayman, P.; Lebron, L.L.; Tremblay, R.L.; Lodge, D.J. Variation in endophytic fungi from roots and leaves of Lepanthes (Orchidaceae). New Phytol. 1997, 135, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Okane, I.; Toyama, K.; Nakagiri, A.; Suzuki, K.; Srikitikulchai, P.; Sivichai, S.; Hywel-Jones, N.; Potacharoen, W.; Læssøe, T. Study of endophytic Xylariaceae in Thailand: Diversity and taxonomy inferred from rDNA sequence analyses with saprobes forming fruit bodies in the field. Mycoscience 2008, 49, 359–372. [Google Scholar] [CrossRef]
- Jiang, W.M.; Yang, G.M.; Zhang, C.L.; Fu, C.X. Species composition and molecular analysis of symbiotic fungi in roots of Changnienia amoena (Orchidaceae). Afr. J. Microbiol. Res. 2011, 5, 222–228. [Google Scholar]
- Sawmya, K.; Vasudevan, T.G.; Murali, T.S. Fungal endophytes from two orchid species pointer towards organ specificity. Czech Mycol. 2013, 65, 89–101. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Miadlikowska, J.; Zimmerman, N.B.; Lutzoni, F.; Stajich, J.E.; Arnold, A.E. Contributions of north American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenet. Evol. 2016, 98, 210–232. [Google Scholar] [CrossRef] [Green Version]
- Carmona, A.; Fournier, J.; Williams, C.; Piepenbring, M. New records of Xylariaceae from Panama. N. Am. Fungi 2009, 4, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Rogers, J.D.; Ju, Y.M. The Xylariaceae of the Hawaiian Islands. N. Am. Fungi 2012, 7, 1–35. [Google Scholar] [CrossRef]
- Chacón, S.; González, D. A new species of penzigioid Xylaria (Xylariaceae) from the cloud forest in eastern Mexico revealed through morphological and phylogenetic analyses. Botany 2019, 97, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Petrini, L.; Petrini, O. Xylariaceous fungi as endophytes. Sydowia 1985, 38, 216–234. [Google Scholar]
- Stadler, M.; Lambert, C.; Wibberg, D.; Kalinowski, J.; Cox, R.J.; Kolařík, M.; Kuhnert, E. Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycol. Prog. 2020, 19, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Wibberg, D.; Stadler, M.; Lambert, C.; Bunk, B.; Spröer, C.; Rückert, C.; Kalinowski, J.; Cox, R.J.; Kuhnert, E. High quality genome sequences of thirteen Hypoxylaceae (Ascomycota) strengthen the phylogenetic family backbone and enable the discovery of new taxa. Fungal Divers. 2021, 106, 7–28. [Google Scholar] [CrossRef]
- Rogers, J.D. Xylaria cubensis and its anamorph Xylocoremium flabelliforme, Xylaria allantoidea, and Xylaria poitei in continental United States. Mycologia 1984, 76, 912–923. [Google Scholar] [CrossRef]
- Hsieh, H.M.; Chou, J.C.; Ju, Y.M. Xylaria insolita and X. subescharoidea: Two newly described species collected from a termite nesting site in Hua-lien, Taiwan. Bot. Stud. 2020, 61, 1–9. [Google Scholar] [CrossRef]
- Garcia-Aroca, T.; Price, P.P.; Tomaso-Peterson, M.; Allen, T.W.; Wilkerson, T.H.; Spurlock, T.N.; Faske, T.R.; Bluhm, B.; Conner, K.; Sikora, E.; et al. Xylaria necrophora sp. nov., is an emerging root-associated pathogen responsible for taproot decline of soybean in the southern United States. Mycologia 2021, 113, 326–347. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, Z.; Liu, Y.; Hu, X.; Guo, Y.; Li, J. Application of high-throughput internal transcribed spacer rRNA metagenomics analysis in deciphering endophytic fungi diversity of Dendrobium officinale. J. Biobased Mater. Bioenergy 2017, 11, 106–118. [Google Scholar] [CrossRef]
- Stranneheim, H.; Lundeberg, J. Stepping stones in DNA sequencing. Biotechnol. J. 2012, 7, 1063–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Razgour, O.; Clare, E.L.; Zeale, M.R.; Hanmer, J.; Schnell, I.B.; Rasmussen, M.; Gilbert, T.P.; Jones, G. High-throughput sequencing offers insight into mechanisms of resource partitioning in cryptic bat species. Ecol. Evol. 2011, 1, 556–570. [Google Scholar] [CrossRef]
- Johnston, P.R.; Park, D.; Smissen, R.D. Comparing diversity of fungi from living leaves using culturing and high-throughput environmental sequencing. Mycologia 2017, 109, 643–654. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, A.R. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Promputtha, I.; Jeewon, R.; Lumyong, S.; McKenzie, E.H.; Hyde, K.D. Ribosomal DNA fingerprinting in the identification of non-sporulating endophytes from Magnolia liliifera (Magnoliaceae). Fungal Divers. 2005, 20, 167–186. [Google Scholar]
- Ma, X.Y.; Nontachaiyapoom, S.; Jayawardena, R.S. Endophytic Colletotrichum species from Dendrobium spp. in China and Northern Thailand. MycoKeys 2018, 43, 23–37. [Google Scholar] [CrossRef] [Green Version]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tang, Q.; Fang, L. First report of Nigrospora sphaerica causing leaf blight on Camellia sinensis in China. Plant Dis. 2016, 100, 221. [Google Scholar] [CrossRef]
- Ondeyka, J.G.; Helms, G.L.; Hensens, O.D.; Goetz, M.A.; Zink, D.L.; Tsipouras, A.; Shoop, W.L.; Slayton, L.; Dombrowski, A.W.; Polishook, J.D. Nodulisporic acid A, a novel and potent insecticide from a Nodulisporium sp. isolation, structure determination, and chemical transformations. J. Am. Chem. Soc. 1997, 119, 8809–8816. [Google Scholar] [CrossRef]
- Sudheep, N.M.; Sridhar, K.R. Non-mycorrhizal fungal endophytes in two orchids of Kaiga forest (Western Ghats), India. J. For. Res. 2012, 23, 453–460. [Google Scholar] [CrossRef]
- Poornima, S.; Ponmurugan, P.; Gnanamangai, B.M.; Gayathri, G.; Dheenadhayalan, K.; Ayyappadasan, G. Screening of biologically potent endolichenic fungi isolated from selected lichens habitat on silver oak tree. Vegetos 2018, 31, 89–94. [Google Scholar] [CrossRef]
- Promputtha, I.; Lumyong, S.; Dhanasekaran, V.; McKenzie, E.H.C.; Hyde, K.D.; Jeewon, R. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb. Ecol. 2007, 53, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Eaton, C.J.; Cox, M.P.; Scott, B. What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci. 2011, 180, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Mohr, K.I.; Stadler, M.; Dickschat, J.S. Volatiles from the tropical ascomycete Daldinia clavata (Hypoxylaceae, Xylariales). Beilstein J. Org. Chem. 2018, 14, 135–147. [Google Scholar] [CrossRef] [Green Version]







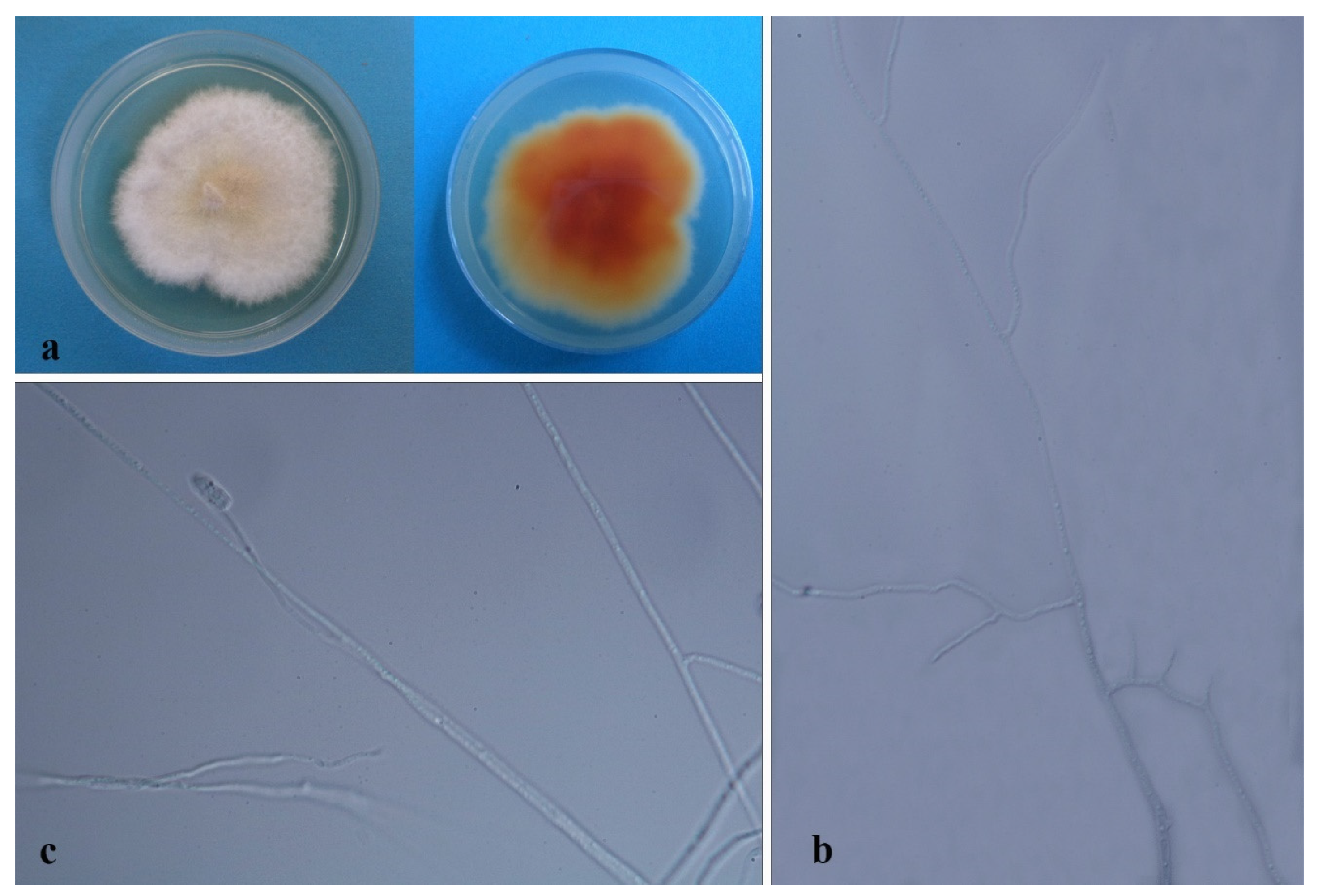
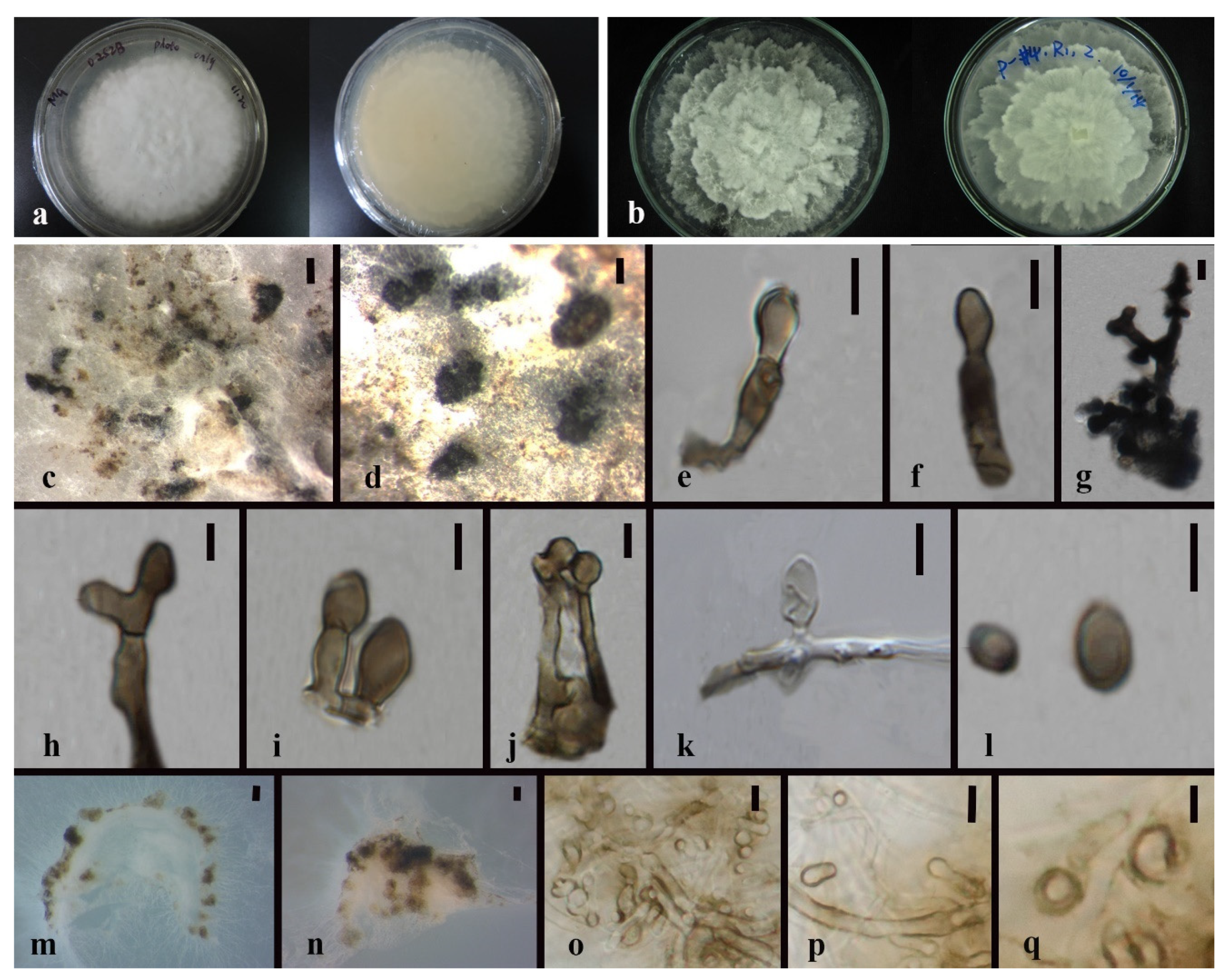

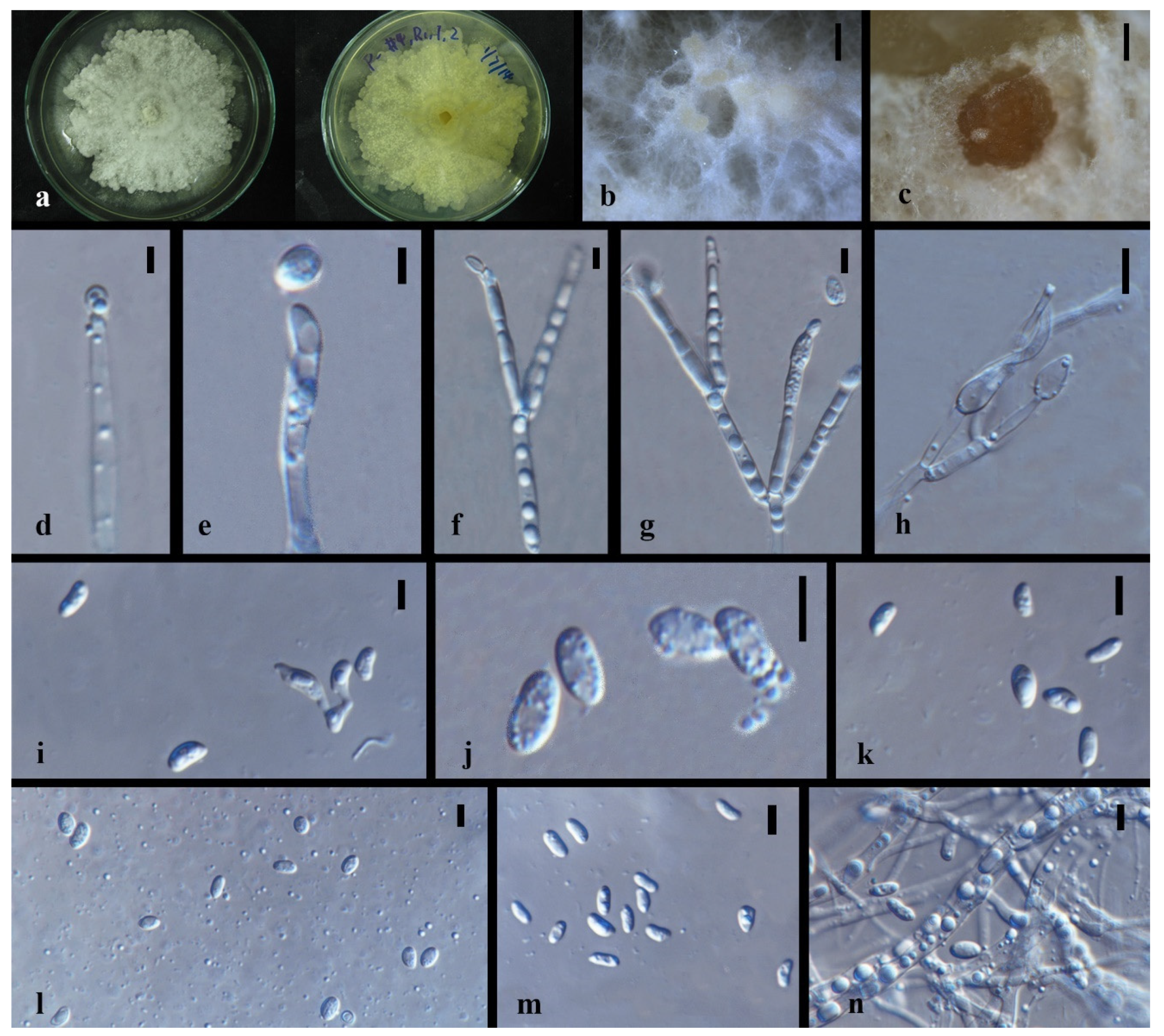
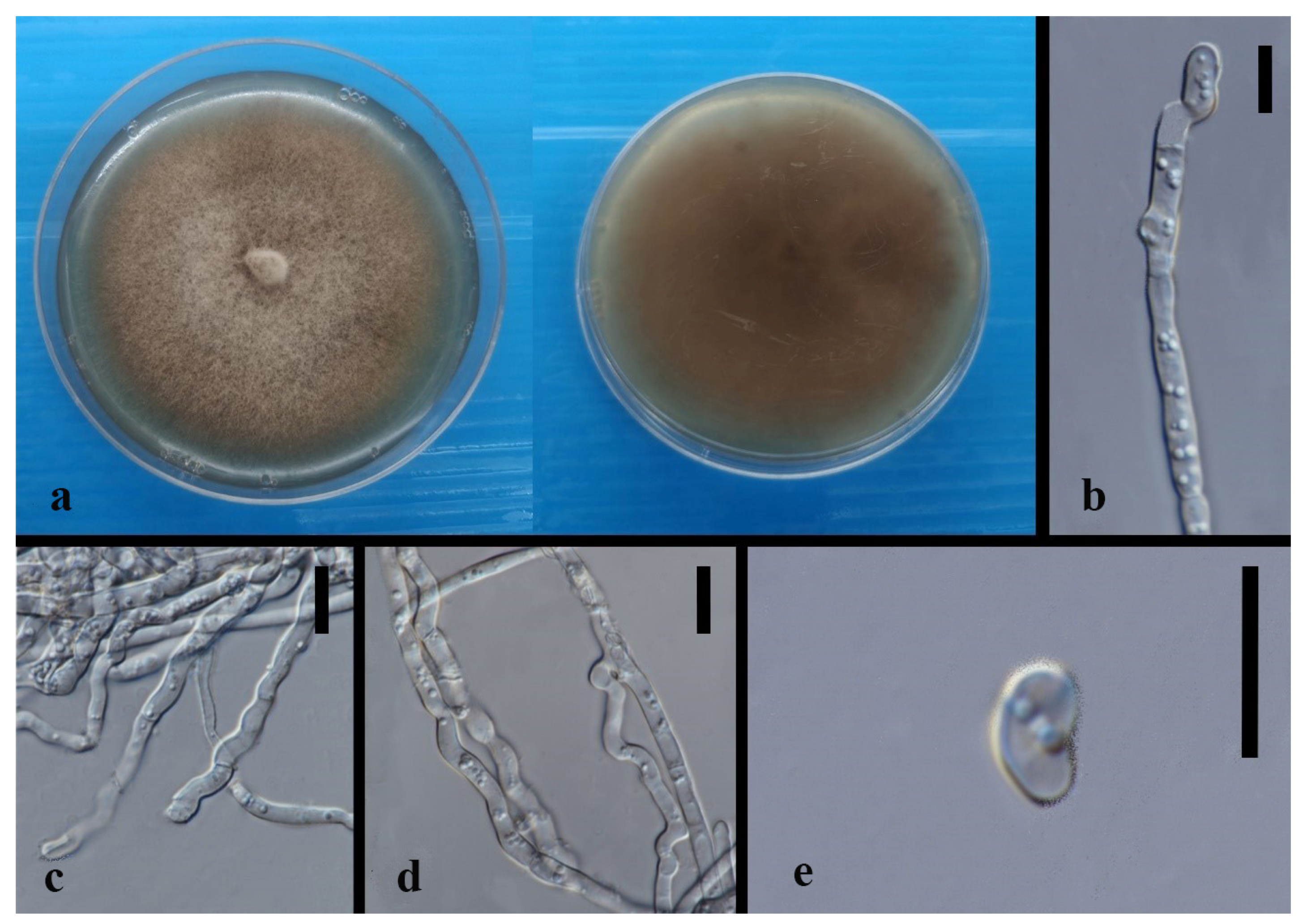
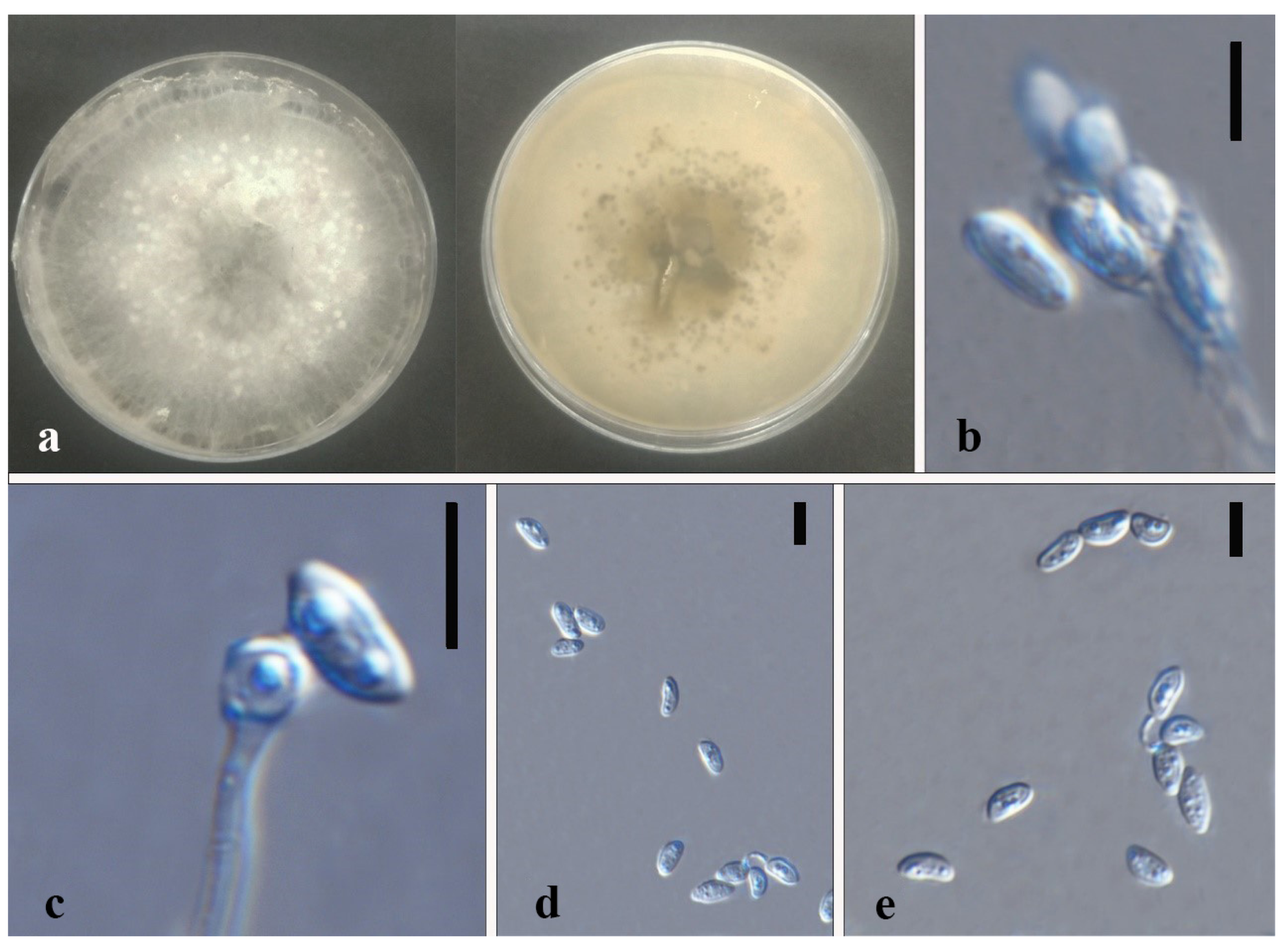


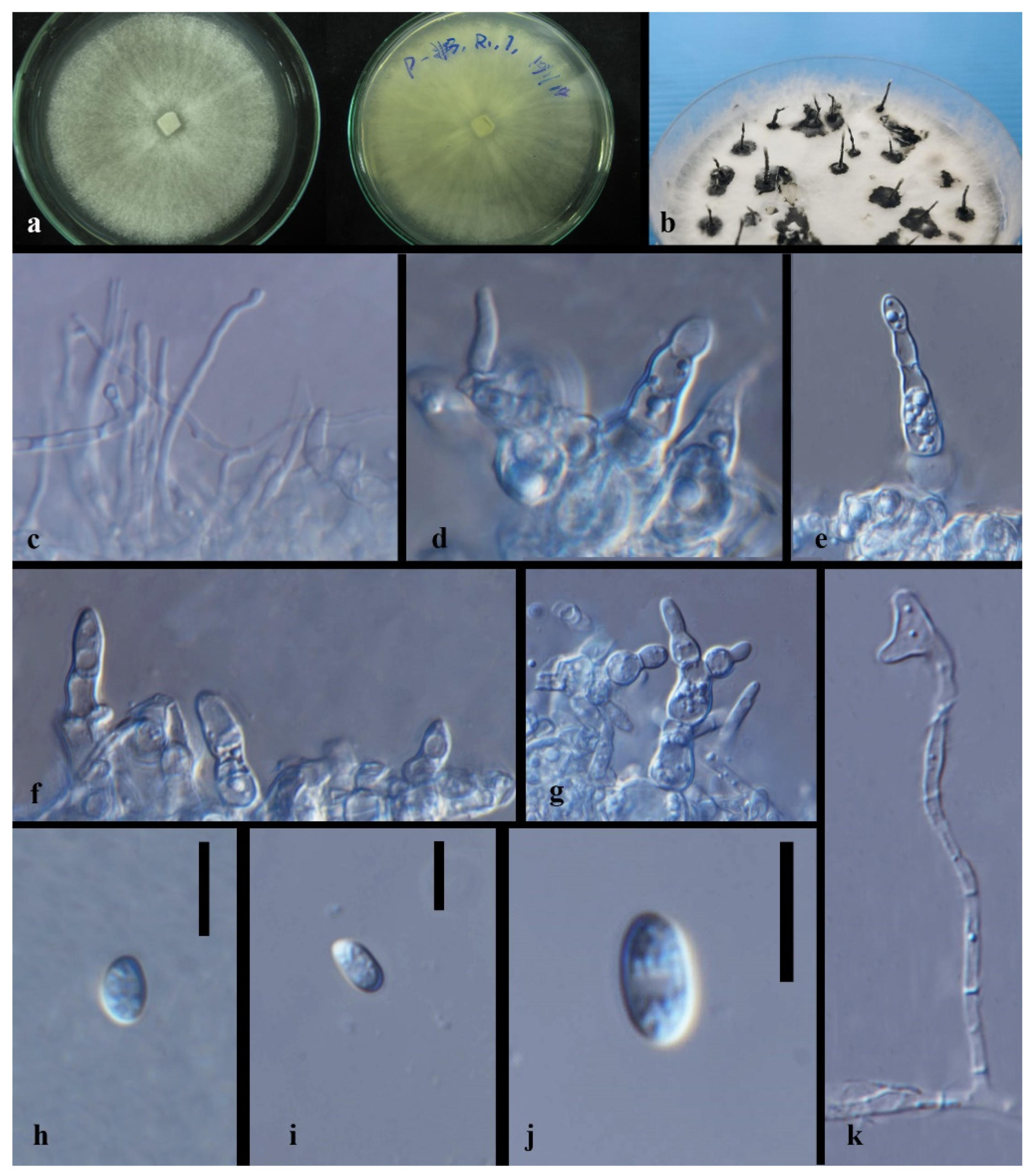
| Code | Sampling Site | Dendrobium Species |
|---|---|---|
| 1 | Animal Husbandry and Veterinary Institute, Guiyang, Guizhou, China | Dendrobium chrysanthum Lindl. |
| 2 | Orchid nursery 1, Luodian, Guizhou, China | Dendrobium officinale Kimura & Migo |
| 3 | Orchid nursery 2, Luodian, Guizhou, China | Dendrobium aphyllum (Roxb.) C. E. |
| Dendrobium aurantiacum Rchb. f. var. denneanum | ||
| Dendrobium chrysotoxum Lindl. | ||
| Dendrobium fimbriatum Hook. | ||
| Dendrobium hercoglossum Rchb. f. | ||
| Dendrobium moniliforme (L.) Sw. | ||
| Dendrobium officinale Kimura & Migo | ||
| Dendrobium primulinum Lindl. | ||
| 4 | Orchid nursery, Xingyi, Guizhou, China | Dendrobium loddigesii Rolfe |
| Dendrobium officinale Kimura & Migo | ||
| Dendrobium sp. 5 | ||
| Dendrobium sp. 6 | ||
| Dendrobium sp. 7 | ||
| Dendrobium sp. 8 | ||
| 5 | Steep forest, Doitung, Mae Fah Luang District, Chiang Rai, Thailand | Dendrobium sp. 4 |
| 6 | Wat Phra That Doi Tung (Temple of Doi Tung Pagodas), Mae Fah Luang District, Chiang Rai, Thailand | Dendrobium cariniferum Rchb. f. |
| Dendrobium harveyanum Rchb.f. | ||
| Dendrobium moschatum (Buch.Ham.) Sw | ||
| Dendrobium sp. 1 | ||
| Dendrobium sp. 2 | ||
| Dendrobium sp. 3 |
| Region/Gene | Primers | Cycle Number | Denaturation | Annealing | Elongation | Reference |
|---|---|---|---|---|---|---|
| ITS | ITS1/ITS4 | 30 | 95 °C 1 min | 53 °C 1 min | 72 °C 1 min | White et al., 1990 |
| LSU | LR0R/LR5 | 35 | 94 °C 1 min | 53 °C 50 s | 72 °C 1 min 30 s | Vilgalys and Hester 1990 |
| RPB2 | Frpb-5f/frpb-7cr | 35 | 95 °C 45 s | 57 °C 50 s | 72 °C 1 min 30 s | Liu et al., 1999 |
| TEF-1α | 728F/986R | 36 | 95 °C 45 s | 56 °C 1 min | 72 °C 1 min | Carbone and Kohn 1999 |
| TUB2 | Bt2A/Bt2B | 35 | 94 °C 1 min | 55 °C 55 s | 72 °C 1 min | Glass and Donaldson 1995 |
| Xylariales Species | Strain Code | Host Dendrobium Orchids | Tissues |
|---|---|---|---|
| Annulohypoxylon moniliformis sp. nov. | MFLUCC 18-1214 HT | Dendrobium moniliforme | Leaf |
| Apiospora dendrobii sp. nov. | MFLUCC 14-0152 HT | Dendrobium harveyanum | Root |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1206 HT | Dendrobium aphyllum | Root |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1209 | Dendrobium huoshanense | Stem |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1211 | Dendrobium chrysotoxum | Leaf |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1208 | Dendrobium sp. 5 | Root |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1210 | Dendrobium loddigesii | Root |
| Hypoxylon endophyticum sp. nov. | MFLUCC 18-1207 | Dendrobium hercoglossum | Leaf |
| Hypoxylon officinalis sp. nov. | MFLUCC 14-0075 HT | Dendrobium sp. 1 | Root |
| Hypoxylon officinalis sp. nov. | MFLUCC 14-0078 | Dendrobium sp. 1 | Root |
| Hypoxylon officinalis sp. nov. | MFLUCC 21-0060 | Dendrobium officinale | Root |
| Hypoxylon investiens (Schwein.) M.A. Curtis | MFLUCC 15-1155 | Dendrobium moschatum * | Stem |
| Hypoxylon pulicicidum J. Fournier, Polishook & Bills | GZAC O37S13 | Dendrobium hercoglossum * | Root |
| Hypoxylaceae sp. | MFLUCC 14-0141 | Dendrobium sp. 3 | Root |
| Induratia sp. | MFLUC C 15-1218 | Dendrobium sp. 4 | Stem |
| Nemania dendrobii sp. nov. | MFLUCC 18-1213 HT | Dendrobium sp. 7 | Stem |
| Nemania dendrobii sp. nov. | MFLUCC 18-1212 | Dendrobium sp. 6 | Root |
| Nemania diffusa (Sowerby) Gray | MFLUCC 14-0139 | Dendrobium sp. 2 | Root |
| Nemania bipapillata (Berk. & M.A. Curtis) Pouzar | MFLUCC 14-0138 | Dendrobium sp. 3 | Root |
| Nemania bipapillata (Berk. & M.A. Curtis) Pouzar | MFLUC C 14-0105 | Dendrobium cariniferum * | Stem |
| Nigrospora chinensis Mei Wang & L. Cai | MFLUCC 14-0109 | Dendrobium cariniferum * | Stem |
| Nigrospora chinensis Mei Wang & L. Cai | MFLUCC 18-1215 | Dendrobium officinale | Root |
| Nigrospora sphaerica (Sacc.) E.W. Mason | GZAC O37S13 | Dendrobium hercoglossum * | Leaf |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0095 | Dendrobium cariniferum * | Root |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0102 | Dendrobium cariniferum | Stem |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0143 | Dendrobium sp. | Stem |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0117 | Dendrobium sp. 2 | Root |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0126 | Dendrobium sp. 2 | Leaf |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLU CC 14-0143 | Dendrobium sp. 3 | Stem |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0150 | Dendrobium harveyanum * | Root |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 14-0158 | Dendrobium harveyanum | Leaf |
| Xylaria berteri (Mont.) Cooke ex J.D. Rogers & Y.M. Ju | MFLUCC 21-0061 | Dendrobium sp. 2 | Leaf |
| Xylaria curta Fr. | GZAC O36L23 | Dendrobium officinale | Leaf |
| Xylaria feejeensis (Berk.) Fr. | GZAC O30S21 | Dendrobium aphyllum * | Stem |
| Xylaria grammica (Mont.) Mont. | MFLUCC 14-0093 | Dendrobium sp. 1 | Leaf |
| Xylaria grammica (Mont.) Mont. | MFLUCC 14-0146 | Dendrobium sp. 3 | Leaf |
| Xylaria laevis Lloyd | GZAC O33L12 | Dendrobium aurantiacum var. denneanum * | Leaf |
| Xylaria laevis Lloyd | GZAC O6LA2 | Dendrobium officinale | Leaf |
| Xylaria papulis Lloyd | GZAC O32S24 | Dendrobium chrysotoxum | Stem |
| Xylaria sp.1 | MFLUCC 14-0137 | Dendrobium sp. 3 | Root |
| Xylaria sp.1 | MFLUCC 14-0110 | Dendrobium cariniferum | Leaf |
| Xylaria sp.2 | MFLUCC 21-0014 | Dendrobium chrysanthum | Stem |
| Xylaria sp.3 | MFLUCC 21-0059 | Dendrobium aphyllum | Root |
| Xylaria venosula Speg. | MFLUCC 14-0114 | Dendrobium sp. 2 | Root |
| Xylaria venosula Speg. | MFLUCC 21-0013 | Dendrobium fimbriatum * | Root |
| Xylaria venosula Speg. | MFLUCC 21-0015 | Dendrobium aurantiacum var. denneanum * | Root |
| Xylaria venosula Speg. | MFLUCC 21-0016 | Dendrobium primulinum * | Stem |
| Xylaria venosula Speg. | MFLUCC 21-0017 | Dendrobium sp. 8 | Stem |
| New Species Names | Type Species | Morphological Differences | Phylogenetic Differences (Exclude Gaps) | References |
|---|---|---|---|---|
| Annulohypoxylon moniliformis MFLUCC 18-1214 | Annulohypoxylon annulatum CBS 140775 | No difference observed | Close to Annulohypoxylon annulatum CBS 140775 (98% ML/1.0BPP). Differ by 1.53% (13/847 bp) of ITS and 7.92% (42/530 bp) of TUB2 | Ju and Rogers (1996), Sir et al. (2016) |
| Apiospora dendrobii MFLUCC 14-0152 | Apiospora xenocordella CBS 478.86 | Conidiogenous cell: straight (A. dendrobii) vs. verruculose, globose to clavate to doliiform (A. xenocordella). | Close to Apiospora xenocordella (100% ML/1.0BPP). Differ by 1.95% (13/666 bp) of ITS, 3.24% (7/216 bp) of TUB2 | Crous and Groenewald (2013) |
| Hypoxylon endophyticum MFLUCC 18-1206 | No ex-type available (Compared with Hypoxylon investiens YMJ 89062905) | No difference observed | Close to Hypoxylon investiens (100%ML/1.0BPP). Differ by 3.54% (30/847 bp) of ITS and 1.89% (15/530 bp) of TUB2 | Ju and Rogers (1996), Platas et al. (2009) |
| Hypoxylon investiens CBS 118185 | No difference observed | Different lineages. Differ by 2.13% (18/847 bp) of ITS, 13.8% (198/1432 bp) of LSU, 4.93% (64/1298 bp) of RPB2 and 2.83% (15/530 bp) of TUB2 | Ju and Rogers (1996), Platas et al. (2009) | |
| Hypoxylon officinalis MFLUCC 14-0075 | Hypoxylon lateripigmentum MUCL 53304 | No periconiella-like conidiogenous structure was observed in H. officinalis | Close to H. lateripigmentum (100%ML/1BPP). Differ by 5.08% (43/847 bp) of ITS, 2.03% (29/1432 bp) of LSU, 8.3% (44/530 bp) of TUB2 | Kuhnert et al. (2014) |
| Nemania dendrobii MFLUCC 18-1213 | No ex-type available (Compared with Nemania bipapillata 90080610 (HAST)) | No difference observed | Close to N. bipapillata 90080610 (100% ML/1.0BPP). Differ by 2.48% (21/847 bp) of ITS; 4.15% (22/530 bp) of TUB2, 4.93% (64/1298 bp) of RPB2 | Ju and Rogers (1999), Hsieh et al. (2010), Database for Rice Seed-borne Fungi-Nemania sp. 15008 |
| Xylaria sp.1 MFLUCC 14-0137 | No ex-type available (Compared with Xylaria cubensis JDR 860) | In X. cubensis, Conidiogenous cell has round to denticulate secession scars; Conidia have flat basal abscission scar | Close to X. cubensis. Differ by 4.37% (37/847 bp) of ITS and 5.28% (28/530 bp) of TUB2 | Rogers 1984, Rodrigues et al. (1993), Hsieh et al. (2010) |
| No ex-type available (Compared with Xylaria cubensis GENT 159 | X. cubensis has smaller conidia size: (3.6–)5–6.3 × 1.8–3.6 μm vs. 4.5–9 × 2.5–4.5 μm | Close to X. cubensis GENT 159 (86% ML/0.99BPP). Differ by 1.13% (6/530 bp) of TUB2 | Rogers 1984, Rodrigues et al. (1993), Hsieh et al. (2010) | |
| Xylaria sp.2 MFLUCC 21-0014 | No ex-type available (Compared with Xylaria phyllocharis 528 (HAST)) | Morphological characters are not sufficient for characterisation | Close to X. phyllocharis (75% ML/0.97BPP). Differ by 6.85% (58/847 bp) of ITS; 16.6% (215/1298 bp) of RPB2; 7.17% (38/530 bp) of TUB2 | Rodrigues et al. (1993), Hsieh et al. (2010) |
| Xylaria sp.3 MFLUCC 21-0059 | Xylaria bambusicola WSP 205 | Morphological characters are not sufficient for characterisation | Close to X. bambusicola (100%ML/1BPP). Differ by 5.31% (45/847 bp) of ITS, 6.04% (32/530 bp) of TUB2 and 4.93% (64/1298 bp) of RPB2 | Rodrigues et al. (1993), Hsieh et al. (2010), Dai et al. (2016) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Chomnunti, P.; Doilom, M.; Daranagama, D.A.; Kang, J. Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand. J. Fungi 2022, 8, 248. https://doi.org/10.3390/jof8030248
Ma X, Chomnunti P, Doilom M, Daranagama DA, Kang J. Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand. Journal of Fungi. 2022; 8(3):248. https://doi.org/10.3390/jof8030248
Chicago/Turabian StyleMa, Xiaoya, Putarak Chomnunti, Mingkwan Doilom, Dinushani Anupama Daranagama, and Jichuan Kang. 2022. "Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand" Journal of Fungi 8, no. 3: 248. https://doi.org/10.3390/jof8030248
APA StyleMa, X., Chomnunti, P., Doilom, M., Daranagama, D. A., & Kang, J. (2022). Multigene Phylogeny Reveals Endophytic Xylariales Novelties from Dendrobium Species from Southwestern China and Northern Thailand. Journal of Fungi, 8(3), 248. https://doi.org/10.3390/jof8030248







