Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of BbSirT2
2.2. BbSirT2 Subcellular Localization
2.3. Construction of BbSirT2 Deleted and Complemented Strains
2.4. Histone Acetylation Detection
2.5. Phenotypic Experiments
2.6. Investigation of Hyphal Septation, Cell Size, and Cell Cycle Progression
2.7. Quantification of Intracellular Mitochondrial and ATP Content
2.8. Gene Expression Analyses of BbSirT2 and Development/Septation Genes
2.9. Global Transcriptomic Analysis
3. Results
3.1. Bioinformatic Analysis of BbSirT2 and Construction of Deleted and Complemented Strains
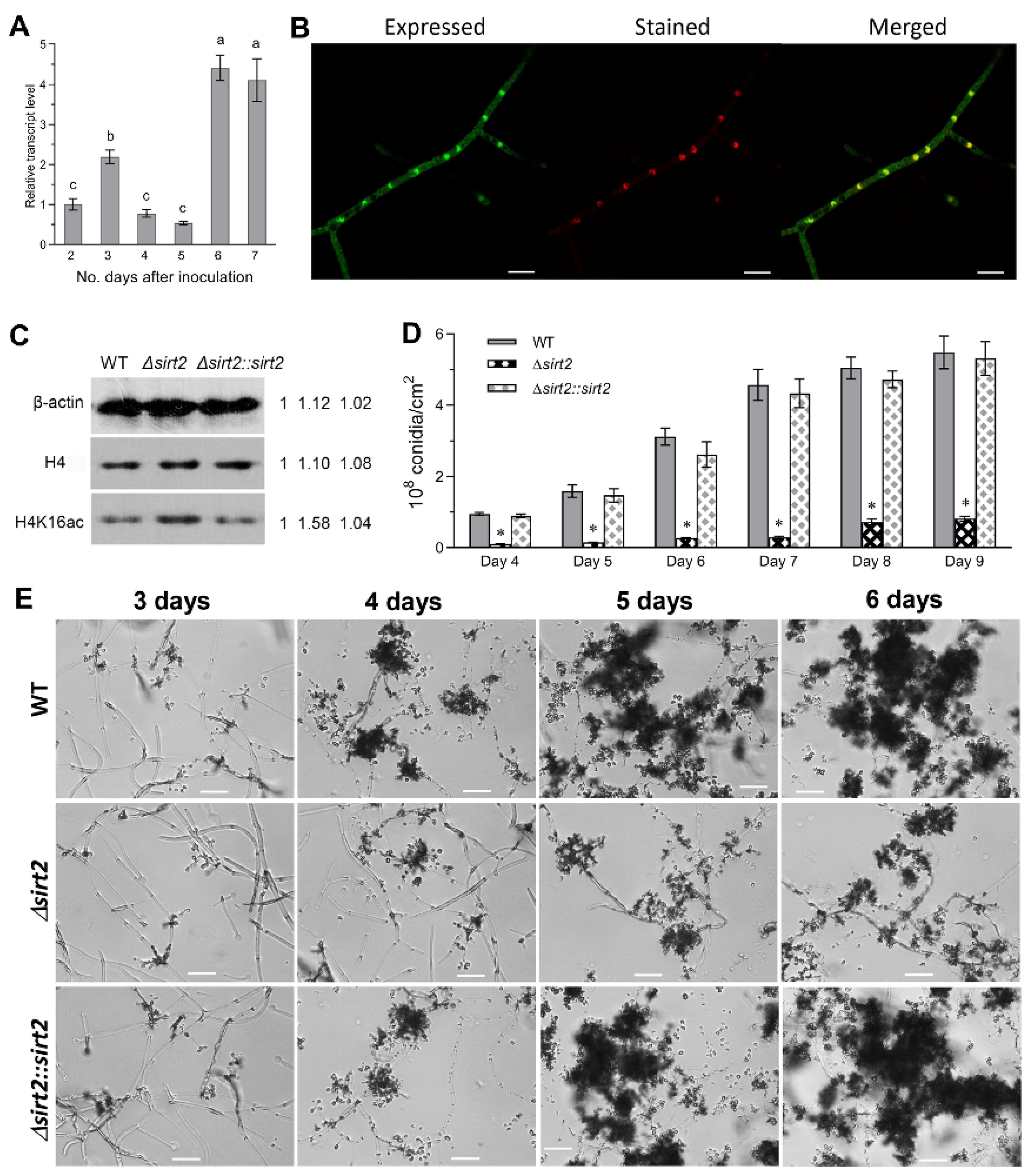
3.2. BbSirT2 Nuclear Localization, Histone Deacetylation Target, and Contributions to Conidiation
3.3. Role of SirT2 in Carbon/Nitrogen Metabolism and Stress Responses
3.4. Contribution of BbSirT2 to B. bassiana Virulence
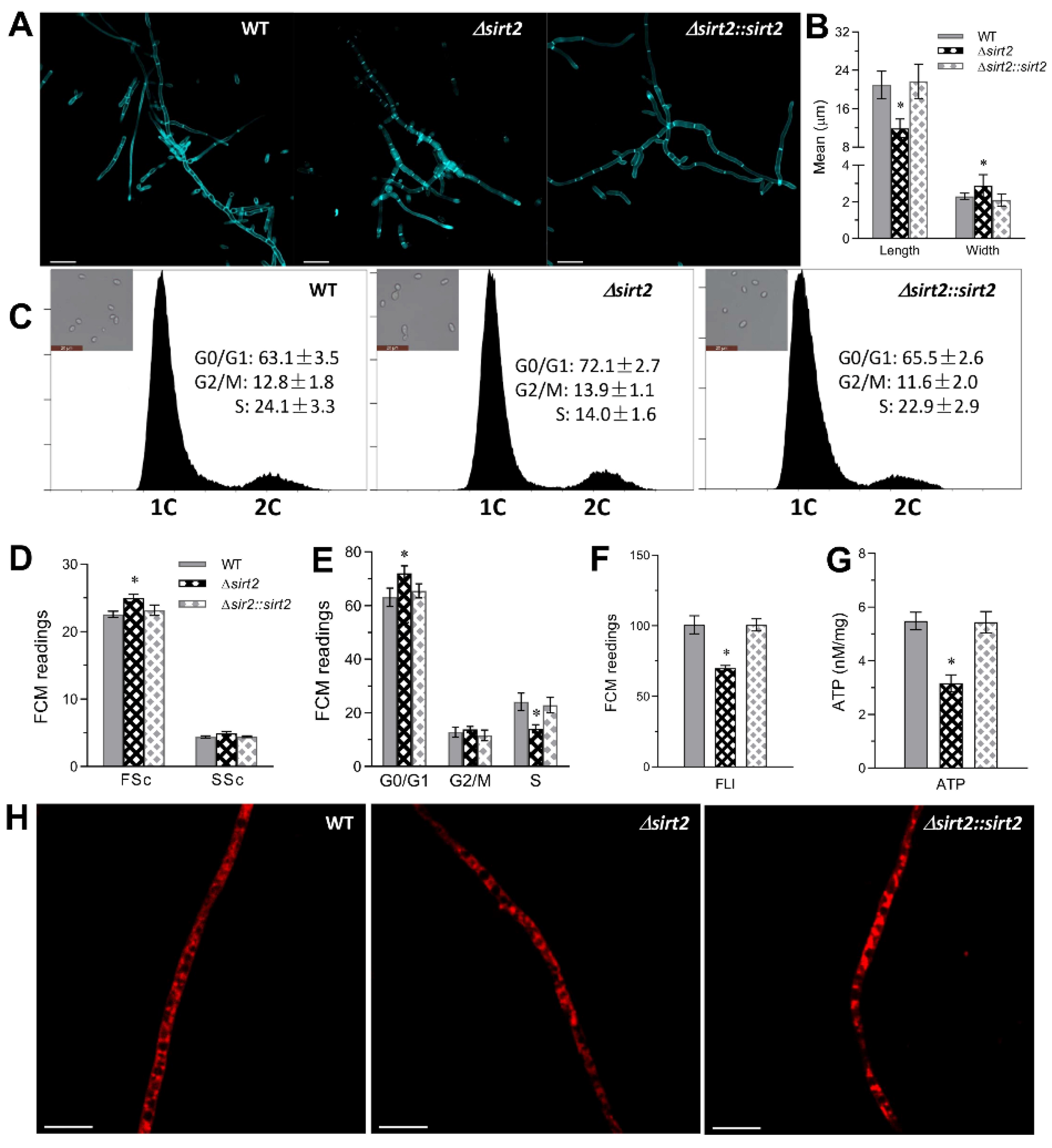
3.5. Role of BbSirT2 in Cell Cycle and Hyphal Septation Regulation
3.6. Global Gene Expression Changes in the ΔBbSirT2 Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shen, Y.; Wei, W.; Zhou, D.X. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015, 20, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, F.; Nicastro, R.; Reghellin, V.; Coccetti, P. Post-translational modifications on yeast carbon metabolism: Regulatory mechanisms beyond transcriptional control. Biochim. Biophys. Acta 2015, 1850, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.L.; Xiong, Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 2010, 36, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, B.E.; Tong, J.K.; Schreiber, S.L. Genome wide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13708–13713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trojer, P.; Brandtner, E.M.; Brosch, G.; Loidl, P.; Galehr, J.; Linzmaier, R.; Haas, H.; Mair, K.; Tribus, M.; Graessle, S. Histone deacetylases in fungi: Novel members, new facts. Nucleic Acids Res. 2003, 31, 3971–3981. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brosch, G.; Loidl, P.; Graessle, S. Histone modifications and chromatin dynamics: A focus on filamentous fungi. FEMS Microbiol. Rev. 2008, 32, 409–439. [Google Scholar] [CrossRef] [Green Version]
- Imai, S.; Guarente, L. Ten years of NAD-dependent SIR2 family deacetylases: Implications for metabolic diseases. Trends Pharmacol. Sci. 2010, 31, 212–220. [Google Scholar] [CrossRef]
- Zhao, K.; Harshaw, R.; Chai, X.; Marmorstein, R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. USA 2004, 101, 8563–8568. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 Regulates TNF-α Secretion through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef]
- Landry, J.; Sutton, A.; Tafrov, S.T.; Heller, R.C.; Stebbins, J.; Pillus, L.; Sternglanz, R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 2000, 97, 5807–5811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, K.G.; Landry, J.; Sternglanz, R.; Denu, J.M. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 2000, 97, 14178–14182. [Google Scholar] [CrossRef] [Green Version]
- Greiss, S.; Gartner, A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 2009, 28, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoggard, T.; Müller, C.A.; Nieduszynski, C.A.; Weinreich, M.; Fox, C.A. Sir2 mitigates an intrinsic imbalance in origin licensing efficiency between early- and late-replicating euchromatin. Proc. Natl. Acad. Sci. USA 2020, 117, 14314–14321. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Sherman, J.M.; Devine, S.E.; Cameron, E.E.; Pillus, L.; Boeke, J.D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995, 9, 2888–2902. [Google Scholar] [CrossRef] [Green Version]
- Derbyshire, M.K.; Weinstock, K.G.; Strathern, J.N. HST1, a new member of the SIR2 family of genes. Yeast 1996, 12, 631–640. [Google Scholar] [CrossRef]
- Perrod, S.; Cockell, M.M.; Laroche, T.; Renauld, H.; Ducrest, A.L.; Bonnard, C.; Gasser, S.M. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001, 20, 197–209. [Google Scholar] [CrossRef] [Green Version]
- Halme, A.; Bumgarner, S.; Styles, C.; Fink, G.R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 2004, 116, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Gasser, S.M.; Cockell, M.M. The molecular biology of the SIR proteins. Gene 2001, 279, 1–16. [Google Scholar] [CrossRef]
- Smith, J.S.; Brachmann, C.B.; Celic, I.; Kenna, M.A.; Muhammad, S.; Starai, V.J.; Avalos, J.L.; Escalante-Semerena, J.C.; Grubmeyer, C.; Wolberger, C.; et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 2000, 97, 6658–6663. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Chai, X.; Clements, A.; Marmorstein, R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 2003, 10, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.M.; Le, V.Q.; Zimmerman, C.; Marmorstein, R.; Pillus, L. Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 2006, 7, 1247–1251. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Lamming, D.W.; Latorre-Esteves, M.; Medvedik, O.; Wong, S.N.; Tsang, F.A.; Wang, C.; Lin, S.J.; Sinclair, D.A. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 2005, 309, 1861–1864. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.C.; Chiu, S.W.; Lee, H.Y.; Leu, J.Y. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell 2012, 23, 1231–1242. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Sinha, I.; Fagerström-Billai, F.; Bonilla, C.; Wright, A.; Grunstein, M.; Ekwall, K. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. EMBO J. 2007, 26, 2477–2488. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Rusche, L.N. Genetic Analysis of Sirtuin Deacetylases in Hyphal Growth of Candida Albicans. Msphere 2021, 6, e00053-21. [Google Scholar] [CrossRef]
- Arras, S.D.M.; Chitty, J.; Wizrah, M.S.I.; Erpf, P.E.; Schulz, B.; Tanurdzic, M.; Fraser, J.A. Sirtuins in the phylum Basidiomycota: A role in virulence in Cryptococcus neoformans. Sci. Rep. 2017, 7, 46567. [Google Scholar] [CrossRef] [Green Version]
- Boivin, A.; Gaumer, S.; Sainsard-Chanet, A. Life span extension by dietary restriction is reduced but not abolished by loss of both SIR2 and HST2 in Podospora anserina. Mech. Ageing Dev. 2008, 129, 714–721. [Google Scholar] [CrossRef]
- de Faria, M.R.; Wraight, S.P. Mycoinsecticides and mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Köhl, J.; Marrone, P.; Morin, L.; Stewart, A. Have biopesticides come of age? Trends Biotechnol. 2012, 30, 250–258. [Google Scholar] [CrossRef]
- Ortiz-Urquiza, A.; Luo, Z.B.; Keyhani, N.O. Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biotechnol. 2015, 99, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Stress response signaling and virulence: Insights from entomopathogenic fungi. Curr. Genet. 2015, 61, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Molecular genetics of Beauveria bassiana infection of insects. Adv. Genet. 2016, 94, 165–249. [Google Scholar]
- Cai, Q.; Tian, L.; Xie, J.T.; Huang, Q.Y.; Feng, M.G.; Keyhani, N.O. A fungal sirtuin modulates development and virulence in the insect pathogen, Beauveria bassiana. Environ. Microbiol. 2021, 23, 5164–5183. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Sanstrum, B.J.; Liu, Y.; Kwon, S.H. Distinct role of Sirtuin 1 (SIRT1) and Sirtuin 2 (SIRT2) in inhibiting cargo-loading and release of extracellular vesicles. Sci. Rep. 2019, 9, 20049. [Google Scholar] [CrossRef]
- Manjula, R.; Anuja, K.; Alcain, F.J. SIRT1 and SIRT2 Activity Control in Neurodegenerative Diseases. Front. Pharmacol. 2021, 11, 585821. [Google Scholar] [CrossRef]
- De Oliveira, R.M.; Sarkander, J.; Kazantsev, A.G.; Outeiro, T.F. SIRT2 as a Therapeutic Target for Age-Related Disorders. Front. Pharmacol. 2012, 3, 82. [Google Scholar] [CrossRef] [Green Version]
- Xiao, G.H.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.W.; Xie, X.U.; Shang, Y.; Leger, R.J.S.; Zhao, G.u.; Wang, C.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, S.H.; Feng, M.G. Novel blastospore-based transformation system for easy integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl. Microbiol. Biotechnol. 2006, 72, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fan, Y.; Xia, Y.X.; Keyhani, N.O. Sulfonylurea resistance as a new selectable marker for the entomopathogenic fungus Beauveria bassiana. Appl. Microbiol. Biotechnol. 2010, 87, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.P.; Bateman, R.; Charnley, A.K. Role of cuticle-degrading proteases in the virulence of Metarhizium spp. for the desert locust, Schistocerca gregaria. J. Invertebr. Pathol. 1998, 71, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Donatti, A.C.; Furlaneto-Maia, L.; Fungaro, M.H.P.; Furlaneto, M.C. Production and regulation of cuticle-degrading proteases from Beauveria bassiana in the presence of Rhammatocerus schistocercoides cuticle. Curr. Microbiol. 2008, 56, 256–260. [Google Scholar] [CrossRef]
- Wang, J.; Ying, S.H.; Hu, Y.; Feng, M.G. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ. Microbiol. 2016, 18, 1037–1047. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Wang, D.Y.; Ying, S.H.; Feng, M.G. Miro GTPase controls mitochondrial behavior affecting stress tolerance and virulence of a fungal insect pathogen. Fungal Genet. Biol. 2016, 93, 1–9. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans asexual development: Making the most of cellular modules. Trends Microbiol. 2010, 18, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Zeng, L.; Huang, X. Protein Acetylation/Deacetylation: A Potential Strategy for Fungal Infection Control. Front. Microbiol. 2020, 11, 574736. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Liu, W.; Wang, G.; Kang, Z.; Kistler, H.; Xu, J.-R. The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum. Mol. Plant. Microbe Interact. 2011, 24, 487–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobile, C.J.; Fox, E.P.; Hartooni, N.; Mitchell, K.; Hnisz, D.; Andes, D.; Kuchler, K.; Johnson, A.D. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 2014, 5, e01201-14. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, J.E.; Lee, J.S. Histone deacetylase-mediated morphological transition in Candida albicans. J. Microbiol. 2015, 53, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Garnaud, C.; Champleboux, M.; Maubon, D.; Cornet, M.; Govin, J. Histone deacetylases and their inhibition in candida species. Front. Microbiol. 2016, 7, 1238. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Wang, Z.K.; Shao, W.; Ying, S.H.; Feng, M.G. Essential role of Rpd3-dependent lysine modification in the growth, development and virulence of Beauveria bassiana. Environ. Microbiol. 2018, 20, 1590–1606. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tong, S.M.; Shao, W.; Ying, S.H.; Feng, M.G. Pleiotropic effects of the histone deacetylase Hos2 linked to H4-K16 deacetylation, H3-K56 acetylation and H2A-S129 phosphorylation in Beauveria bassiana. Cell. Microbiol. 2018, 20, e12839. [Google Scholar] [CrossRef]
- Li, F.; Shi, H.Q.; Ying, S.H.; Feng, M.G. WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl. Microbiol. Biotechnol. 2015, 99, 10069–10081. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xia, Y.X.; Kim, B.; Keyhani, N.O. Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol. Microbiol. 2011, 80, 811–826. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, J.J.; Ying, S.H.; Feng, M.G. Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle-required cyclin-dependent kinase 1 activity. Environ. Microbiol. 2015, 17, 1119–1133. [Google Scholar] [CrossRef]
- Perez-Martin, J.; Uria, J.A.; Johnson, A.D. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999, 18, 2580–2592. [Google Scholar] [CrossRef] [Green Version]
- Freire-Benéitez, V.; Gourlay, S.; Berman, J.; Buscaino, A. Sir2 regulates stability of repetitive domains differentially in the human fungal pathogen Candida albicans. Nucleic Acids Res. 2016, 44, gkw594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouklas, T.; Jain, N.; Fries, B.C. Modulation of replicative lifespan in Cryptococcus neoformans: Implications for virulence. Front. Microbiol. 2017, 8, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, J.; Marroquin-Guzman, M.; Nandakumar, R.; Shijo, S.; Cornwell, K.M.; Li, G.; Wilson, R.A. Plant defence suppression is mediated by a fungal sirtuin during rice infection by Magnaporthe oryzae. Mol. Microbiol. 2014, 94, 70–88. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.J.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. Subtilisin-like Pr1 proteases marking the evolution of pathogenicity in a wide-spectrum insect-pathogenic fungus. Virulence 2020, 11, 365–380. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.W.; Robalino, I.V.; Keyhani, N.O. Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3110–3120. [Google Scholar] [CrossRef] [Green Version]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3121–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cen, K.; Li, B.; Lu, Y.; Zhang, S.; Wang, C. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017, 13, e1006604. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Liu, X.; Keyhani, N.O.; Tang, G.; Pei, Y.; Zhang, W.; Tong, S. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl. Acad. Sci. USA 2017, 114, E1578–E1586. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and AbaA Govern Virulence-Required Dimorphic Switch, Conidiation, and Pathogenicity in a Fungal Insect Pathogen. mSystems 2019, 4, e00140-19. [Google Scholar] [CrossRef] [Green Version]
- Mou, Y.N.; Gao, B.J.; Ren, K.; Tong, S.M.; Ying, S.H.; Feng, M.G. P-type Na+/K+ ATPases essential and nonessential for cellular homeostasis and insect pathogenicity of Beauveria bassiana. Virulence 2020, 11, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Song, T.T.; Zhao, J.; Ying, S.H.; Feng, M.G. Differential contributions of five ABC transporters to mutidrug resistance, antioxidion and virulence of Beauveria bassiana, an entomopathogenic fungus. PLoS ONE 2013, 8, e62179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Q.; Tian, L.; Xie, J.-T.; Jiang, D.-H.; Keyhani, N.O. Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence. J. Fungi 2022, 8, 236. https://doi.org/10.3390/jof8030236
Cai Q, Tian L, Xie J-T, Jiang D-H, Keyhani NO. Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence. Journal of Fungi. 2022; 8(3):236. https://doi.org/10.3390/jof8030236
Chicago/Turabian StyleCai, Qing, Li Tian, Jia-Tao Xie, Dao-Hong Jiang, and Nemat O. Keyhani. 2022. "Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence" Journal of Fungi 8, no. 3: 236. https://doi.org/10.3390/jof8030236
APA StyleCai, Q., Tian, L., Xie, J.-T., Jiang, D.-H., & Keyhani, N. O. (2022). Contributions of a Histone Deacetylase (SirT2/Hst2) to Beauveria bassiana Growth, Development, and Virulence. Journal of Fungi, 8(3), 236. https://doi.org/10.3390/jof8030236






